Abstract
G-quadruplexes are a family of four-stranded DNA structures, stabilized by G-quartets, that form in the presence of monovalent cations. Efforts are currently being made to identify ligands that selectively bind to G-quadruplex motifs as these compounds may interfere with the telomere structure, telomere elongation/replication and proliferation of cancer cells. The kinetics of quadruplex–ligands interactions are poorly understood: it is not clear whether quadruplex ligands lock into the preformed structure (i.e. increase the lifetime of the structure by lowering the dissociation constant, koff) or whether ligands actively promote the formation of the complex and act as quadruplex chaperones by increasing the association constant, kon. We studied the effect of a selective quadruplex ligand, a bisquinolinium pyridine dicarboxamide compound called 360A, to distinguish these two possibilities. We demonstrated that, in addition to binding to and locking into preformed quadruplexes, this molecule acted as a chaperone for tetramolecular complexes by acting on kon. This observation has implications for in vitro and in vivo applications of quadruplexes and should be taken into account when evaluating the cellular responses to these agents.
INTRODUCTION
G-quadruplexes are a family of secondary nucleic acids structures stabilized by G-quartets that form in the presence of monovalent cations (1,2). The level of interest in these structures has recently increased due to hypotheses that G-quadruplex structures play roles in key biological processes (3–9) and recent demonstrations of their existence in vivo (10–12). G-quadruplexes may have applications in areas ranging from supramolecular chemistry and bio- and nanotechnology (13–15) to medicinal chemistry [for recent reviews: (2,16,17)]. Efforts have been made to identify ligands that selectively bind to G-quadruplex motifs, as they may interfere with telomere structure, elongation and replication, oncogene expression and proliferation of cancer cells (18,19). These compounds may be of natural origin [such as cryptolepine (20), berberine (21) and telomestatin (22,23)] or synthetic [such as BSU1051 (18), RHPS4 (24), TMPyP4 (25), pyridine or phenanthroline dicarboxamides (26,27), triazines (28), PIPER (29,30) or bi- and trisubstituted acridines (31,32)]. It is important to understand the rules that govern the formation of G-quadruplexes and determine the stability and folding kinetics of the structures. Unfortunately, with a few notable exceptions (29), little is known concerning these parameters. Han et al. presented the first example of a small ligand (a perylene derivative, PIPER) that drives the assembly of bimolecular G-quadruplex structures (29). The presence of 10 µM PIPER accelerates the assembly of varied dimeric G-quadruplexes an estimated 100-fold (29). However, the analysis of the results was complicated by the existence of a number of different G-quadruplex structures, including dimeric and tetrameric species. Furthermore, the end product structure and molecularity were different in the presence and in the absence of the ligand, indicating that the molecule favors some conformers over the others and displaces the thermodynamic equilibria.
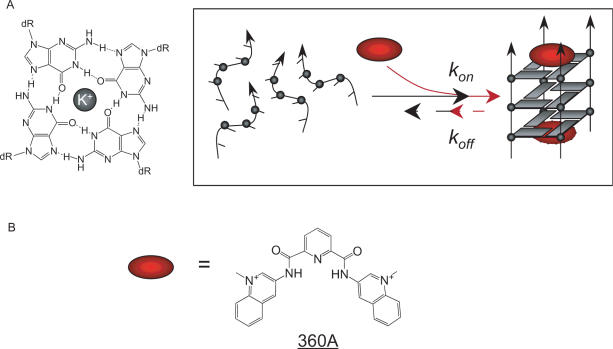
Parallel-stranded, tetramolecular quadruplexes (Figure 1A) offer a unique opportunity to study kinetic effects for several reasons. First, our understanding of their kinetic properties increased, thanks to the seminal work by Wyatt et al. that described the properties of simple short segments such as T2G4T2 (33). More recently, we (34,35) and others (36) analyzed the kinetics of quadruplex formation with short sequences. From a kinetic point of view, the association reaction strongly depends on strand concentration with an experimentally determined order approaching four (33,34,36); an exception being the four-stranded structures formed by the sugar-modified oligonucleotides known as locked nucleic acids (37). Second, the kinetic inertia of these complexes, rather than being a hindrance, facilitates the study of the uncoupling of the folding and unfolding phenomenon. One may deconvolute the effect of a parameter and determine whether this factor mainly affects association or dissociation—or both. We and others successfully used this property to analyze the effects of temperature, sequence, ionic strength, introduction of a modified backbone or modified quartet on the kinetics of G-quadruplex formation. Third, in contrast to structures adopted by bi- and intramolecular quadruplexes, which are highly polymorphic, tetramolecular complexes are structurally well defined. They are formed by short oligodeoxynucleotides bearing a block of three of more guanines. In this configuration (also called G4-DNA), all strands are parallel and all guanines are in the anti conformation (Figure 1A). Although direct evidence for the formation of tetramolecular structures in vivo is currently lacking, these quadruplexes have been proposed to play a role during meiosis (38,39).
