Abstract
The exosome is a complex of 3′–5′ exoribonucleases and RNA-binding proteins, which is involved in processing or degradation of different classes of RNA. Previously, the characterization of purified exosome complexes from yeast and human cells suggested that C1D and KIAA0052/hMtr4p are associated with the exosome and thus might regulate its functional activities. Subcellular localization experiments demonstrated that C1D and KIAA0052/hMtr4p co-localize with exosome subunit PM/Scl-100 in the nucleoli of HEp-2 cells. Additionally, the nucleolar accumulation of C1D appeared to be dependent on PM/Scl-100. Protein–protein interaction studies showed that C1D binds to PM/Scl-100, whereas KIAA0052/hMtr4p was found to interact with MPP6, a previously identified exosome-associated protein. Moreover, we demonstrate that C1D, MPP6 and PM/Scl-100 form a stable trimeric complex in vitro. Knock-down of C1D, MPP6 and KIAA0052/hMtr4p by RNAi resulted in the accumulation of 3′-extended 5.8S rRNA precursors, showing that these proteins are required for rRNA processing. Interestingly, C1D appeared to contain RNA-binding activity with a potential preference for structured RNAs. Taken together, our results are consistent with a role for the exosome-associated proteins C1D, MPP6 and KIAA052/hMtr4p in the recruitment of the exosome to pre-rRNA to mediate the 3′ end processing of the 5.8S rRNA.
INTRODUCTION
The human exosome is a complex with 3′–5′ exoribonuclease activity, consisting of nine core components and the stably associated component PM/Scl-100, which is more abundant in but not restricted to the nucleus (1,2). Six of the core components (hRrp41p, hRrp42p, hRrp46p, hMtr3p, OIP2 and PM/Scl-75) contain an RNase PH domain, which assemble into a hexameric ring structure, whereas the three remaining components (hRrp4p, hRrp40p and hCsl4p) containing an S1 (and KH) RNA-binding domain are positioned on top of this ring (3–8).
A variety of nuclear functions have been described for the yeast exosome, including 3′ end processing of sn(o)RNAs (9,10) and the degradation of aberrant pre-mRNAs (11–13) and pre-tRNAs (14,15). In the nucleolus of eukaryotes, the 18S, 5.8S and 25S/28S rRNAs are transcribed as a single precursor by RNA polymerase I. After transcription, maturation of rRNAs is achieved by a complex processing pathway in which many nucleotide-modification events occur and in which the two internally transcribed spacers (ITS1 and ITS2) and two externally transcribed spacers (5′-ETS and 3′-ETS) are removed (16,17). In yeast, depletion of exosome components as well as several associated proteins such as Rrp47p and Mtr4p resulted in the accumulation of 3′-extended 5.8S rRNAs, elevated levels of 5′ ETS fragments, and polyadenylated (pre-)rRNAs. Exosome depletion also indirectly affected early pre-rRNA cleavage events at A(0), A(1), A(2) and A(3) leading to a reduction of the mature 18S and 25S rRNAs (9,18–25).
Besides the core exosome components, several cytoplasmic and nuclear exosome-associated proteins have been identified which are most likely required for the recruitment of the exosome to specific substrate RNAs, its association with other processing complexes, or the modulation of its in vivo activity. Nuclear exosome-associated proteins include human MPP6, a protein required for the maturation of the 5.8S rRNA, and yeast Rrp47p (also known as C1D and Lrp1p), which functions in sn(o)RNA and pre-rRNA processing (2,26,27). The putative human homolog of yeast Rrp47p/C1D/Lrp1p is the C1D protein, which has previously been identified as a DNA-binding protein involved in DNA double-strand break repair by activating a DNA-dependent protein kinase (28,29). The putative RNA helicase Mtr4p is another auxiliary protein required for most of the functions of the nuclear exosome in yeast (9,30). The Mtr4p protein is also part of the recently identified TRAMP complex, which is required for the activation of the nuclear exosome in vivo by polyadenylation of target RNAs (15,21). The KIAA0052 protein is the putative human homolog of Mtr4p and was found to co-purify with the human exosome (31).
In this study, we investigated the exosome association and function of C1D and KIAA0052, which hereafter will be referred to as hMtr4p, in human cells. Our data indicate that PM/Scl-100 mediates the association of hMtr4p, C1D and MPP6 with the exosome and that hMtr4p and the complex formed by PM/Scl-100, C1D and MPP6 are required for the maturation of 5.8S rRNA.
MATERIALS AND METHODS
cDNA cloning
The cDNA of human C1D was obtained by a PCR-based approach using a teratocarcinoma cDNA library and oligonucleotides C1D-forward, 5′-CGTCGACTTCTCGAGATGGCAGGTGAAGAAATTAATG-3′ and C1D-reverse, 5′-AGCGGCCGCTTACCCGGGACTTTTACTTTTTCCTTTATTGG-3′. The human hMtr4p cDNA sequence was isolated by PCR from clone IRATp970F0129D6 provided by the IMAGE consortium using oligonucleotides hMtr4p-forward, 5′-GCGACGATATCCTCGAGCATGGCGGACGCATTCGGAGA-3′ and hMtr4p-reverse 5′-GCGTCGGTACCCTACAAGTAGAGGCTGGCA-3′. The resulting PCR products were cloned into the pCR4-TOPO vector according to the manufacturer's procedure (Invitrogen).
Immunoprecipitation
Polyclonal rabbit anti-EGFP antibodies were coupled to protein A-agarose beads (Kem-En-Tec) in IPP500 [(500 mM NaCl, 10 mM Tris-HCl, pH 8.0 and 0.05% Nonidet P-40 (NP-40)] at room temperature for 1 h. Beads were washed once with IPP500 and twice with IPP150 (same as IPP500, but containing 150 mM NaCl). For each immunoprecipitation, cell extract was incubated with the antibody-coupled beads for 2 h at 4°C. After washing the beads four times with IPP150, the precipitated proteins were separated by 10% SDS-PAGE and analyzed by immunoblotting.
