Abstract
Current literature describes several methods for the design of efficient siRNAs with 19 perfectly matched base pairs and 2 nt overhangs. Using four independent databases totaling 3336 experimentally verified siRNAs, we compared how well several of these methods predict siRNA cleavage efficiency. According to receiver operating characteristics (ROC) and correlation analyses, the best programs were BioPredsi, ThermoComposition and DSIR. We also studied individual parameters that significantly and consistently correlated with siRNA efficacy in different databases. As a result of this work we developed a new method which utilizes linear regression fitting with local duplex stability, nucleotide position-dependent preferences and total G/C content of siRNA duplexes as input parameters. The new method's discrimination ability of efficient and inefficient siRNAs is comparable with that of the best methods identified, but its parameters are more obviously related to the mechanisms of siRNA action in comparison with BioPredsi. This permits insight to the underlying physical features and relative importance of the parameters. The new method of predicting siRNA efficiency is faster than that of ThermoComposition because it does not employ time-consuming RNA secondary structure calculations and has much less parameters than DSIR. It is available as a web tool called ‘siRNA scales’.
INTRODUCTION
This work addresses chemically synthesized siRNAs. After introduction of siRNAs into cells in the form of synthetic oligonucleotide duplexes, there is specific cleavage and subsequent degradation of target mRNAs, but the mRNA cleavage efficacy varies. Predicting the cleavage efficacy of siRNAs is an important task, the success of which influences the prospects of using this comparatively new method for genetic studies. The first attempt to compare siRNA efficacy predictors was done in 2004 by Sætrom (1). The efficacy of siRNAs is now predicted using several approaches and webtools developed in the last two years following analyses of experimental data. There is no clear indication yet as to which approach or web tool is optimal (2–22) and what siRNA features are crucial for the optimal prediction (23,24).
All parameters (See Appendix note 1) in the methods currently in use are either related (group 1), or unrelated (group 2), to the stability of the siRNA duplex termini. Group 1 includes the duplex termini stability (as calculated by  ), or the presence or absence of a specific nucleotide at a certain duplex terminal position. Group 2 includes, for example, the percentage of a particular nucleotide in the sense or antisense strand of siRNA, the presence or absence of a specific nucleotide at a certain internal position, the stability of secondary structures of target mRNA or the stability of the siRNA antisense strand.
), or the presence or absence of a specific nucleotide at a certain duplex terminal position. Group 2 includes, for example, the percentage of a particular nucleotide in the sense or antisense strand of siRNA, the presence or absence of a specific nucleotide at a certain internal position, the stability of secondary structures of target mRNA or the stability of the siRNA antisense strand.
The rules for group 1 relate to siRNA strand entry into the RNA-induced silencing complex (RISC); entry of the siRNA antisense strand is critical for mRNA cleavage. The siRNA duplex strand with the least stable 5′ end enters RISC faster than the other strand (2,4). Accordingly, the 5′ end of the strand that is antisense to the target, has to be AU-rich and its 3′ end GC-rich. This is more formally captured by the 5′ terminal free energy ( ) of the antisense strand and the difference in 5′ terminal free energy (
) of the antisense strand and the difference in 5′ terminal free energy (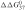 ) of the sense and antisense strand.
) of the sense and antisense strand.
Since different researchers have used different parameters and lengths of terminal sequences ranging from 1 to 7 nt (2,4–7), there is no uniformity in the current rules for group 1. Also there is no general agreement about the best way to calculate and apply the parameters for group 2. Consequently, the currently developed webtools for efficient siRNA design frequently employ variable, user adjustable settings for specifying parameters and selection rules in their algorithms. To make users better equipped to choose the best design algorithm and parameter settings, we compared several existing approaches on the several thousand experimental data points recently generated (2,4,8,25). Furthermore, we used the largest of these datasets to develop a new method with optimal group 1 and group 2 parameters.
RESULTS
We collected siRNA experimental databases using previously described siRNA silencing experiments performed at Isis Pharmaceuticals (25), Amgen (4), Dharmacon (2), Sloan Kettering Cancer Center (9) and Novartis (8). The data produced by Amgen and Dharmacon were pooled together in one database for the present work. All databases are presented in Table 1 and are available for downloading from http://gesteland.genetics.utah.edu/members/olgaM/siRNA_database_September_2006.xls
Table 1.
Summary of features of experimental databases
| Base | Database name | Concentration of siRNAs (nM) | Success rate in database (efficient siRNAs versus total amount) | Total Number of siRNAs tested (n) |
|---|---|---|---|---|
| 1 | Isis Pharmaceuticals | 100 | 9 | 67 |
| 2 | Amgen/Dharmacon | 100 | 56 | 238 |
| 3 | Sloan Kettering | 100 | 30 | 601 |
| 4 | Novartis | 50 | 50 | 2430 |
For success rate calculations, siRNAs that yielded at least 70% target gene silencing, were considered to be efficient. Other siRNAs were considered to be ‘inefficient’.
Comparison and performance evaluation of published siRNA design algorithms
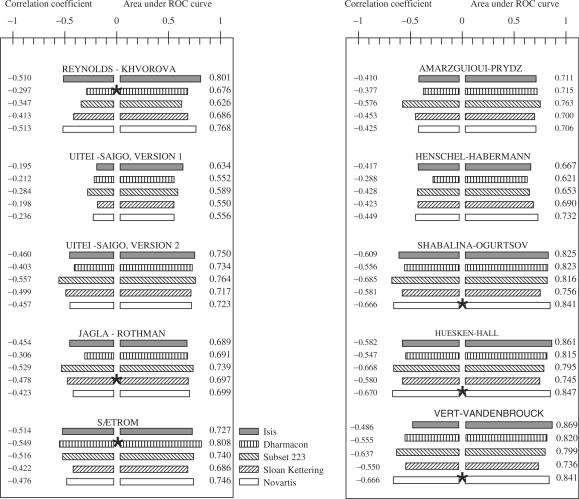
Our first goal was to compare the siRNA design approaches described in published literature. Two criteria were used for choosing approaches for the comparison study. The approach had to be either easily reproduced by us or its authors had to be willing to cooperate by calculating predicted siRNA efficacy values for the four experimental databases compiled for this study (Table 1). The approaches used in this work are named according to the first and last authors of the relevant publications (Figure 1).
Figure 1.