Figure 1.
Quadruplexes and quadruplex ligands. (A) Schematic of a quadruplex (left) and tetramolecular G-quadruplex formation/dissociation (right). This scheme simply presents the initial and final states of the reaction studied here, and assumes terminal stacking of the ligand; it does not claim to reflect the pathway of association. Are quadruplex ligands (red ovals) simply able to lock the preformed structure (i.e. increase the lifetime of the structure by lowering its dissociation constant koff), or do ligands actively promote the formation of the complex and act as quadruplex chaperones by increasing its association constant kon? (B) Formula of the quadruplex ligand 360A.
In this report, we unambiguously demonstrate that a well-characterized quadruplex ligand is able to accelerate the association of single strands into quadruplexes. Therefore, this molecule acts as a molecular chaperone for the formation of tetramolecular quadruplexes. This observation has implications for in vitro and in vivo applications of quadruplexes and should be taken into account when interpreting the cellular responses to these agents.
EXPERIMENTAL PROCEDURES
Oligonucleotides and compounds
Oligonucleotides were synthesized by Eurogentec (Seraing, Belgium). Concentrations of all oligodeoxynucleotides were estimated using published sequence-dependent extinction coefficients (40). Compounds 360A, 307A, BRACO19, BSU1051 and 12 459 were kind gifts from P. Mailliet (Sanofi-Aventis, Vitry/Seine, France). These compounds were stored at 1 or 2 mM in DMSO, further dilution being made in ddH20. TMPyP4 and PIPER were purchased from Calbiochem and solubilized in ddH20 and dilute acetic acid, respectively. Telomestatin was a kind gift of Dr K. Shin-ya; its isolation and purification was previously described (22). Telomestatin was stored at 1 mM at −20°C in the dark in 50% DMSO/50% MeOH; fresh dilutions were made for each series of experiments. The formulae of all tested ligands are shown in Supplementary Figure S1; some of their properties are shown in Supplementary Table S1.
Gel electrophoresis
Non-denaturing gel electrophoresis allows separation of single-stranded oligonucleotides from tetramolecular G-quadruplex structures (34,35). The oligonucleotides (5–200 µM final strand concentrations) were heat denatured in a buffer containing 10 mM lithium cacodylate (pH 7.2) and 110 mM KCl (or NaCl for the oligonucleotide T2G5T2). Immediately after denaturation, oligonucleotides were mixed with the compounds (5 × concentrate solutions) to reach the indicated final concentrations, in a volume of 10–20µl, and incubated at 4°C for times varying from 2 min to several days.