Western blot analysis
For western blot analysis, proteins were separated by SDS-PAGE and transferred to nitrocellulose membranes. After blocking, the blots were incubated with autoimmune patient antisera or monoclonal anti-hRrp4p antibodies (culture supernatant) (ModiQuest, Nijmegen, The Netherlands), diluted 1000- and 25-fold, respectively, in blocking buffer (5% skimmed milk, 0.05% NP-40 in PBS). As secondary antibodies, horseradish peroxidase-conjugated rabbit anti-human IgG or goat anti-mouse IgG (Dako Immunoglobulins) were used, 2500-fold diluted in blocking buffer. Bound antibodies were visualized by chemiluminescence detection.
Expression and purification of recombinant proteins
For prokaryotic expression, the C1D and MPP6 cDNAs were cloned into the pGEX4 vector, resulting in sequences encoding glutathione S-transferase (GST)-tagged recombinant proteins. The GST fusion proteins were expressed and purified essentially as described previously (32). After induction of protein expression with IPTG, the bacteria were grown at room temperature to enhance the production of soluble protein. The proteins were purified using a reduced glutathione affinity resin in the presence of 0.2% empigen and stored at −70°C after the addition of glycerol (final concentration 10%).
In vitro transcription and translation
The open reading frames of PM/Scl-100, MPP6 and C1D were cloned into the pCI-neo vector (Promega), in frame with the vesicular stomatitis virus G epitope (VSV-G tag). The resulting pCI-neo5′VSV constructs of PM/Scl-100, MPP6 and C1D as well as a pCR4-TOPO-hMtr4p construct were transcribed and translated in the presence of 35S-methionine using the TnT-coupled transcription/translation kit (Promega).
Transient transfection and fluorescence microscopy
The cDNAs were cloned into suitable pEGFP vectors (Clontech) allowing expression of the protein fused to the C-terminus of the EGFP protein. HEp-2 cells were grown to 70% confluency in DMEM containing 10% fetal calf serum (FCS) (DMEM+). For immunoprecipitation, ∼10 × 106 cells were transfected with 30 µg of plasmid DNA in 1600 µl of DMEM+ by electroporation at 260 V and 950 µF using a Gene-Pulsar II (Bio-Rad). After transfection, cells were seeded in 75-cm2 culture flasks and cultured overnight. The trypsinized cells were washed with PBS, resuspended in 750 µl of lysis buffer (25 mM Tris-HCl, pH 7.5, 150 mM KCl, 2 mM EDTA, 1 mM dithiotreitol (DTT), 0.5 mM PMSF and 0.05% NP-40) and homogenized by sonication using a Branson micro-tip. For fluorescence microscopy ∼3 × 106 cells were transfected with 10 µg of plasmid DNA in 800 µl DMEM+ by electroporation as described above. The cells were seeded on coverslips and cultured overnight. Subsequently, the cells were washed twice with PBS and fixed with 4% paraformaldehyde in PBS for 20 min. After fixation, cells were washed three times with PBS and mounted with 50% glycerol in PBS. The EGFP-fusion proteins were visualized by fluorescence microscopy using a Leica DM IRBE confocal microscope. For co-localization experiments, HEp-2 cells were transfected with pEGFP-C1D or pEGFP-hMtr4p and grown on coverslips for 24 h. Cells were fixed with methanol for 5 min at −20°C, briefly rinsed in acetone, air dried and incubated for 1 h at room temperature with anti-PM/Scl-100 rabbit serum (diluted 1:100 in PBS), followed by washing and incubation with secondary Texas-Red-conjugated donkey anti-rabbit IgG. The cells were analyzed by confocal fluorescence microscopy.
siRNA transfection experiments
The 21-nt siRNA duplexes used in this study are based on the coding region of the gene of interest, containing dTdT overhangs and were obtained from Eurogentec (Belgium): siC1D, 5′-GUUGGAUCCACUUGAACAATT-3′; siPM/Scl-75, 5′-GCCAAGAUGCUCCCAUAAUTT–3′; sihMtr4p, 5′-GCCUAUGCACUUCAAAUGATT-3′. The siRNAs against human MPP6 and PM/Scl-100 were described previously (1,2). The siRNA negative control (OR-0030-NEG05) was obtained from Eurogentec, Belgium. For each transfection, ∼3 × 105 HEp-2 cells were transfected with 100 pmol of each siRNA using Oligofectamine transfection reagent (Invitrogen), as described by the manufacturer with the exception that during transfection 10% FCS was present in the medium. Total RNA was extracted 48 h after transfection using the TRIzol reagent (Invitrogen). For each sample, 5 µg of total RNA was separated on a denaturing 7% polyacrylamide gel, and after transfer to Hybond N+ membranes (Amersham Biosciences) the blots were hybridized with 32P-labeled antisense RNA probes in hybridization solution (6× SSC, 10× Denhardt's, 100 µg/ml sheared herring sperm DNA and 0.1% SDS). After overnight incubation at 65°C, the blots were washed three times with 2× SSC/0.1% SDS and analyzed by autoradiography. To study the subcellular localization of C1D in siRNA-treated cells, HEp-2 cells were transfected with C1D-EGFP as described above. After 16 h, cells were treated with 100 pmol of siRNA and cultured overnight. Subsequently, the cells were washed twice with PBS, fixed with 4% paraformaldehyde in PBS for 20 min and mounted in PBS/glycerol. Expressed proteins were visualized by fluorescence microscopy using a Leica DM4000B microscope.
GST pull-down
GST-fusion protein (∼1 μg) was immobilized on glutathione-Sepharose beads (Amersham Pharmacia Biotech), washed with pull-down buffer PB-100 (PB-100: 20 mM HEPES-KOH, pH 7.6, 100 mM KCl, 0.5 mM EDTA, 0.05% NP-40, 1 mM DTT, 5 mM MgCl2, 0.02% BSA, 0.5 mM PMSF) and incubated with in vitro translated, 35S-labeled protein at 4°C for 2 h under continuous agitation. After incubation, the beads were washed three times with PB-150 (composition like PB-100, but containing 150 mM KCl), and the bound proteins were analyzed by SDS-PAGE and autoradiography. In case of protein–RNA interactions, immobilized GST-C1D was resuspended in 200 µl of PB-200 (composition like PB-100, but containing 200 mM KCl and lacking MgCl2). After addition of the 32P-labeled RNAs and 20 U of RNasin (Promega), the mixture was incubated for 1 h at 4°C under continuous agitation. The beads were washed four times with PB-200, and the co-precipitating RNAs were extracted and analyzed by denaturing PAGE and autoradiography.