A comparison of siRNA efficacy predictors on four experimental databases (9 published approaches). The list of analyzed approaches included: ‘8 criteria algorithm’ by Reynolds–Khvorova (2), four rules algorithm by Ui Tei-Saigo (6) version 1, four rules scoring algorithm by Ui Tei-Saigo version 2 modified by us (described in this study, see Appendix note 2), a decision tree based algorithm by Jagla–Rothman (9), genetic programming algorithm Gpboost by Sætrom (3), 6 criteria algorithm by Amarzguioui-Prydz (5), algorithm created by Henschel-Habermann (12) used by program DEQOR, BioPredsi algorithm by Huesken-Hall (8), ThermoComposition by Shabalina-Ogurtsov (10) and DSIR by Vert-Vandenbrouck (11). The histogram was created on the basis of the analysis of four databases and one data subset using published and unpublished siRNA design algorithms. For each database and each algorithm, the area under a corresponding ROC curve, as well as the correlation coefficient between experimental and theoretically predicted siRNA efficacy, were calculated. The star indicates that the relevant databases were used for creating the algorithm. So, the statistical characteristics (correlation coefficient and area under ROC curve values) might be positively overestimated for these cases. The right part of the histogram shows the areas under the ROC curves. The left part of the histogram shows correlation coefficients between experimentally obtained and predicted values of siRNA efficacy. The significance values for correlation and ROC analyses are reported in ‘Supplementary Table 1 online’. The columns with variable filling indicate different databases.
We compared the approaches by using receiver operating characteristics (ROC) and correlation analyses (Figure 1). An ROC curve describes the relationship between specificity and the sensitivity of an approach (see Methods section). The area under the ROC curve captures the approach's overall performance such that an area of 1 indicates a perfect classification, and an area of 0.5 indicates a random classification. We also performed correlation analysis between experimentally determined and predicted siRNA efficacy (Figure 1). For correlation and ROC analyses, the levels of mRNA or proteins remaining in cells after siRNA treatments were expressed as percentages of the control levels. For ROC analysis, siRNAs that yielded at least 70% target gene knockdown were considered to be efficient; other siRNAs were considered inefficient.
All analyzed approaches can, to some degree, discriminate between efficient and inefficient siRNAs. The correlation coefficients are significant and the areas under the ROC curves are higher than 0.5 for all approaches and for all databases. All ROC values obtained with all the approaches are significant for the three largest databases (Amgen/Dharmacon, Sloan-Kettering, Novartis). BioPredsi by Huesken-Hall (8), ThermoComposition by Shabalina-Ogurtsov (10) and DSIR by Vert-Vandenbrouck (11) have better performance than the others. These three approaches have the highest absolute correlation coefficient values and the largest areas under their ROC curves on all datasets (Figure 1). Furthermore, whereas the performance of the other approaches varies on the different datasets, BioPredsi, ThermoComposition and DSIR have statistically similar performance on all datasets (performance measured in terms of ROC-area and sensitivity at highly specific algorithm thresholds; see Supplementary Tables 6–10 online).
The neural network BioPredsi approach by Huesken-Hall was trained using siRNAs in which the antisense strands (21-mers) were completely complementary to mRNA. In the databases from Isis, Amgen/Dharmacon and SloanKettering, mainly 19 mRNA nucleotides were targeted. Only a small subset of the data (223 data points) was represented by siRNAs in which dTdT overhangs in the siRNA antisense strands were complementary to ‘AA’ dinucleotides in mRNAs. For fairly evaluating the performance of the BioPredsi algorithm, we chose this subset of data and calculated the areas under the ROC curves and correlation coefficients for the predicted and experimentally obtained data. For this subset, the BioPredsi algorithm shows a better correlation coefficient, but the area under the ROC curve did not improve (Figure 1).
We used two versions of the ThermoComposition algorithm in this work. One version was trained on a heterogeneous set of 653 siRNAs to classify 19-mers (10). The other version was trained on the Novartis database to classify 21-mers. Both versions used correlation and Student's t-test analyses to determine position-dependent nucleotide preferences and avoidances, and used linear regression to combine these into siRNA efficacy predictors. As we used the Novartis database to train the 21-mer version, the reported prediction results on this dataset are from a standard non-overlapping cross-validation experiment. Both the 19-mer and 21-mer classifiers are available at ftp://ftp.ncbi.nlm.nih.gov/pub/shabalin/siRNA/ThermoComposition
It was shown that the performance of approaches based on both thermodynamic and composition features is almost identical for neural networks and linear regression (10). Our next goals were (1) to analyze parameters used in the described approaches and (2) to create a regression method based on the best parameters.
Parameter selection
Using correlation analyses, we studied the influence of such parameters as siRNA local duplex stability (measured as  for two neighboring nucleotides), nucleotide position preferences and total G/C content on siRNA efficacy in the four experimental databases.
for two neighboring nucleotides), nucleotide position preferences and total G/C content on siRNA efficacy in the four experimental databases.
Duplex stability profile
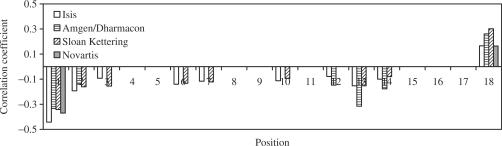
We analyzed the correlation between observed siRNA efficacy and thermodynamic stability ( ) calculated for every two base pairs along the duplex siRNA antisense strand. Thermodynamic parameters for the calculations were published earlier (26). Only correlation coefficients that are significant (P < 0.05) at least in two databases, are shown (Figure 2). Relationships between siRNA efficacy and
) calculated for every two base pairs along the duplex siRNA antisense strand. Thermodynamic parameters for the calculations were published earlier (26). Only correlation coefficients that are significant (P < 0.05) at least in two databases, are shown (Figure 2). Relationships between siRNA efficacy and  are strongest and most consistent for the first and last two base pairs. The second and third two base pairs demonstrate weaker correlations and less consistency. These observations provide evidence that the thermodynamic stability in the first two base pairs is a good indicator of siRNA efficacy and thermodynamic consideration of four terminal nucleotides provides poorer correlation with siRNA efficacy (see Table 2). Figure 2 also shows that some instability for two base pairs at the 6th, 7th, 10th, 12th and 13th positions from the 5′ end of antisense strand might be related to siRNA efficacy.
are strongest and most consistent for the first and last two base pairs. The second and third two base pairs demonstrate weaker correlations and less consistency. These observations provide evidence that the thermodynamic stability in the first two base pairs is a good indicator of siRNA efficacy and thermodynamic consideration of four terminal nucleotides provides poorer correlation with siRNA efficacy (see Table 2). Figure 2 also shows that some instability for two base pairs at the 6th, 7th, 10th, 12th and 13th positions from the 5′ end of antisense strand might be related to siRNA efficacy.