For the association of these short oligonucleotides, at concentrations ranging from 50 to 500 µM, UV shadow at 254 nm was a convenient method to quantify with fair precision the amount of single-strand versus G-quadruplex. The use of dyes, like methylene blue, SYBR green I or gold (Molecular Probes) and acridine orange, are often misleading as stainings and quantum yields may vary when bound to single-strands versus G-quadruplexes (not shown). About 1 nmol of oligonucleotide was loaded with 10% final sucrose concentration on a non-denaturing gel containing 20% acrylamide-bisacrylamide (19:1), 20 mM KCl and 1 × TBE. About the same amount (1 nmol) of oligothymidylate markers (dT6, dT12 or dT24) were also loaded on the gel. The TG3T sample incubated without any potassium and heat denatured just before loading was used as a control for single-strand migration. One should note that the migration of the oligothymidylate markers (short 5′dTn oligonucleotides) does not necessarily correspond to single-strands (41): these oligonucleotides were chosen here to provide an internal migration standard, not to identify single-stranded or higher-order structures. The gel was electrophoresed at 4°C in 20 mM KCl, 1 × TBE. Bands were detected by UV-shadow at 254 nm using a fluorescent silica screen (Whatman).
When using the UV-shadow approach to quantitate, one must assume that the extinction coefficients of the oligonucleotide in single-stranded and quadruplex forms are identical. Absorbance spectroscopy confirmed that these two coefficients were indeed very close. Unfortunately, compound 360A also absorbs light in the same wavelength range, and its molar extinction coefficient at 260 nm (40 000 M−1 cm−1) cannot be neglected as compared to TG3T (47 700 M−1 cm−1). 360A binding may lead to an overestimation of the fraction of oligonucleotide in the G-quadruplex structure. Nevertheless, when the same amounts were loaded on a gel, we did not observe huge variations of band intensity from induced G-quadruplex with or without any compound. Moreover, we confirmed and quantified the induction of G-quadruplex by 360A using radioactivity.
For induction of G-quadruplexes at low oligonucleotide concentrations, 20 pmol of TG3T was radiolabeled in the 5′ position using 10 µCi of γ-32P-ATP and 5 units of T4 polynucleotide kinase (New England Biolabs) in a buffer containing 70 mM Tris-HCl (pH 7.6), 10 mM MgCl2 and 5 mM DTT for 40 min at 37°C. The short radiolabeled oligonucleotide was gel-purified using standard protocols and precipitated by 2.5 M ammonium acetate and a large excess of ethanol. Finally, ammonium acetate was removed by lyophilization to avoid a possible effect of NH4+ on the oligonucleotide association kinetics. The purified, labeled 32P-TG3T was then mixed with unlabeled phosphorylated oligonucleotide to reach a final strand concentration of 5–200 µM. The amount of radiolabeled oligonucleotide was negligible compared to the quantities of unlabeled oligonucleotide added. Complex was separated from single-strand using non-denaturing gel electrophoresis (acrylamide:bis, 19:1; 20% in 1 × TBE, 20 mM KCl at 4°C) and the radioactive signal was detected after overnight exposure to a phosphorimager screen (Molecular Dynamics) at −20°C and scanning with a Typhoon apparatus (Amersham Bioscience). Under certain conditions, two retarded bands are observed in the presence of a ligand. These two species correspond to the migration of the tetramolecular quadruplex without ligand, and to the quadruplex + ligand, respectively (the latter being more retarded than the former). This behavior may be found when using sub-saturating amounts of ligand.
Absorbance measurements
Comparison of the absorbance spectra of the oligonucleotide under conditions where it should be totally unfolded (at t = 0) and completely folded allowed us to generate an isothermal differential absorbance spectrum. The shape of this curve is specific for quadruplexes (Supplementary Figure S3) (42). Isothermal and melting experiments were recorded as previously described (34). For isothermal experiments, starting from completely unfolded strands, absorbance was recorded at regular time intervals (150 s) at 245 (or 240), 295 and 488 nm. Oligonucleotide strand concentration was 100 µM for TG3T and 300 µM for PO4-TG3T in a buffer containing 10 mM lithium cacodylate (pH 7.2) and 110 mM KCl. After overnight association, a UV-melting experiment was performed on the same samples with a heating gradient of 0.5°C/min, recording the absorbance at 240, 295 and 490 nm. This melting temperature depends on the heating rate (34,43).