RESULTS
C1D and hMtr4p co-localize with PM/Scl-100 in the nucleoli of HEp-2 cells
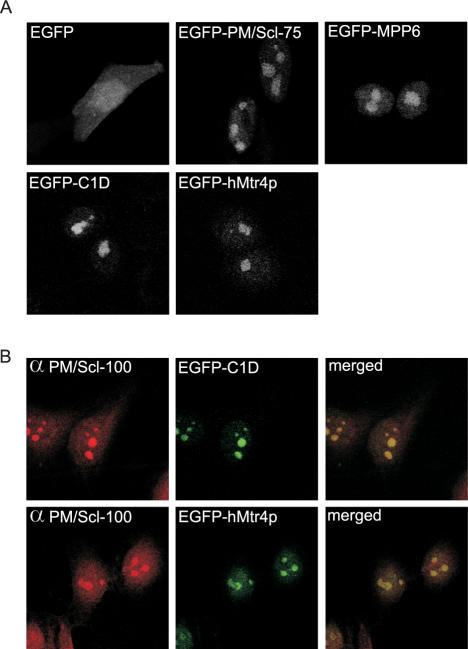
To determine the subcellular localization of C1D and hMtr4p, HEp-2 cells were transiently transfected with constructs encoding EGFP-tagged C1D, hMtr4p, MPP6 or the core exosome component PM/Scl-75. After 24 h, the EGFP fusion proteins were visualized by confocal fluorescence microscopy (see Figure 1). In agreement with previous observations, EGFP-PM/Scl-75 and EGFP-MPP6 accumulated in the nucleoli, whereas also a weak but significant staining of the nucleoplasm was observed for both proteins (2,33) A very similar staining pattern was observed for both EGFP-C1D and EGFP-hMtr4p. Thus, the subcellular distribution of C1D and hMtr4p is consistent with their putative exosome association.
Figure 1.
C1D and hMtr4p co-localize with PM/Scl-100 in the nucleoli of HEp-2 cells. (A) HEp-2 cells were transiently transfected with cDNA constructs encoding EGFP alone, EGFP-PM/Scl-75, EGFP-MPP6, EGFP-C1D or EGFP-hMtr4p. Twenty-four hours after transfection, the cells were fixed and EGFP or EGFP-fusion proteins were visualized by confocal fluorescence microscopy. (B) HEp-2 cells were transfected with cDNA constructs encoding C1D and hMtr4p fused to the C-terminus of EGFP; 24 h after transfection, cells were fixed and incubated with rabbit anti-PM/Scl-100 antibodies, which were visualized by Texas-Red-conjugated secondary antibodies (left panels). The localization of EGFP-tagged C1D and hMtr4p is shown in the middle panels, and the corresponding merged images are shown on the right.
To investigate this more thoroughly, HEp-2 cells were transfected with either EGFP-C1D or EGFP-hMtr4p and also stained with an antibody to the exosome subunit PM/Scl-100. As shown in Figure 1B, a nearly complete co-localization of EGFP-C1D and EGFP-hMtr4p with PM/Scl-100 in the nucleoli of these cells was observed. In the nucleoplasm, there appears to be less extensive co-localization, which might be due to the low levels of protein present in this subcellular compartment and/or to overexpression of the EGFP-fusion proteins.
Interaction of C1D and hMtr4p with the exosome
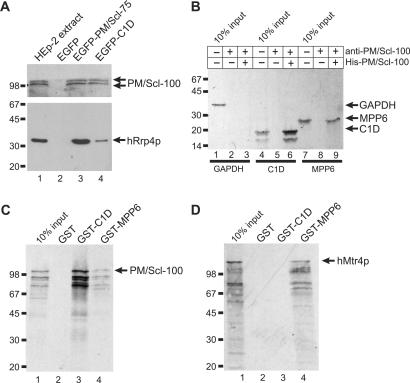
To investigate whether C1D is indeed associated with the human exosome, immunoprecipitations with anti-EGFP antibodies were performed with lysates from HEp-2 cells expressing EGFP-tagged C1D, PM/Scl-75 or EGFP alone as a negative control. Co-precipitated material was analyzed by western blotting using a human patient serum (known to be reactive with PM/Scl-100) and a monoclonal antibody to the human exosome component hRrp4p. As expected, both PM/Scl-100 and hRrp4 were co-precipitated with the EGFP-PM/Scl-75 protein (Figure 2A, lane 3). PM/Scl-100 and hRrp4p also appeared to be co-precipitated with EGFP-C1D (lane 4), although the efficiency of hRrp4p co-precipitation was much lower than that observed with EGFP-PM/Scl-75. With the EGFP protein alone no co-precipitation of hRrp4p and PM/Scl-100 was detected (lane 2). This result suggests that C1D is preferentially associated with exosome complexes containing PM/Scl-100. To substantiate the direct interaction of C1D and MPP6, another protein reported to interact with PM/Scl-100 (5), with PM/Scl-100, immunoprecipitations were performed using recombinant His-tagged PM/Scl-100 and in vitro translated glyceraldehyde-3-phosphate dehydrogenase (GAPDH), C1D and MPP6. As shown in Figure 2B, precipitation of His-PM/Scl-100 by anti-PM/Scl-100 antibodies resulted in the efficient co-precipitation of C1D (lane 6) and, albeit to a lesser extent, MPP6 (lane 9), but not GAPDH (lane 3). In lanes 2, 5 and 8, anti-PM/Scl-100 antibodies were incubated with the in vitro translated proteins in the absence of His-PM/Scl-100 demonstrating that the antibodies are not reactive with either of these proteins. In the reciprocal experiment, precipitation of GST-fusion proteins using glutathione-Sepharose beads resulted in the co-precipitation of in vitro translated PM/Scl-100 with both GST-C1D and, although less efficiently, with GST-MPP6, but not with GST alone (Figure 2C).