Figure 2.
Correlation between the siRNA efficacy in cell and  values relevant to siRNA local duplex stability from position 1 to 18 in the antisense strand.
values relevant to siRNA local duplex stability from position 1 to 18 in the antisense strand.  values were calculated for each of two neighboring base pairs in siRNA duplexes starting from the 5′ end of the antisense strand. siRNA efficacy values were measured as remaining levels of mRNA or normalized protein products levels. Only correlation coefficients that are significant (P < 0.05) for at least in two databases, are shown.
values were calculated for each of two neighboring base pairs in siRNA duplexes starting from the 5′ end of the antisense strand. siRNA efficacy values were measured as remaining levels of mRNA or normalized protein products levels. Only correlation coefficients that are significant (P < 0.05) for at least in two databases, are shown.
Table 2.
Evaluation of the relationships between siRNA efficacy and theoretically calculated values
| Base | Database name | ΔG values of the 5′ duplex terminal base pairs (5′ antisense strand) | ΔΔG values of the duplex 5′ two base pairs (ΔG 5′ of antisense strand minus ΔG of 5′ of sense strand) | Output values from model 1. | |
|---|---|---|---|---|---|
| Two | Four | ||||
| 1 | Isis Pharmaceuticals | −0.414 | N.S. | −0.450 | −0.475 |
| 2 | Amgen/Dharmacon | −0.355 | N.S. | −0.457 | −0.477 |
| 3 | Sloan Kettering | −0.370 | −0.344 | −0.477 | −0.487 |
| 4 | Novartis | −0.469 | −0.378 | −0.475 | −0.578 |
Correlation coefficients calculated for  and
and 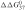 parameters as well as for model 1 output values. The significance values for relevant correlation coefficients are shown in Supplementary Table 2 online.
parameters as well as for model 1 output values. The significance values for relevant correlation coefficients are shown in Supplementary Table 2 online.
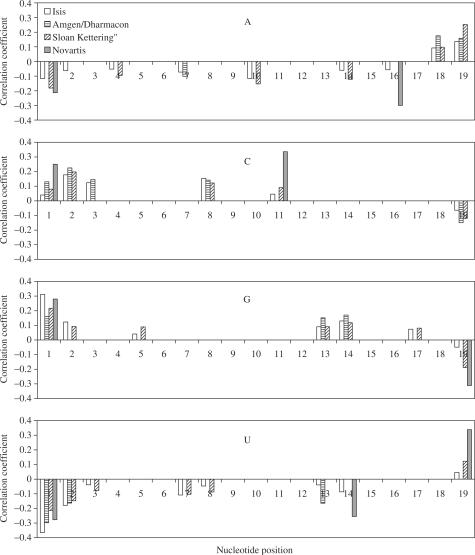
Position-dependent consensus
To determine position-dependent nucleotide preferences and avoidances, we computed the correlation coefficients between siRNA efficacy and the presence of A, G, C or U in the different positions in the antisense strand. Those coefficients that are significant (P < 0.05) at least in two databases, are shown in Figure 3. A positive correlation means that the corresponding nucleotide is less frequently found in efficient siRNAs, while negative correlation means that the corresponding nucleotide is more frequently found in efficient siRNAs. The strongest correlations with siRNA efficacy were found for U/A presence at the 5′ end and for G/C presence at the 3′ end of the antisense strand. The preferences and avoidance of other nucleotides at different positions of siRNA antisense strands which are significant (P < 0.05) for at least two databases, are summarized in Table 3.
Figure 3.
Correlation between the siRNA efficacy in cell and nucleotide occurrence in the antisense strand of siRNA duplex. Nucleotide presence or absence was registered for each position starting from the 5′ end of the antisense strand. siRNA efficacy values were measured as the remaining levels of mRNA or normalized protein products levels. Only correlation coefficients that are significant (P < 0.05) at least in two databases, are shown.
Table 3.
Nucleotides preferences and avoidance in the different positions of antisense strand staring from 5′ end
| Nucleotides AS/SS | Positions | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Antisense strand (AS) | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 |
| Sense Strand (SS) | 19 | 18 | 17 | 16 | 15 | 14 | 13 | 12 | 11 | 10 | 9 | 8 | 7 | 6 | 5 | 4 | 3 | 2 | 1 |
| A/U | + | + | + | + | + | + | − | − | |||||||||||
| G/C | − | − | − | − | − | − | + | ||||||||||||
| C/G | − | − | − | − | − | + | |||||||||||||
| U/A | + | + | + | + | + | + | + | − | |||||||||||
Plus and minus signs indicate that the presence or absence of relevant nucleotides at corresponding positions is related to siRNA efficacy in a significant way at least in two experimental datasets. Plus sign means that the nucleotide is preferred in efficient siRNAs; minus sign indicates nucleotide avoidance.
Total nucleotide content
We were unable to find any significant correlation between siRNA efficacy and total nucleotide content of siRNA antisense strand (total A, G, C and U), which are present in at least two experimental datasets. So we focused our study on total G + C content.
It was shown before that G + C nucleotide content for siRNA sequences is related to siRNA efficacy (2). Here we found a weight coefficient (Supplementary Table 3 online) for this parameter and used it in our new model. This weight coefficient is assigned for siRNA sequences with G + C content being in the interval from 20 to 53%; for those that are outside the interval, a weight equal to 0 is assigned.
Model selection
In this study, we chose to work with linear regression rather than with neural networks or other data mining techniques that can pick up nonlinearity in relationships between siRNA activity and a model's parameters. It is likely that linear relationships between model parameters and siRNA efficacy values are more common and consequently, nonlinearity is rare (10). In addition, linear regression models generate lists of relative weighting of the significant model parameters. So, for similar parameters such as nucleotide identity at certain positions, their relative importance for predicting siRNA efficacy can be established. This is much more difficult to achieve using a neural network approach (8), which does not reveal the relevant importance of its input parameters. Linear regression models can provide lists of weights of significant model parameters; so for similar parameters such as nucleotide identity at certain positions, their relative importance in the prediction of siRNA efficacy can be established. This can help in planning future experiments for finding functional meaning of nucleotide preferences at certain positions. The relative importance of specific nucleotides for siRNA function is not easy to estimate using a neural network approach.