Analysis of quadruplex formation
Kinetic experiments followed by UV-absorbance or gel electrophoresis allowed determination of the fraction of quadruplex formed as a function of time. In the absence of ligand, experimental points were fitted to a kinetic model using Kaleidagraph 3.6.4 according to published procedures (34,35) and assuming an experimental order of four. However, in the presence of non-saturating amounts of G4 ligand, kinetic curves could not be fitted with this model, as a concomitant decrease in free-ligand concentration leads to a more complex profile. Therefore, a comparison of the kon values (in M−3 s−1) in the absence or presence of the ligands was not possible except when the ligand was present in large excess. In that case, the free ligand concentration was considered to be invariant during the time course of the kinetics experiment. However, one may always compare the experimental time required for 50% quadruplex formation (t1/2) under a given set of conditions.
RESULTS
Presentation of the system
All oligonucleotides studied here contain a single short block of three to five guanines and may form a tetramolecular complex. We chose TG3T as a model sequence. Several other sequences, such as T2G5T2 and the human telomeric oligonucleotides TTAG3 and TTAG3T, were also tested. Formation of these quadruplexes is slow, even in potassium (34); therefore, ligands that increase the kinetics of association are relatively easy to identify.
Comparison of various DNA ligands
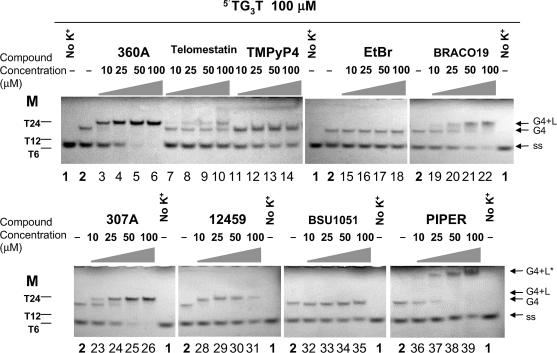
Formation of a tetrameric quadruplex may be evidenced by non-denaturing gel electrophoresis. In the absence of ligand and potassium, no quadruplex is formed, and a single band is obtained on the gel, corresponding to the migration of a single-stranded species (Figure 2, lane 1). When 110 mM K+ is present, a partial conversion of the single-strand to the quadruplex was observed (Figure 2, lane 2). Increasing the concentration of ethidium bromide, a ligand that has a high affinity for duplexes, but a weak affinity for quadruplexes (44–46) had no effect on the quadruplex:single-strand ratio, even at 100 µM (Figure 2, lanes 15–18). On the other hand, increasing concentrations of a variety of known G-quadruplex ligands had rather diverse effects: BSU1051 (Figure 2, lanes 32–35) and 12 459 (Figure 2, lanes 28–31) had limited (if any) effect on the relative amounts of single-strands over G-quadruplex species; telomestatin (Figure 2, lanes 7–10) and TMPyP4 (Figure 2, lanes 11–14), altered slightly the relative intensities of the bands. BRACO19 (Figure 2, lanes 19–22) induced the formation of a tetramolecular quadruplex, whereas PIPER (lanes 36–39) led to the formation of a higher molecular weight species with very low mobility; however, significant amounts of single-strands were present even at the highest ligand concentration. In contrast, in the presence of the pyridine dicarboxamide derivatives, 360A or 307A, known for their high affinity and selectivity for quadruplexes (26,47,48), only quadruplex was observed (Figure 2, lanes 3–6 and 23–26). The 360A (or 307A)/quadruplex complex was slightly retarded compared to the quadruplex formed in the absence of any ligand (Figure 2, lane 2), and a similar phenomenon was observed for telomestatin (lanes 8–10), BRACO19 (lanes 20–22) and 12459 (lanes 28–31). This slightly altered migration is probably due to the molecular weight/charge contribution of the ligand to the complex, which suggests that the quadruplex–ligand complex is maintained during electrophoresis (this may not be the case for BSU1051, ethidium bromide or TMPyP4). For PIPER, the migration is drastically modified and, although we cannot exclude that this simply results from PIPER binding to the tetramolecular quadruplex, it is more likely that higher molecular weight species are induced in the presence of this compound as already reported (29).