Figure 2.
Association of C1D, MPP6 and hMtr4p with components of the exosome. (A) Co-immunoprecipitations were performed using anti-EGFP antibodies and extracts of HEp-2 cells transiently transfected with expression constructs for either EGFP alone (lane 2), EGFP-PM/Scl-75 (lane 3) or EGFP-C1D (lane 4). In the first lane, total extract from non-transfected HEp-2 cells was separated. The precipitated proteins were analyzed by western blotting, using anti-PM/Scl-positive patient serum R212 (upper part of the blot) or a monoclonal antibody to hRrp4p (lower part of the blot). Arrows indicate the positions of PM/Scl-100 and hRrp4p. The positions of molecular weight markers are indicated on the left. (B) Bacterially expressed, recombinant His-tagged PM/Scl-100 was immobilized using anti-PM/Scl-100 antibodies and incubated with either 35S-labeled, in vitro translated GAPDH (lane 1), C1D (lane 4) or MPP6 (lane 7). Co-precipitated proteins were analyzed by SDS-PAGE and autoradiography. The positions of these proteins are indicated with arrows, and the positions of molecular weight markers are indicated on the left. Lanes 3, 6 and 9 show proteins co-precipitated with His-PM/Scl-100. In lanes 2, 5 and 8, material from control incubations, in which no recombinant His-PM/Scl-100 was added, was analyzed. (C) Glutathione-Sepharose beads were used to precipitate GST (lane 2), GST-C1D (lane 3) or GST-MPP6 (lane 4), which were incubated with 35S-labeled, in vitro translated PM/Scl-100. After precipitation, bound PM/Scl-100 was analyzed by SDS-PAGE and autoradiography. In lane 1, 10% of the amount of labeled PM/Scl-100 used per incubation was loaded. On the left, the positions of molecular weight markers are indicated. (D) Similar experiments as described in (C), but now with 35S-labeled, in vitro translated hMtr4p instead of PM/Scl-100.
To identify protein–protein interactions of hMtr4p with the exosome, GST pull-down assays were performed using GST-fusion proteins of several exosome components and auxiliary proteins and in vitro translated hMtr4p. The only protein found to specifically interact with hMtr4p was GST-MPP6 as shown in Figure 2D, whereas none of the other proteins analyzed (C1D, hRrp4p, hRrp40p, hRrp41p, hRrp42p, hRr46p, hMtr3p, OIP2 and hCsl4p) were able to precipitate hMtr4p (data not shown). Because the in vitro translated protein samples contain RNA, the possibility existed that the observed protein–protein interactions between GST-MPP6 and hMtr4p as well as GST-C1D and PM/Scl-100 might be RNA-mediated. Similar GST pull-down experiments were performed with in vitro translated protein samples that were treated with RNase A and RNase T1. The results demonstrated that these protein–protein interactions were not affected by RNase treatment, even though control experiments showed that the RNAs were efficiently degraded (data not shown).
PM/Scl-100, C1D and MPP6 form a heterotrimeric complex
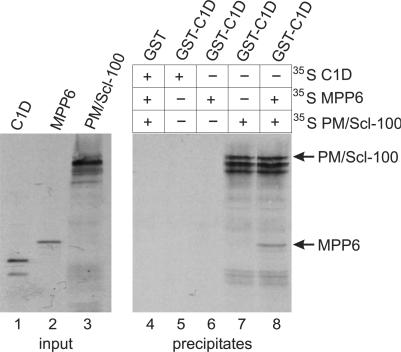
The finding that both MPP6 and C1D interact with PM/Scl-100 prompted us to examine the possibility that MPP6 and C1D bind simultaneously to PM/Scl-100. GST pull-down experiments were performed using GST-C1D and in vitro translated C1D, MPP6 and PM/Scl-100 proteins. As can be seen in Figure 3, no detectable interaction was found between GST-C1D and in vitro translated C1D or MPP6 (lanes 5 and 6). In contrast, GST-C1D efficiently interacted with PM/Scl-100 as observed before (lane 7). When both radiolabeled PM/Scl-100 and MPP6 were incubated with GST-C1D, not only PM/Scl-100 but also MPP6 was recovered in the precipitate (lane 8), demonstrating that C1D, MPP6 and PM/Scl-100 form a heterotrimeric complex in vitro. With the control, GST alone, no detectable interaction was observed with radiolabeled C1D, MPP6 or PM/Scl-100 (lane 4).
Figure 3.
C1D, MPP6 and PM/Scl-100 form a trimeric complex in vitro. GST-tagged C1D was incubated with either 35S-labeled, in vitro translated C1D (lane 5), MPP6 (lane 6), PM/Scl-100 (lane 7) or MPP6 and PM/Scl-100 (lane 8). As a control, GST alone was incubated with all three labeled proteins (lane 4). After incubation, GST(-C1D)-containing complexes were precipitated with glutathione-Sepharose beads and analyzed by SDS-PAGE and autoradiography. In lanes 1–3, the in vitro translated C1D, MPP6 and PM/Scl-100 proteins were analyzed.