A new method for siRNA design
Linear regression model with group 1 parameters
To create a new method, we started to analyze group 1 parameters, which are related to the stability of siRNA duplex termini. This stability is responsible for the rate of entry of the duplex strands into RISC. Since we found that  of the first two base pairs in the antisense strand correlates most strongly with siRNA efficacy, we used this parameter in our model. We also included
of the first two base pairs in the antisense strand correlates most strongly with siRNA efficacy, we used this parameter in our model. We also included  of the last two base pairs in the antisense strand as it also showed consistent and significant correlation with siRNA efficacy (Figure 2).
of the last two base pairs in the antisense strand as it also showed consistent and significant correlation with siRNA efficacy (Figure 2).
The  values need to be adjusted for dangling end effects. The identity of the terminal 5′ paired nucleotide of siRNA duplexes and composition of 3′ non-paired overhangs are related to these effects. Including these factors can improve the relationship of calculated terminal siRNA duplex stability with siRNA efficacy. Dangling end parameters have been published for DNA and RNA (27–30), but we are dealing with RNA sequences which have DNA overhangs. Parameters for such hybrid DNA–RNA sequences have not been published. Addition of published DNA or RNA parameters did not improve our model. Therefore, we made an attempt to add the identity of nucleotides located at positions 1, 19, 20 and 21 in the antisense strand into the linear regression model. Accordingly, the model with group 1 parameters used siRNA efficacy value as the dependant variable and local stability of siRNA duplex ends (
values need to be adjusted for dangling end effects. The identity of the terminal 5′ paired nucleotide of siRNA duplexes and composition of 3′ non-paired overhangs are related to these effects. Including these factors can improve the relationship of calculated terminal siRNA duplex stability with siRNA efficacy. Dangling end parameters have been published for DNA and RNA (27–30), but we are dealing with RNA sequences which have DNA overhangs. Parameters for such hybrid DNA–RNA sequences have not been published. Addition of published DNA or RNA parameters did not improve our model. Therefore, we made an attempt to add the identity of nucleotides located at positions 1, 19, 20 and 21 in the antisense strand into the linear regression model. Accordingly, the model with group 1 parameters used siRNA efficacy value as the dependant variable and local stability of siRNA duplex ends ( ), as well as nucleotides located at positions 1, 19, 20, 21 in siRNA duplex antisense strands, as independent variables. We trained the model using the Novartis database and cross-validated it using the remaining experimental databases.
), as well as nucleotides located at positions 1, 19, 20, 21 in siRNA duplex antisense strands, as independent variables. We trained the model using the Novartis database and cross-validated it using the remaining experimental databases.
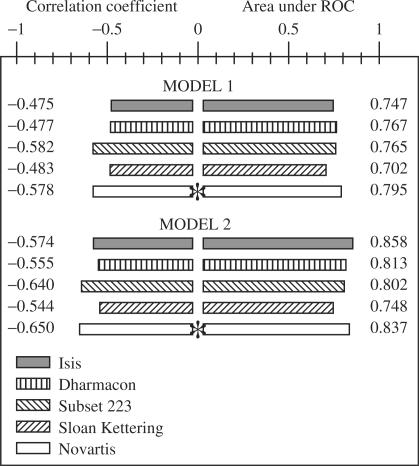
The statistical values related to the model's input are shown in Supplementary Table 3 online. The statistical values related to the model's output are shown in Figure 4. The correlation from the cross-validation study demonstrates that the model output can predict better siRNA efficacy than  or
or 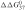 calculated for two terminal nucleotide base pairs (Table 2).
calculated for two terminal nucleotide base pairs (Table 2).
Figure 4.
Statistical characteristics obtained for model 1 and 2. The figure was created on the basis of the analysis of four databases. For each database and approach, the area under a corresponding ROC curve, as well as the correlation coefficient between experimental and theoretically predicted siRNA efficacy, were calculated. The star near the columns indicates that the relevant databases were used for approach creation. The left part of the histogram shows correlation coefficients between experimentally obtained and predicted values of siRNA efficacy. The right part of the histogram shows values for the areas under the ROC curves. The significance values for correlation coefficients and ROC curve areas are shown in Supplementary Table 5 online.
Linear regression model with group 2 parameters
It is known that local duplex stability values ( ) and certain nucleotides at specific internal positions of siRNA antisense strands can influence siRNA efficacy. To more accurately analyze these features and the G + C composition factor, a linear model with group 1 and 2 parameters was created. The model integrated the parameters found to be significant for model 1 and the new group 2 parameters. These new parameters are described in the ‘Methods’ section. For the best correlation between siRNA efficacy and G + C content, an input weighting value of 1 point was assigned for GC content in the interval from 20 to 52% in 21-mers, with 0 being used outside this range. The other parameters found to be significant for model input are shown in Figures 2 and 3.
) and certain nucleotides at specific internal positions of siRNA antisense strands can influence siRNA efficacy. To more accurately analyze these features and the G + C composition factor, a linear model with group 1 and 2 parameters was created. The model integrated the parameters found to be significant for model 1 and the new group 2 parameters. These new parameters are described in the ‘Methods’ section. For the best correlation between siRNA efficacy and G + C content, an input weighting value of 1 point was assigned for GC content in the interval from 20 to 52% in 21-mers, with 0 being used outside this range. The other parameters found to be significant for model input are shown in Figures 2 and 3.
The model with group 2 parameters’ output demonstrated that some nucleotides and some internal local stability values, as well as G + C composition, are factors that influence siRNA efficacy prediction. At certain positions of the siRNA duplex, local stability and identity of nucleotides are more important than in others (Table 3). Model 2 was cross-validated using the remaining experimental datasets (correlation coefficients and ROC curve areas were calculated for training and testing datasets). A summary of the comparative results for two models is shown in Figure 4. Group 2 parameters improve the predictability of siRNA efficacy. According to the method's correlation and sensitivity at highly specific thresholds, the linear regression model, based on the current study's parameters from groups 1 and 2, predicts siRNA efficacy with comparable performance to the earlier neural network models (8,10).
siRNA–scale web program
An interactive tool siRNA-scale was created using JavaScript. The tool takes an mRNA sequence as an input and splits the sequence into 21-mers. Antisense strands of these 21-mers include 3′ overhangs (two nucleotides that are complementary to target mRNA). For each potential siRNA candidate, the tool outputs the sense 19-mer (overhanging two nucleotides are not shown) and antisense 21-mer (overhanging two nucleotides are shown) of the corresponding siRNA and the predicted value of the siRNA efficacy (remaining amount of non cleaved mRNA in percentage towards control). In the siRNA-scale output, the sequences of siRNA candidates that are expected to lead to efficient cleavage of target mRNA are highlighted in gold. Other siRNA candidates are not highlighted. siRNA-scale can also use as input a list of siRNA duplex antisense strands from an experimentally obtained database, in which efficacy of each siRNA molecule is determined. This option allows verification of a method's discriminatory activity with newly produced experimental databases. SiRNA-scales is located at http://gesteland.genetics.utah.edu/siRNA scales.