Figure 2.
Effects of various compounds on the association of a tetramolecular G-quadruplex from the oligonucleotide TG3T. These molecules are known to bind to G-quadruplexes (360A, telomestatin, TMPyP4, BRACO19, BSU1051, 307A, PIPER and 12 459, formulae shown as Supplementary Figure S1) or duplexes (ethidium bromide (EtBr)). The oligonucleotide TG3T was heat denatured before incubation at 100 µM (strand concentration) in 10 mM lithium cacodylate (pH 7.2), 110 mM KCl buffer (at 4°C for 4.5 h) with or without any compound at the indicated concentrations. Samples (1 nmol of oligonucleotide loaded) were loaded on a 20% non-denaturing acrylamide gel containing 20 mM KCl and 1× TBE, and detection was performed using UV-shadow at 254 nm. Oligonucleotide markers (M) were short 5′dTn sequences. A sample incubated without any potassium and heat denatured just before loading was used as a control for single-strand migration (ss) on the gel (lanes 1), whereas incubation without any compound indicated position of the migration of the tetramolecular G-quadruplex (G4) (lanes 2). Some gel-shifts can be observed depending on the ligand used (G4 + L). G4 + L* stands for the very slow mobility product formed with PIPER.
Kinetics of quadruplex formation in the presence of 360A
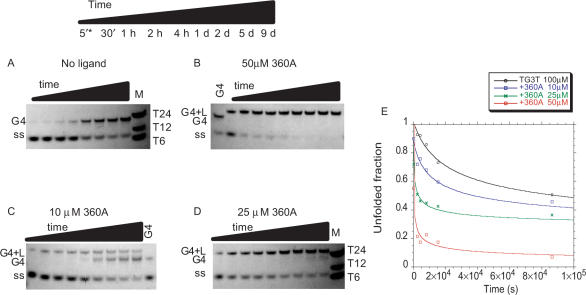
In the experiment presented in Figure 3A, solutions of TG3T were incubated for various amounts of time, between 5 min and 9 days, before separation of single-strand from quadruplex on a non-denaturing gel. In the absence of the ligand, a significant but still incomplete conversion into quadruplex was observed after 9 days; half conversion required approximately 1 day at 100 µM strand concentration. In contrast, addition of various concentrations of 360A had a dramatic effect on the association kinetics. In the presence of 50 µM 360A, half conversion required only 5 min, and the single-stranded species completely disappeared after a few days (Figure 3B). The effects of intermediate concentrations, 10 and 25 µM 360A, are shown in Figure 3, panels C and D, respectively. The relative amount of the single-stranded species was determined from a quantitative analysis of the band intensities; the unfolded fraction is plotted versus time in Figure 3E. Starting from a completely unfolded oligonucleotide in each case, the differences between curves at 0, 10, 25, and 100 µM are striking, especially for short incubation times.
Figure 3.
Association kinetics of TG3T with various concentrations of 360A ligand. (A-D) Non-denaturing gel electrophoresis was used to separate single strand from quadruplex, and species were detected by UV-shadow at 254 nm. Incubations were carried out in 10 mM lithium cacodylate (pH 7.2), 110 mM KCl at 4°C for the indicated time at a 100 µM strand concentration with or without 360A at the indicated concentration. Time is given in days (d), hours (h), and minutes (′). * no time point at 5' for panel A. (E) Quantification of the single-stranded fraction using ImageQuant software. Black triangles: without compound; blue squares: with 10 µM 360A; green crosses: with 25 µM 360A; red squares: with 50 µM 360A.