PM/Scl-100 is required for the nucleolar localization of C1D
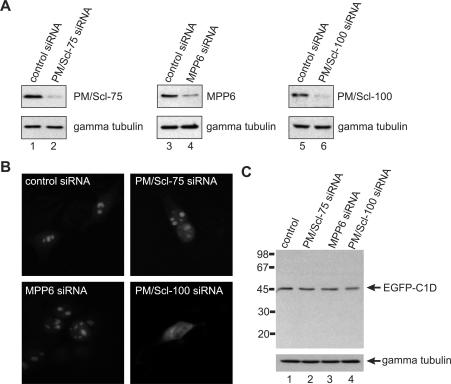
Both MPP6 and PM/Scl-100 contain a putative nuclear localization signal. C1D on the other hand does not have such a sequence motif raising the question whether PM/Scl-100 and/or MPP6 is/are required for the nuclear localization of C1D. To investigate this possibility, we transfected HEp-2 cells with a C1D-EGFP construct and after overnight incubation the cells were treated with siRNAs directed against PM/Scl-75, MPP6 and PM/Scl-100, or a control siRNA. The reduction in protein levels was monitored by western blotting, which confirmed the efficiency of the siRNAs used (Figure 4A). The results shown in Figure 4B demonstrate that upon knock-down of PM/Scl-100 the nucleolar accumulation of C1D is abrogated, whereas its localization was not affected upon knock-down of PM/Scl-75 or MPP6. Also, when the HEp-2 cells were transfected with the control siRNA, no change in the localization of C1D was observed. Although these results suggest that PM/Scl-100 is required for nucleolar localization of C1D, it is also possible that PM/Scl-100 knock-down reduced the stability of C1D-EGFP which might result in a cleaved EGFP fragment and the fluorescence pattern as observed in Figure 4B, panel PM/Scl-100 siRNA. To rule out the latter possibility, cell extracts of the transfected HEp-2 cells were analyzed by western blotting using anti-EGFP antibodies. The results shown in Figure 4C demonstrate that knock-down of PM/Scl-100 did neither result in the detection of EGFP-containing degradation products nor in a significant change of the amount of full-length C1D-EGFP. In this regard, it is also important to note that knock-down of PM/Scl-100 did not result in co-depletion of other exosome components, such as hRrp4, hRrp41, hRrp46 or PM/Scl-75. Taken together, these data show that the nucleolar accumulation of C1D is depending on its interaction with PM/Scl-100. Knock-down of exosome components PM/Scl-75 or PM/Scl-100 had no effect on the subcellular localization of hMtr4p and MPP6 in similar experiments (data not shown).
Figure 4.
Nucleolar accumulation of C1D requires PM/Scl-100. HEp-2 cells were transfected with a construct encoding C1D fused to the N-terminus of EGFP and 16 h after transfection cells were treated with a control siRNA (lanes 1, 3 and 5), or siRNAs targeting PM/Scl-75 (lane 2), MPP6 (lane 4) or PM/Scl-100 (lane 6). (A) After 24 h, cells were harvested, and total cell extracts were analyzed by western blotting using a polyclonal anti-PM/Scl-75 serum (lanes 1 and 2), a polyclonal anti-MPP6 serum (lanes 3 and 4) and a polyclonal anti-PM/Scl-100 serum (lanes 5 and 6). A mouse monoclonal antibody to gamma tubulin was used as a control. (B) Twenty-four hours after siRNA treatment, cells were fixed and the expressed C1D-EGFP fusion protein was visualized by fluorescence microscopy. Panel control siRNA, cells treated with a control siRNA; panel PM/Scl-75 siRNA, cells treated with the siRNA targeting PM/Scl-75; panel MPP6 siRNA, cells treated with the siRNA targeting MPP6; panel PM/Scl-100 siRNA, cells treated with the siRNA targeting PM/Scl-100. (C) To investigate the integrity of the C1D-EGFP fusion protein upon PM/Scl-100 siRNA-mediated knock-down, extracts were prepared from the transfected HEp-2 cells and analyzed by western blotting using a polyclonal anti-EGFP antiserum. A mouse monoclonal antibody directed against gamma tubulin was used as a control.
C1D and hMtr4p are required for the maturation of the 5.8S rRNA
The nucleolar accumulation of both C1D and hMtr4p, as well as their association with the exosome, suggested that both proteins might play a role in the processing of pre-rRNAs. In yeast, it has been demonstrated that the maturation of the 3′ end of the 5.8S rRNA requires the exosome as well as several auxiliary proteins including Rrp47p and Mtr4p (9,26,30). Recently, we demonstrated that the exosome and MPP6 are involved in this process in human cells (2).
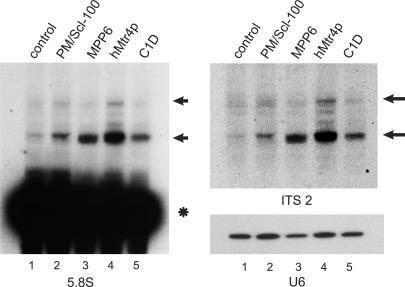
To investigate the involvement of C1D and hMtr4p in pre-rRNA processing, RNAi experiments were performed by transfection of HEp-2 cells with siRNAs directed against PM/Scl-100, MPP6, hMtr4p or C1D. Two days later RNA was isolated and analyzed by northern blot hybridization using a radiolabeled probe specific for the 5.8S rRNA (Figure 5A). The results demonstrate that knock-down of all four proteins leads to the accumulation of similar 5.8S rRNA precursors. The size of the faster migrating processing intermediate seemed to differ slightly between the RNAs from the cells in which these proteins were depleted. The same processing intermediates were detected with a probe specific for ITS2, which indicates that these 5.8S rRNA precursors are extended at their 3′ end (Figure 5B). Probing for other ribosomal processing intermediates such as 5′ ETS fragments did not result in detectable accumulation of processing intermediates corresponding to this part of the primary transcript (data not shown).
Figure 5.
Knock-down of hMtr4p and C1D leads to the accumulation of 5.8S rRNA precursors. HEp-2 cells were transiently transfected with siRNAs directed to PM/Scl-100 (lane 2), MPP6 (lane 3), hMtr4p (lane 4), C1D (lane 5) or with a control siRNA (lane 1). Cells were harvested 2 days after transfection and 2.5 µg of total RNA was analyzed by northern blot hybridization using radiolabeled probes specific for 5.8S rRNA (left) or ITS2 (upper right). As a control, a U6 snRNA probe was used (lower right). The positions of the 5.8S rRNA precursors are indicated by arrows and the position of mature 5.8S rRNA is indicated with an asterisk.