DISCUSSION
The previous comparative analyses of siRNA design approaches were performed when a smaller number of approaches had been developed (1,15). With the intervening expansion of databases and approaches, the comparison performed here proved to be informative, with three of the ten pre-existing approaches (8,10,11) faring better than the others. The goals for performing this analysis were (1) to compare the previously developed methods and (2) to create a new, fast, transparent and user friendly method for web-based siRNA design which is convenient for different groups of researchers.
Comparison of newly created siRNA design method with previously reported methods
In this study we found that the newly created method is comparable in its prediction efficiency to the three most efficient earlier methods. However the new method has certain advantages. First, it is faster than the ThermoComposition method by Shabalina-Ogurtsov because it does not employ time-consuming RNA secondary structure calculations (10,31). Second, linear regression models generate lists of relative weighting of the significant model parameters, so, for similar parameters such as nucleotide identity at certain positions, their relative importance for predicting siRNA efficacy can be established. This is much more difficult to achieve using a neural network approach (8), which does not reveal the relevant importance of its input parameters.
Third, the new method is transparent and this permits insight to the underlying physical features of siRNA-directed target cleavage. The improved transparency of the new method compared with neural networks, for example, should facilitate further experimentation, which is ultimately expected to lead to the design of better algorithms and software.
Our work confirms recent findings reported after we completed our main analyses (11). We show that correlations between siRNA efficacy and local duplex stability calculated as  values are strongest and most significant for the first and last two duplex base pairs. The recently published study demonstrates the heaviest weighting for both these relevant
values are strongest and most significant for the first and last two duplex base pairs. The recently published study demonstrates the heaviest weighting for both these relevant  variables in the linear regression model (11). We show here that some instability for the two base pairs at the 6th and 13th positions from the 5′ end of the antisense strand correlates significantly with siRNA efficacy while the other study also shows elevated weighting for the relevant variables. In addition, while creating model 2 we found that the nucleotide composition of overhangs of siRNA duplex is related to siRNA-directed cleavage efficiency. This finding challenges the standard siRNA design approach of introducing the dinucleotide ‘AA’ into the overhang sequence. The recently published study (11) reached the same conclusion.
variables in the linear regression model (11). We show here that some instability for the two base pairs at the 6th and 13th positions from the 5′ end of the antisense strand correlates significantly with siRNA efficacy while the other study also shows elevated weighting for the relevant variables. In addition, while creating model 2 we found that the nucleotide composition of overhangs of siRNA duplex is related to siRNA-directed cleavage efficiency. This finding challenges the standard siRNA design approach of introducing the dinucleotide ‘AA’ into the overhang sequence. The recently published study (11) reached the same conclusion.
Analysis of four different databases allows us to reveal universal features for efficient siRNA prediction; however, some findings were not mentioned in previous studies. For example, we find that local instability for the two base pairs at the 7th, 10th and 12th positions correlates significantly with siRNA efficacy. The preference of A at positions 4, 7, 10, 14, 16, U at positions 2, 3, 7, 8, 13, 14, avoidance of G at positions 2, 5, 13, 14, 17 and C at positions 2, 3, 12, 13, 14 from the 5′ end of the antisense strand is not wholly consistent with the previous findings (2,6,9,11). Because the correlations between siRNA efficiency and nucleotide presence or absence are significant for at least two independent experimental databases, we consider that our findings are reliable and will be helpful for future regression model improvements.
The advantage of our model 2 compared to the ‘composite model’ of Vert et al. (11), is its much lower number of input parameters. The ‘composite model’ uses preferences for particular nucleotides at certain siRNA antisense strand positions and total numbers of di- and tri-nucleotide motifs in this strand as input parameters. The total number of significant input parameters for this model is 115. Our model 2 uses thermodynamic values related to local siRNA duplex stability, preferences for particular nucleotides at certain antisense strand positions and total GC content as input parameters. Its total number of input parameters is 22. The significant decrease in the number of input parameters, also called dimension reduction, is fundamental to multivariate statistical model building. As the performance of our method is identical to that of Vert and collegues (11), it is likely that these additional parameters are redundant. These parameters do not contribute to siRNA efficacy prediction, but instead obfuscate the truly important parameters for siRNA efficacy. Thus, our model generally provides a better understanding of the underlying mechanism of the process related to experimental data generation.
Relationship of group 1 and 2 input parameters with the mechanism of siRNA-directed target cleavage
The process of siRNA-directed target cleavage involves several reactions starting with RISC interacting with one of the siRNA strands. Cleavage of the strand complementary to that selected by RISC and release of the fragments generated ensues. RISC loaded with the uncleaved guide strand interacts with the target, which is then cleaved and released. The overall efficiency of the process depends on the sum of the rates of each reaction. The parameters of our models are related to these rates.
The algorithm's group 1 parameters are related to the chemical reaction that takes place before enzymatic cleavage of target mRNA. One of the individual parameters that correlates most strongly with siRNA efficacy is  calculated for the two 5′ terminal nucleotide base pairs. This
calculated for the two 5′ terminal nucleotide base pairs. This  value most likely relates to the rate of siRNA antisense strand entry into RISC. Another parameter,
value most likely relates to the rate of siRNA antisense strand entry into RISC. Another parameter, 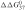 , likely relates to the ratio of rates of the antisense and sense strands for RISC entry (see ‘Binding model for siRNA-RISC entry’ in Supplementary Data, Materials section). Sense strand entry into RISC is undesirable; it can cause competition for RISC with the antisense strand and unspecific cleavage of mRNAs partially complementary to the sense strand.