Influence of a 5′ phosphate on quadruplex association and dissociation
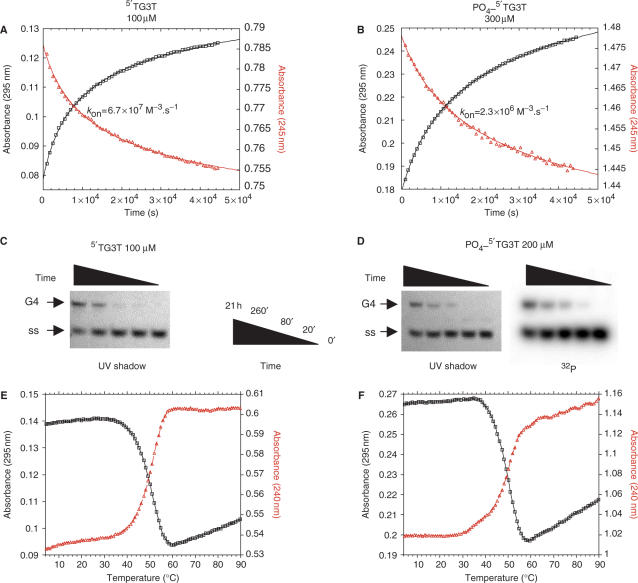
The single strands and complexes in the gels presented in Figures 2 and 3 were revealed by a relatively insensitive technique, UV-shadowing. In order to study quadruplex formation at lower strand and ligand concentrations, a more sensitive approach was required. We visualized the oligonucleotides on some of the gels by methylene blue staining or by the use of fluorescent dyes such as SYBR or acridine orange. Unfortunately, the former technique is not sensitive enough, whereas the fluorescent dyes have different quantum yields and/or affinity for quadruplex and single-strands, complicating a quantitative analysis (data not shown). For these reasons, we followed quadruplex formation using a 5′-radiolabeled oligonucleotide. Our initial experiments were done by mixing a 5′ phosphate radiolabeled oligonucleotide, mixed with a large excess of unlabeled, non-phosphorylated oligonucleotide. We noticed that the amount of quadruplex formed as determined by the radioactive-labeling method leads to an underestimation of the total quadruplex, indicating that the radiolabeled strand was underrepresented in the complex as a result of the extra 5′ phosphate group (data not shown). In agreement with the results of Uddin et al. (49), it appeared that the presence of one or two extra negative charges per strand affected the stability of the complex and/or association rate of the quadruplex formation. Figure 4 and Supplementary Figure S4 present a comparison of the kinetics of association and dissociation of the G-quadruplexes formed by 5′-OH and 5′-phosphate TG3T oligonucleotide at various salt concentrations. As shown in Figure 4A and B, quadruplex formation was hampered by the presence of the phosphate group. Higher strand concentrations were required to form a quadruplex overnight and, at 110 mM ionic strength, the corresponding association rate constant was 29 times lower for the oligonucleotide bearing a terminal phosphate [kon = 67 × 106 and 2.3 × 106 M−3 s−1 for TG3T and PO4-TG3T, respectively; for a discussion and comparison of these absolute values, see (34,35)]. However, the difference of kon values between the 5′-phosphate and the 5′ OH oligonucleotides was slightly decreasing when working at higher salt concentrations (Supplementary Figure S4B), confirming that an electrostatics phenomenon is taking part in this unfavorable effect. This effect was also confirmed by non-denaturing gel electrophoresis, as higher strand concentrations were required to form similar amounts of quadruplexes for the phosphorylated oligonucleotide compared to the non-phosphorylated (Figure 4C and 4D), as revealed by UV-shadow analysis. For oligonucleotides bearing a 5′ phosphate, this analysis was also performed by radioactivity, using a 32P-radiolabeled oligonucleotide mixed with the 5′phosphate oligonucleotide. A quantitative analysis of the gel experiments (revealed by radioactivity or UV-shadow) is presented in Supplementary Figure S4A. Similar results were obtained when amounts were quantitated using UV-shadow or radioactivity, arguing that the fraction of radiolabeled oligonucleotides involved accurately reflects the total amount of quadruplex formed. The association constants for PO4-TG3T (kon = 2 × 106 and 1.4 × 106 M−3 s−1 calculated using values from UV-shadow and radioactivity, respectively) are in excellent agreement with the absorbance spectroscopy results (kon = 2.3 × 106 M−3 s−1) and confirm that the kinetics of association are negatively affected by the presence of a terminal phosphate. The association rate constant of the 5′-OH oligonucleotide determined using radioactively labeled oligonucleotide was calculated to be 4 × 107 M−3 s−1, which is also in good agreement with the value found by absorbance spectroscopy (considering 30% precision is given on these values). Association constant values are summarized in Table 1.