C1D is an RNA-binding protein
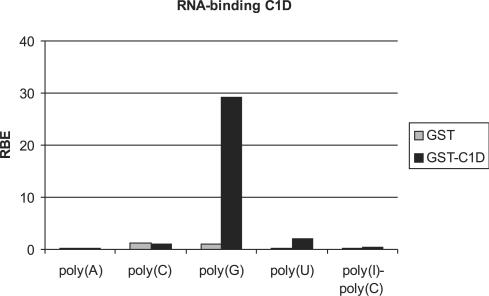
The results of the experiments described above demonstrate that the exosome together with MPP6, C1D and hMtr4p forms a multi-subunit complex involved in the 3′–5′ processing of nucleolar and possibly also nuclear RNA. This raises the question how these different components cooperate in this process. MPP6 has been shown to contain RNA-binding activity with preference for pyrimidine-rich sequences, which is predicted to be involved for the recruitment of the exosome to RNA substrates (2). The Mtr4p protein is a putative RNA helicase which is most likely required for resolving secondary structures that inhibit nucleolytic processing by the exosome (30). The C1D protein has been reported to be a member of the family of non-histone polypeptides involved in higher order chromatin folding (28), of which some have been found to be associated with highly repetitive DNA sequences (34). We conducted GST pull-down experiments to investigate whether C1D is also able to bind to RNA. GST-C1D was immobilized using glutathione-Sepharose beads, incubated with different 32P-labeled homopolynucleotides and the bound RNAs were quantified in a scintillation counter. The results in Figure 6 show that GST-C1D binds efficiently to poly(G), whereas no binding to poly(A), poly(C), poly(U) or poly(I)–poly(C) was observed. Additional experiments showed that C1D indeed binds to G-rich RNAs such as the ITS1, but also to tRNAs, which lack G-rich sequences, suggesting that the structure of the RNA may be important for C1D binding as well (data not shown).
Figure 6.
C1D binds RNA in vitro. GST and GST-C1D were incubated with radiolabeled homopolynucleotides and bound RNAs were quantified in a scintillation counter. The binding efficiency of GST and GST-MPP6 to poly(A), poly(C), poly(G), poly(U) and poly(I)–poly(C) is depicted as a percentage of input RNA (RBE: relative binding efficiency). These results are the average values of two independent experiments.
DISCUSSION
Although the molecular mechanisms by which the exosome is recruited to different RNA substrates is poorly understood, it is believed that in most cases auxiliary proteins are involved. Also, the processing or degradation of RNAs by the exosome may require additional proteins, such as helicases to resolve stable structures in the substrates. Here, we report the characterization of the association of the human C1D and hMtr4p proteins with the exosome and their involvement in pre-rRNA processing. Our data demonstrate that both C1D and hMtr4p accumulate in the nucleus with the highest concentrations found in the nucleoli. In addition, the results of our experiments provide insight into the way these proteins associate with the exosome. hMtr4p interacts with MPP6, which in turn binds to PM/Scl-100, C1D appeared to bind directly to PM/Scl-100. The binding of MPP6 and C1D to PM/Scl-100 is not mutually exclusive. The involvement of hMtr4p, MPP6 and C1D with exosome-mediated processing of pre-rRNA may be explained by the putative RNA helicase activity of hMtr4p and the RNA-binding capacity of MPP6 and C1D.
Identification of C1D and hMtr4p as auxiliary proteins for the human exosome
Strong support for the association of hMtr4p with the human exosome was already obtained several years ago by the co-purification of this protein with the TAP-tagged exosome complex from human cells (31). The exosome association of C1D was predicted based on the interaction of its yeast counterpart Rrp47p with the yeast exosome (26,35). The relatively low level of co-precipitation of hRrp4p with EGFP-C1D in comparison with PM/Scl-100 (Figure 2A) suggests that C1D is preferentially associated with exosome complexes containing PM/Scl-100. This is further supported by the direct interaction between C1D and PM/Scl-100 in both GST pull-down experiments. Moreover, in a mammalian two-hybrid system C1D interacted with PM/Scl-100, but failed to interact with all other exosome subunits (data not shown). The binding of MPP6 to both hMtr4p and PM/Scl-100 is consistent with previously published yeast two-hybrid data (5). In combination with the observation that C1D and MPP6 can bind simultaneously to PM/Scl-100, these data strongly suggest that the complex of C1D, MPP6 and PM/Scl-100 interacts with the exosome by virtue of the binding of PM/Scl-100 with the exosome. In this model, hMtr4p associates via its interaction with MPP6, although we have not yet succeeded in demonstrating the simultaneous binding of PM/Scl-100 and hMtr4p to MPP6. In agreement with this mode of assembly, the depletion of C1D or MPP6 did not interfere with the ability of PM/Scl-100 to interact with the core exosome, as monitored by Rrp4p co-immunoprecipitation (data not shown). The PM/Scl-100 protein is a putative 3′–5′ exoribonuclease with a conserved RNase D and HRDC (helicase and RNase D C-terminal) domain (36). Although, no known protein motifs can be identified in the primary structure of C1D, the protein was proposed to bind to the 3′-overhanging ends of DNA during repair (28,29). Here, we demonstrate that the nucleic-acid-binding capacity of C1D is not restricted to DNA, but that it also efficiently binds to structured RNAs including poly(G). Interestingly, we recently reported that MPP6 is also a RNA-binding protein, which preferentially binds to pyrimidine homopolymers (2). Thus, various RNA-binding specificities are associated with the exosome via PM/Scl-100 and therefore it will be interesting to investigate whether these contribute to the selectivity of PM/Scl-100 and/or the exosome for substrate RNAs.