, likely relates to the ratio of rates of the antisense and sense strands for RISC entry (see ‘Binding model for siRNA-RISC entry’ in Supplementary Data, Materials section). Sense strand entry into RISC is undesirable; it can cause competition for RISC with the antisense strand and unspecific cleavage of mRNAs partially complementary to the sense strand.
The majority of the algorithm's group 2 parameters are also likely related to reaction rates. Target interaction with RISC loaded with the guide strand and subsequent enzymatic target cleavage would be too slow if the duplexes formed between siRNA antisense strands and mRNA are insufficiently stable. However, target mRNA and siRNA antisense strand secondary structures should not be too stable. Otherwise target or siRNA antisense strand unwinding will delay their subsequent interaction. An optimal GC content should provide some trade-off between optimal duplex stability and secondary structures of target or siRNA antisense strand.
The potential significance of the preference for particular nucleotides at certain positions of the siRNA antisense strand is most likely related to the rates of RISC binding or Argonaute 2 cleavage of the complementary strand.
Input parameters for our model are represented by the combination of thermodynamic evaluations of local duplex stability, preference for particular nucleotides at certain strand positions and total GC content. During the process of model fitting some parameters are selected and others are discarded as insignificant. At certain positions, the nucleotide-related parameters are selected whereas at others, local duplex stability related parameters are selected. It is likely that the identity of the nucleotide is more important than local duplex for the efficiency of siRNA cleavage at some strand positions but at others, the converse pertains. For example, the model fitting process prefers local stability values ΔG7 and ΔG13 in the antisense strand over relevant nucleotide identities. So it is likely that for efficient siRNA-directed cleavage low duplex stability at these positions is more important than the presence of particular nucleotides. The rates of melting of siRNA complementary target fragments generated in the cleavage process are most likely related to these local duplex stabilities.
Model transparency can improve siRNA design
Model 1 was created using group 1 parameters on their own and is almost as good as the other complete approaches. It is the most important component of our method. The underlying basis for its efficiency is understood, and there is a known way to design any siRNA duplex with superior antisense strand entry into RISC. This can be achieved by introducing a mismatched nucleotide near the 3′ position of the siRNA sense strand and chemical modification of the 5′ position of the sense strand that blocks RISC entry (32,33). This experimental approach can be used to create the next generation of databases in which factors unrelated to model 1 (group 2 parameters) can be unmasked and easier to study. Dissection of group 1 and 2 parameters will allow further optimization and use of only group 2 parameters for prediction of siRNA efficacy.
Combination of our group 1 and 2 parameters with a recently published scheme for optimal detection of oligonucleotide hybridization targets common to families of aligned sequences (34), permits prediction of optimal targets for families of aligned viral RNA or DNA sequences.
Finally, model transparency combined with comparative analyses can, as shown here, reveal redundant parameters in existing design algorithms. Redundant parameters make models more complex without contributing to model predictions and may obscure the biological mechanisms for siRNA efficacy.
Implications for this work on the design of shRNA
The field of RNA interference (RNAi) has continued to advance rapidly; vector-based short hairpin RNA (shRNA) technologies have evolved into an alternative method of inducing RNAi. The transparency of our method will facilitate the transfer of siRNA design rules to shRNA. At least three considerations have to be taken into account during this transfer. (1) Since shRNA-derived siRNAs are likely present at a lower cellular concentration than exogenously introduced siRNAs, the optimal duplex stability and relevant GC content for shRNA derivatives, can be different. (2) Dicer processing generates a few species of shRNAs from one predecessor molecule of dsRNA or pre-microRNA (35), so the rules of siRNA design may be applied to all possible species. (3) Dangling end thermodynamic parameters for shRNAs are most likely different from those for siRNAs since in the former they are represented by RNA and in the latter, by DNA nucleotides. So the accumulation and study of experimental shRNA databases currently being generated by the scientific community should allow careful consideration of these issues and relevant parameter optimization.
In summary, a number of previously developed approaches for efficient siRNA design were compared, and a new, transparent and efficient method with low number of input parameters was created together with an accompanying web tool, ‘siRNA scale’, for its use.
METHODS
Parameters for the models
To create models, we used two groups of parameters. Group 1 parameters are related to terminal siRNA duplex stability. Group 1 parameters include (1)  values which provide an estimate of the terminal stability of the two nucleotides at siRNA duplex ends, (2) identity of both nucleotides in 3′ overhangs of an siRNA antisense strand (20th and 21st positions) and (3) identity of the 1st and 19th nucleotides of the antisense strand. Group 2 parameters include (1) G + C percentage of the antisense strand, (2) identity of nucleotides from the 2nd to 18th positions in the siRNA antisense strand and (3) local duplex stability values from the 2nd to 17th positions in the siRNA antisense strand.
values which provide an estimate of the terminal stability of the two nucleotides at siRNA duplex ends, (2) identity of both nucleotides in 3′ overhangs of an siRNA antisense strand (20th and 21st positions) and (3) identity of the 1st and 19th nucleotides of the antisense strand. Group 2 parameters include (1) G + C percentage of the antisense strand, (2) identity of nucleotides from the 2nd to 18th positions in the siRNA antisense strand and (3) local duplex stability values from the 2nd to 17th positions in the siRNA antisense strand.
Parameter calculations
The  calculations related to duplex terminal and the local stability parameters used were thermodynamic parameters published earlier (26–30,36). The
calculations related to duplex terminal and the local stability parameters used were thermodynamic parameters published earlier (26–30,36). The 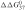 value was calculated for the two terminal nucleotide base pairs of each siRNA duplex by subtracting the
value was calculated for the two terminal nucleotide base pairs of each siRNA duplex by subtracting the  value for the 5′ sense strand from that of the 5′ antisense strand.
value for the 5′ sense strand from that of the 5′ antisense strand.
Statistical analyses
For ROC and regression analyses, we used a set of commercially developed Excel macros that are available from http://www.analyse-it.com. For correlation analyses, the normalized levels of mRNA or protein products remaining in cells after siRNA treatments were expressed as percentage values of the control levels. For categorization analysis, siRNAs that yielded at least 70% target gene knockdown were considered to be efficient. We compared the approaches by analyzing their receiver operating characteristics (ROC) curves. An ROC curve describes the relationship between the specificity Sp = TN/(FP + TN) and the sensitivity Se = TP/(TP + FN) of an algorithm. Here, TP, FP, TN and FN are the number of true positives, false positives, true negatives and false negatives. A prediction is true positive if the siRNA is both predicted and experimentally known to be efficient; true negative if it is both predicted and experimentally known to be inefficient; false positive if it is predicted to be efficient but experimentally found to be inefficient; and false negative if predicted to be inefficient but found to be efficient. In the area tests, we calculate the area under the ROC curve, also known as the ROCscore. An area of 1 indicates a perfect classification, and an area of 0.5 indicates a random classification. The ROCIT software (37) was applied for the calculation of significance levels for ROC area comparisons.