Figure 4.
Effect of a terminal phosphate on G-quadruplex association and dissociation. Results obtained with TG3T and PO4-TG3T are shown in the left and right parts of the figure, respectively. All experiments were performed in 10 mM lithium cacodylate (pH 7.2), KCl 110 mM at 4°C. (A and B) Association kinetics of G-quadruplex formation followed by absorbance at 295 nm (black squares) and 245 nm (red triangles) for (A) TG3T and (B) PO4-TG3T at 100 µM and 300 µM strand concentration, respectively; kon values were calculated using the fourth-order model (34,35). (C and D): Association kinetics of G-quadruplex formation followed by non-denaturing gel electrophoresis (20% acrylamide in 1× TBE, 20 mM KCl at 4°C) at the time points indicated for (C) TG3T at 100 µM strand concentration and (D) PO4-5′dTG3T at 200 µM total strand concentration (mixture of 32P-TG3T labeled at the 5′ end by 32P-γ-ATP and non-radioactive PO4-TG3T). Migration of the associated (G4) and dissociated (ss) forms are indicated; species were revealed using UV-shadowing at 254 nm for non-radioactive species or by detection of the radioactive signal. (E and F) Thermal denaturation of preformed (E) [TG3T]4 and (F) [PO4-TG3T]4 G-quadruplexes (strand concentrations as in panels A and B respectively) followed by absorbance at 295 nm (black squares) and 240 nm (red triangles) at a heating gradient of 0.5°C/min.
Table 1.
Association constants found
| Oligonucleotide sequencea | 5′end | 360Ac | kond (M−3 s−1) | T1/2e (°C) | Method | Reference |
|---|---|---|---|---|---|---|
| TGGGT | OH | – | 5.2 × 107 | 48 | UV-absorbance | (34) |
| TGGGT | OH | – | 6.7 × 107 | 50 | UV-absorbance | This study |
| TGGGT | OH | – | 4.0 × 107 | n/a | Gel; UV shadow | This study |
| TGGGT | Pb | – | 2.3 × 106 | 49.5 | UV-absorbance | This study |
| TGGGT | Pb | – | 2.0 × 106 | n/a | Gel; UV shadow | This study |
| TGGGT | Pb | – | 1.4 × 106 | n/a | Gel; radioactivity | This study |
| TGGGT | Pb | 50 µM | 3.4 × 1012 | n/a | Gel; radioactivity | This study |
| TGGGT | Pb | 100 µM | 1.4 × 1013 | n/a | Gel; radioactivity | This study |
| TTAGGG | OH | – | 1.8 × 107 | 50 | UV-absorbance | (34)f |
| TTAGGGT | OH | – | 2.1 × 106 | 55 | UV-absorbance | (34)f |
| TTGGGGGTT | OH | – | 6.1 × 107 | >90 | UV-absorbance | (34)f |
a5′ to 3′ direction. bP for 5′-phosphate group. cConcentration of Quadruplex ligand (360A) added.—: no ligand. din 110 mM KCl (or NaCl for T2G5T2), 10 mM lithium cacodylate pH 7.2 buffer. Values with ±30% accuracy. ein 110 mM KCl, 10 mM lithium cacodylate pH 7.2 buffer, with a temperature gradient of 0.5°C/min. fAlso see Supplementary Figure S6. NA: not applicable (this apparent melting temperature cannot be determined in a gel).