PM/Scl-100 is required for the nucleolar accumulation of C1D
Nuclear entry of exosome-associated proteins may be facilitated by protein–protein interactions occurring in the cytoplasm and may proceed via a piggyback mechanism. Knock-down of exosome components or one of the auxiliary proteins did not affect the subcellular localization of hMtr4p and MPP6 (data not shown). Thus, these proteins seem to be capable to enter the nucleus independently of their association with components of the exosome. Knock-down of PM/Scl-100, on the other hand, led to reduced expression levels of C1D and abrogated the nucleolar accumulation of C1D. The PM/Scl-100 protein contains a putative nuclear localization signal (NLS; aa 752–758, AKKRERA). Since C1D binds very efficiently to PM/Scl-100, it is likely that C1D is dependent on PM/Scl-100 to enter the nuclei and to accumulate in the nucleoli. The MPP6 protein binds less efficiently to PM/Scl-100 in vitro and appears to enter the nucleus independently of PM/Scl-100, which might be mediated by the putative bipartite NLS (aa 116–132) in MPP6. Taken together, these data suggest that the identified exosome-associated proteins are transported to the nuclei via distinct mechanisms.
5.8S rRNA processing requires multiple human exosome auxiliary proteins
In yeast, the exosome was initially identified as a complex required for the exonucleolytic 3′ end maturation of the 5.8S rRNA (22). Later, it became evident that even for this relatively simple processing step the core exosome requires additional protein factors, such as Rrp6p, Rrp47p and Mtr4p (9,19,26,30). Here, we have shown that besides the core exosome and PM/Scl-100 at least three additional proteins, C1D, MPP6 and hMtr4p, are involved in 5.8S rRNA maturation in human cells. In yeast, the depletion of core exosome components, Rrp6p, Rrp47p or Mtr4p results in the accumulation of different 5.8S rRNA-processing intermediates (18,19,26,30). In the human system, no major differences in the accumulation of 5.8S rRNA precursors were observed after knock-down of PM/Scl-100, C1D and hMtr4p. Previously, accumulation of similar precursors was also observed after depletion of hRrp41p and the exosome-associated protein MPP6 (2). The observed differences between the yeast and human systems may be explained by the depletion of the whole exosome from the site of processing upon knock-down of auxiliary proteins of the exosome in the human system.
Besides rRNA processing yeast Rrp47p and Mtr4p were shown to be involved in the 3′ end processing of many small nuclear and nucleolar RNAs (9,21,25,26,30,37,38). Although we have extensively investigated whether similar defects in 3′ end processing of such sn(o)RNAs upon knock-down of MPP6, C1D or hMtr4p occur in human cells, we failed to detect any accumulating precursors. In yeast, it has been shown that for several exosome substrates, such as the U4 and U5 snRNA, other 3′–5′ exoribonucleases can be involved in the 3′ end maturation of these RNAs as well (39). Currently, it is unknown whether the failure to detect such processing intermediates in human cells is due to processing factor redundancy or to the low abundance of sn(o) RNA precursors in these cells.
ACKNOWLEDGEMENTS
We would like to thank Stijn van Dongen for the mammalian two-hybrid experiments, Wiljan Hendriks (Department of Cell Biology, University of Nijmegen, The Netherlands) for the anti-EGFP antibodies and Diarect AG (Freiburg, Germany) for the recombinant PM/Scl-100 protein. This work was supported in part by the Council for Chemical Sciences (NWO-CW) and the Council of Earth and Life Sciences (NWO-ALW) of the Netherlands Organization for Scientific Research. Funding to pay the Open Access publication charges for this article was provided by Radboud University Nijmegen.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lejeune F, Li X, Maquat LE. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol. Cell. 2003;12:675–687. doi: 10.1016/s1097-2765(03)00349-6. [DOI] [PubMed] [Google Scholar]
- 2.Schilders G, Raijmakers R, Raats JMH, Pruijn GJM. MPP6 is an exosome-associated RNA-binding protein involved in 5.8S rRNA maturation. Nucleic Acids Res. 2005;33:6795–6804. doi: 10.1093/nar/gki982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aloy P, Ciccarelli FD, Leutwein C, Gavin AC, Superti-Furga G, Bork P, Bottcher B, Russell RB. A complex prediction: three-dimensional model of the yeast exosome. EMBO Rep. 2002;3:628–635. doi: 10.1093/embo-reports/kvf135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buttner K, Wenig K, Hopfner KP. Structural framework for the mechanism of archaeal exosomes in RNA processing. Mol. Cell. 2005;20:461–471. doi: 10.1016/j.molcel.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 5.Lehner B, Sanderson CM. A protein interaction framework for human mRNA degradation. Genome Res. 2004;14:1315–1323. doi: 10.1101/gr.2122004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lorentzen E, Walter P, Fribourg S, Evguenieva-Hackenberg E, Klug G, Conti E. The archaeal exosome core is a hexameric ring structure with three catalytic subunits. Nat. Struct. Mol. Biol. 2005;12:575–581. doi: 10.1038/nsmb952. [DOI] [PubMed] [Google Scholar]
- 7.Schilders G, van Dijk E, Raijmakers R, Pruijn GJ. Cell and molecular biology of the exosome: how to make or break an RNA. Int. Rev. Cytol. 2006;251:159–208. doi: 10.1016/S0074-7696(06)51005-8. [DOI] [PubMed] [Google Scholar]
- 8.Liu Q, Greimann JC, Lima CD. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:1223–1237. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- 9.Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, Tollervey D. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 1999;18:5399–5410. doi: 10.1093/emboj/18.19.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van-Hoof A, Lennertz P, Parker R. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol. Cell Biol. 2000;20:441–452. doi: 10.1128/mcb.20.2.441-452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bousquet-Antonelli C, Presutti C, Tollervey D. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell. 2000;102:765–775. doi: 10.1016/s0092-8674(00)00065-9. [DOI] [PubMed] [Google Scholar]