SUPPLEMENTARY DATA
Supplementary Data is available at NAR Online.
ACKNOWLEDGEMENTS
We are grateful to D. Hicks and J. Fohrman (DNA_Software, Inc., MI 48104) for creation of Excel Macro ‘RNA_RNA Positional Energy reporter’; to Andy W. Hammer, University of Utah, UT84112 for ‘siRNA_scale’ creation and to Dr B. Jagla (Columbia University, NY 10032) for providing raw experimental data. Our particular thanks to Drs A. Chenchik (System Biosciences, CA 94041) and G. Gursky (Engelhardt Institute of Molecular Biology, 119991 Moscow) for reading and useful comments on the manuscript. This study was supported by NIH SBIR grant R43 HG003355-01 to Dr A. Chenchik, University of Utah ‘Bridge’ and ‘Technology Commercialization’ grants to J.F.A. J.F.A. was personally supported in part by Science Foundation Ireland. Y.N. was personally supported in part by the program from the Presidium of RAS ‘Molecular and cell biology’ and the Russian State Grant ‘Construction of new drugs for therapy and prophylaxis of antiviral diseases by methods of organic chemistry’. This research was also supported by the Intramural Research Program of the NIH, National Center for Biotechnology Information. We would like to express gratitude to Dr R.F. Gesteland for suggesting the idea of this study. Funding to pay the Open Access publication charge was provided by Science Foundation Ireland.
Conflict of interest statement. None declared.
APPENDIX
Note 1
The term ‘parameter’ in our work can be replaced by term ‘argument’, since it is more correct from a formal mathematics perspective. We avoided this replacement, since in biological articles on similar topics the term ‘parameter’ is used in the sense of a mathematical argument.
Note 2
The non-scoring algorithm described by Ui Tei-Saigo (“version 1”) was modified to become a scoring algorithm, which would be more comparable with the other scoring algorithms studied here. This modified algorithm, here called version 2, performs better than the original algorithm. Ui Tei-Saigo's non-scoring algorithm predicted effective if the siRNA satisfied the algorithm's four efficacy parameters. (1) A/U at the 5′ end of the antisense strand; (2) G/C at the 5′ end of the sense strand; (3) at least five A/U residues in the 5′ terminal one-third of the antisense strand; and (4) the absence of any GC stretch of more than 9 nt in length.
The following scores were assigned to this algorithm's parameters: (1) A/U at the 5′ end of the antisense strand was assigned the value of +1 point; (2) G/C at the 5′ end of the sense strand was assigned the value of +1 point, (3) at least five A/U residues in the 5′ terminal one-third of the antisense strand was assigned the value of +1 point; and (4) The presence of any GC stretch of more than 9 nt in length in the siRNA duplex was assigned the value of −3 points.
In addition, we also assigned minus to all positive correlation coefficients derived from the algorithms. The absolute values of the correlation coefficients remained the same. This procedure was done for easier graphical comparison of correlation coefficients as the great majority is negative. This modification did not influence the algorithms’ discriminative capacity.
REFERENCES
- 1.Sætrom P, Snove O., Jr. A comparison of siRNA efficacy predictors. Biochem. Biophys. Res. Commun. 2004;321:247–253. doi: 10.1016/j.bbrc.2004.06.116. [DOI] [PubMed] [Google Scholar]
- 2.Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. Rational siRNA design for RNA interference. Nat. Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 3.Sætrom P. Predicting the efficacy of short oligonucleotides in antisense and RNAi experiments with boosted genetic programming. Bioinformatics. 2004;20:3055–3063. doi: 10.1093/bioinformatics/bth364. [DOI] [PubMed] [Google Scholar]
- 4.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 5.Amarzguioui M, Prydz H. An algorithm for selection of functional siRNA sequences. Biochem. Biophys. Res. Commun. 2004;316:1050–1058. doi: 10.1016/j.bbrc.2004.02.157. [DOI] [PubMed] [Google Scholar]
- 6.Ui-Tei K, Naito Y, Takahashi F, Haraguchi T, Ohki-Hamazaki H, Juni A, Ueda R, Saigo K. Guidelines for the selection of highly effective siRNA sequences for mammalian and chick RNA interference. Nucleic Acids Res. 2004;32:936–948. doi: 10.1093/nar/gkh247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 8.Huesken D, Lange J, Mickanin C, Weiler J, Asselbergs F, Warner J, Meloon B, Engel S, Rosenberg A, Cohen D, et al. Design of a genome-wide siRNA library using an artificial neural network. Nat. Biotechnol. 2005;23:995–1001. doi: 10.1038/nbt1118. [DOI] [PubMed] [Google Scholar]
- 9.Jagla B, Aulner N, Nelly PD, Song D, Volchuk A, Zatorski A, Shum D, Mayer T, De Angelis DA, Ouerfelli O, et al. Sequence characteristics of functional siRNAs. RNA. 2005;11:864–872. doi: 10.1261/rna.7275905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shabalina SA, Spiridonov AN, Ogurtsov AY. Computational models with thermodynamic and composition features improve siRNA design. BMC Bioinformatics. 2005;7:65. doi: 10.1186/1471-2105-7-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vert JP, Foveau N, Lajaunie C, Vandenbrouck Y. An accurate and interpretable model for siRNA efficacy prediction. BMC Bioinformatics. 2006;7:520. doi: 10.1186/1471-2105-7-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henschel A, Buchholz F, Habermann B. DEQOR: a web-based tool for the design and quality control of siRNAs. Nucleic Acids Res. 2004;32:W113–120. doi: 10.1093/nar/gkh408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chalk AM, Wahlestedt C, Sonnhammer EL. Improved and automated prediction of effective siRNA. Biochem. Biophys. Res. Commun. 2004;319:264–274. doi: 10.1016/j.bbrc.2004.04.181. [DOI] [PubMed] [Google Scholar]