- 12.Hilleren P, McCarthy T, Rosbash M, Parker R, Jensen TH. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature. 2001;413:538–542. doi: 10.1038/35097110. [DOI] [PubMed] [Google Scholar]
- 13.Torchet C, Bousquet-Antonelli C, Milligan L, Thompson E, Kufel J, Tollervey D. Processing of 3′-extended read-through transcripts by the exosome can generate functional mRNAs. Mol. Cell. 2002;9:1285–1296. doi: 10.1016/s1097-2765(02)00544-0. [DOI] [PubMed] [Google Scholar]
- 14.Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev. 2004;18:1227–1240. doi: 10.1101/gad.1183804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vanacova S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fatica A, Tollervey D. Making ribosomes. Curr. Opin. Cell Biol. 2002;14:313–318. doi: 10.1016/s0955-0674(02)00336-8. [DOI] [PubMed] [Google Scholar]
- 17.Fromont-Racine M, Senger B, Saveanu C, Fasiolo F. Ribosome assembly in eukaryotes. Gene. 2003;313:17–42. doi: 10.1016/s0378-1119(03)00629-2. [DOI] [PubMed] [Google Scholar]
- 18.Allmang C, Mitchell P, Petfalski E, Tollervey D. Degradation of ribosomal RNA precursors by the exosome. Nucleic Acids Res. 2000;28:1684–1691. doi: 10.1093/nar/28.8.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Briggs MW, Burkard KT, Butler JS. Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8S rRNA 3′ end formation. J. Biol. Chem. 1998;273:13255–13263. doi: 10.1074/jbc.273.21.13255. [DOI] [PubMed] [Google Scholar]
- 20.Fang F, Hoskins J, Butler JS. 5-Fluorouracil enhances exosome-dependent accumulation of polyadenylated rRNAs. Mol. Cell Biol. 2004;24:10766–10776. doi: 10.1128/MCB.24.24.10766-10776.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 22.Mitchell P, Petfalski E, Tollervey D. The 3′ end of yeast 5.8S rRNA is generated by an exonuclease processing mechanism. Genes Dev. 1996;10:502–513. doi: 10.1101/gad.10.4.502. [DOI] [PubMed] [Google Scholar]
- 23.Zanchin NI, Goldfarb DS. The exosome subunit Rrp43p is required for the efficient maturation of 5.8S, 18S and 25S rRNA. Nucleic Acids Res. 1999;27:1283–1288. doi: 10.1093/nar/27.5.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Win TZ, Draper S, Read RL, Pearce J, Norbury CJ, Wang SW. Requirement of fission yeast Cid14 in polyadenylation of rRNAs. Mol. Cell Biol. 2006;26:1710–1721. doi: 10.1128/MCB.26.5.1710-1721.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Regnault B, Devaux F, Namane A, et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 26.Mitchell P, Petfalski E, Houalla R, Podtelejnikov A, Mann M, Tollervey D. Rrp47p Is an exosome-associated protein required for the 3′ processing of stable RNAs. Mol. Cell Biol. 2003;23:6982–6992. doi: 10.1128/MCB.23.19.6982-6992.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hieronymus H, Yu MC, Silver PA. Genome-wide mRNA surveillance is coupled to mRNA export. Genes Dev. 2004;18:2652–2662. doi: 10.1101/gad.1241204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nehls P, Keck T, Greferath R, Spiess E, Glaser T, Rothbarth K, Stammer H, Werner D. cDNA cloning, recombinant expression and characterization of polypeptides with exceptional DNA affinity. Nucleic Acids Res. 1998;26:1160–1166. doi: 10.1093/nar/26.5.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yavuzer U, Smith GC, Bliss T, Werner D, Jackson SP. DNA end-independent activation of DNA-PK mediated via association with the DNA-binding protein C1D. Genes Dev. 1998;12:2188–2199. doi: 10.1101/gad.12.14.2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de-la-Cruz J, Kressler D, Tollervey D, Linder P. Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8S rRNA in Saccharomyces cerevisiae. EMBO J. 1998;17:1128–1140. doi: 10.1093/emboj/17.4.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, Pruijn GJ, Stoecklin G, Moroni C, Mann M, et al. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- 32.Frangioni JV, Neel BG. Solubilization and purification of enzymatically active glutathione S-transferase (pGEX) fusion proteins. Anal. Biochem. 1993;210:179–187. doi: 10.1006/abio.1993.1170. [DOI] [PubMed] [Google Scholar]
- 33.Raijmakers R, Egberts WV, van Venrooij WJ, Pruijn GJ. The association of the human PM/Scl-75 autoantigen with the exosome is dependent on a newly identified N terminus. J. Biol. Chem. 2003;278:30698–30704. doi: 10.1074/jbc.M302488200. [DOI] [PubMed] [Google Scholar]
- 34.Neuer-Nitsche B, Lu XN, Werner D. Functional role of a highly repetitive DNA sequence in anchorage of the mouse genome. Nucleic Acids Res. 1988;16:8351–8360. doi: 10.1093/nar/16.17.8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peng WT, Robinson MD, Mnaimneh S, Krogan NJ, Cagney G, Morris Q, Davierwala AP, Grigull J, Yang X, et al. A panoramic view of yeast noncoding RNA processing. Cell. 2003;113:919–933. doi: 10.1016/s0092-8674(03)00466-5. [DOI] [PubMed] [Google Scholar]
- 36.Midtgaard SF, Assenholt J, Jonstrup AT, Van LB, Jensen TH, Brodersen DE. Structure of the nuclear exosome component Rrp6p reveals an interplay between the active site and the HRDC domain. Proc. Natl. Acad. Sci. USA. 2006;103:11898–11903. doi: 10.1073/pnas.0604731103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Egecioglu DE, Henras AK, Chanfreau GF. Contributions of Trf4p- and Trf5p-dependent polyadenylation to the processing and degradative functions of the yeast nuclear exosome. RNA. 2006;12:26–32. doi: 10.1261/rna.2207206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Houseley J, Tollervey D. Yeast Trf5p is a nuclear poly(A) polymerase. EMBO Rep. 2006;7:205–211. doi: 10.1038/sj.embor.7400612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van-Hoof A, Lennertz P, Parker R. Three conserved members of the RNase D family have unique and overlapping functions in the processing of 5S, 5.8S, U4, U5, RNase MRP and RNase P RNAs in yeast. EMBO J. 2000;19:1357–1365. doi: 10.1093/emboj/19.6.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]