- 14.Gong D, Ferrell JE., Jr. Picking a winner: new mechanistic insights into the design of effective siRNAs. Trends Biotechnol. 2004;22:451–454. doi: 10.1016/j.tibtech.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 15.Yiu SM, Wong PW, Lam TW, Mui YC, Kung HF, Lin M, Cheung YT. Filtering of ineffective siRNAs and improved siRNA design tool. Bioinformatics. 2005;21:144–151. doi: 10.1093/bioinformatics/bth498. [DOI] [PubMed] [Google Scholar]
- 16.Ge G, Wong GW, Luo B. Prediction of siRNA knockdown efficiency using artificial neural network models. Biochem. Biophys. Res. Commun. 2005;336:723–728. doi: 10.1016/j.bbrc.2005.08.147. [DOI] [PubMed] [Google Scholar]
- 17.Levenkova N, Gu Q, Rux JJ. Gene specific siRNA selector. Bioinformatics. 2004;20:430–432. doi: 10.1093/bioinformatics/btg437. [DOI] [PubMed] [Google Scholar]
- 18.Wang L, Mu FY. A Web-based design center for vector-based siRNA and siRNA cassette. Bioinformatics. 2004;20:1818–1820. doi: 10.1093/bioinformatics/bth164. [DOI] [PubMed] [Google Scholar]
- 19.Yuan B, Latek R, Hossbach M, Tuschl T, Lewitter F. siRNA Selection Server: an automated siRNA oligonucleotide prediction server. Nucleic Acids Res. 2004;32:W130–134. doi: 10.1093/nar/gkh366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Naito Y, Yamada T, Ui-Tei K, Morishita S, Saigo K. siDirect: highly effective, target-specific siRNA design software for mammalian RNA interference. Nucleic Acids Res. 2004;32:W124–129. doi: 10.1093/nar/gkh442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Santoyo J, Vaquerizas JM, Dopazo J. Highly specific and accurate selection of siRNAs for high-throughput functional assays. Bioinformatics. 2005;21:1376–1382. doi: 10.1093/bioinformatics/bti196. [DOI] [PubMed] [Google Scholar]
- 22.Arziman Z, Horn T, Boutros M. E-RNAi: a web application to design optimized RNAi constructs. Nucleic Acids Res. 2005;33:W582–588. doi: 10.1093/nar/gki468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aravin A, Tuschl T. Identification and characterization of small RNAs involved in RNA silencing. FEBS Lett. 2005;579:5830–5840. doi: 10.1016/j.febslet.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 24.Silva JM, Sachidanandam R, Hannon GJ. Free energy lights the path toward more effective RNAi. Nat. Genet. 2003;35:303–305. doi: 10.1038/ng1203-303. [DOI] [PubMed] [Google Scholar]
- 25.Vickers TA, Koo S, Bennett CF, Crooke ST, Dean NM, Baker BF. Efficient reduction of target RNAs by small interfering RNA and RNase H-dependent antisense agents. A comparative analysis. J. Biol. Chem. 2003;278:7108–7118. doi: 10.1074/jbc.M210326200. [DOI] [PubMed] [Google Scholar]
- 26.Xia T, SantaLucia J, Jr, Burkard ME, Kierzek R, Schroeder SJ, Jiao X, Cox C, Turner DH. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry. 1998;37:14719–14735. doi: 10.1021/bi9809425. [DOI] [PubMed] [Google Scholar]
- 27.Bommarito S, Peyret N, SantaLucia J., Jr. Thermodynamic parameters for DNA sequences with dangling ends. Nucleic Acids Res. 2000;28:1929–1934. doi: 10.1093/nar/28.9.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Freier SM, Burger BJ, Alkema D, Neilson T, Turner DH. Effects of 3’ dangling end stacking on the stability of GGCC and CCGGdouble helices. Biochemistry. 1983;22:6198–6206. [Google Scholar]
- 29.Freier SM, Alkema D, Sinclair A, Neilson T, Turner DH. Contributions of dangling end stacking and terminal base-pair formation to the stabilities of XGGCCp, XCCGGp, XGGCCYp, and XCCGGYp helixes. Biochemistry. 1985;24:4533–4539. doi: 10.1021/bi00338a008. [DOI] [PubMed] [Google Scholar]
- 30.Sugimoto N, Kierzek R, Turner DH. Sequence dependence for the energetics of dangling ends and terminal mismatches in ribonucleic acid. Biochemistry. 1987;26:4554–4558. doi: 10.1021/bi00388a058. [DOI] [PubMed] [Google Scholar]
- 31.Ogurtsov AY, Shabalina SA, Kondrashov AS, Roytberg MA. Analysis of internal loops within the RNA secondary structure in almost quadratic time. Bioinformatics. 2006;22:1317–1324. doi: 10.1093/bioinformatics/btl083. [DOI] [PubMed] [Google Scholar]
- 32.Kraynack BA, Baker BF. Small interfering RNAs containing full 2′-O-methylribonucleotide-modified sense strands display Argonaute2/eIF2C2-dependent activity. RNA. 2006;12:163–176. doi: 10.1261/rna.2150806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koller E, Propp S, Murray H, Lima W, Bhat B, Prakash TP, Allerson CR, Swayze EE, Marcusson EG, Dean NM. Competition for RISC binding predicts in vitro potency of siRNA. Nucleic Acids Res. 2006;34:4467–4476. doi: 10.1093/nar/gkl589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matveeva OV, Foley BT, Nemtsov VA, Gesteland RF, Matsufuji S, Atkins JF, Ogurtsov AY, Shabalina SA. Identification of regions in multiple sequence alignments thermodynamically suitable for targeting by consensus oligonucleotides: application to HIV genome. BMC Bioinformatics. 2004;5:44. doi: 10.1186/1471-2105-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vermeulen A, Behlen L, Reynolds A, Wolfson A, Marshall WS, Karpilow J, Khvorova A. The contributions of dsRNA structure to Dicer specificity and efficiency. RNA. 2005;11:674–682. doi: 10.1261/rna.7272305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mathews DH, Burkard ME, Freier SM, Wyatt JR, Turner DH. Predicting oligonucleotide affinity to nucleic acid targets. RNA. 1999;5:1458–1469. doi: 10.1017/s1355838299991148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Metz CE, Herman BA, Roe CA. Statistical comparison of two ROC-curve estimates obtained from partially-paired datasets. Med. Decis. Making. 1998;18:110–121. doi: 10.1177/0272989X9801800118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.