Although there were minor differences, both methods indicate that the presence of a 5′ terminal phosphate has a negative impact on the kinetics of association of a tetramolecular quadruplex. In contrast, the presence of a phosphate group had a limited effect on the thermal stability of the quadruplex. As shown in Figures 4E and 4F, the mid-point of the thermal melting transition (T1/2) was 50 for the 5′-OH oligonucleotide and 49.5°C for the 5′PO4-TG3T oligonucleotide. Even if, at lower salt concentrations (Supplementary Figure S4C), a more significant 2°C difference was recorded, suggesting again a role of the negative charges in the destabilization of the structure, these results indicate that the presence of a terminal phosphate significantly affects the association kinetics, but has rather little effect on the thermal stability or dissociation kinetics of the quadruplex.
Kinetics of 5′PO4-TG3T quadruplex formation in the presence of 360A
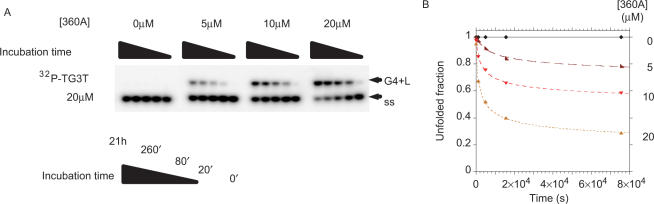
The addition of a terminal phosphate slows, but does not prevent, quadruplex formation. It is, therefore, possible to follow quadruplex formation with oligonucleotides radioactively labeled at the 5′ terminus with 32P and to determine whether a ligand affects association kinetics. In the gel presented in Figure 5A, no significant quadruplex formation was observed after 21 h at 20 µM strand concentration in the absence of a ligand. At this concentration, 50% quadruplex formation (t1/2) would take years, and the reverse reaction would not be negligible. A 10 fold-higher strand concentration leads to partial quadruplex formation, see Figure 4D). Addition of 360A leads to faster association (Figure 5A, right lanes), confirming UV-shadow analysis at higher strand and ligand concentrations. The rate acceleration is quantified in Figure 5B: 0, 22, 42 and 76% quadruplex formation was obtained after 21 h in the presence of 0, 5, 10 and 20 µM 360A, respectively. At the highest ligand concentration tested, t1/2 was 80 min.
Figure 5.
Compound 360A promotes formation of tetramolecular G-quadruplex [PO4-TG3T]4 at 20 µM strand concentration. (A) Association kinetics of the G-quadruplex formed by PO4-TG3T at 20 µM total strand concentration (mixture of 32P-TG3T and non-radioactive PO4-TG3T) alone or in presence of 5–20 µM 360A in 10 mM lithium cacodylate (pH 7.2), 110 mM KCl at 4°C, followed by non-denaturing gel electrophoresis (20% in 1× TBE, 20 mM KCl at 4°C) at the time points indicated. Position of migration of the quadruplex (G4+L) and single-stranded (ss) forms are shown. (B) Quantification of the single-stranded fraction (black diamonds: PO4-TG3T alone; dark red triangles: +5 µM 360A; red triangles: +10 µM 360A; and orange triangles: +20 µM 360A). Data cannot be fitted by the classical fourth-order model (34).
Kinetics of quadruplex formation in the presence of a large molar excess of 360A
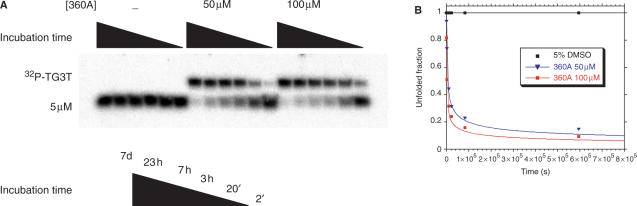
Quantitative analysis of the previous gel suggested that the presence of a quadruplex ligand lead to an initial increase in the apparent association rate. However, the effects were less pronounced at longer times: the quantitative difference between 4.3 h and 21 h incubations was relatively small (Figure 5B). We reasoned that, as more and more quadruplexes were formed, the ligand, which was not in huge molar excess, would be progressively titrated out. In order to simplify the modeling of ligand action, one must work under experimental conditions for which the free ligand concentration can be considered invariant during the reaction. To approach this condition, we chose to lower the oligonucleotide strand concentration to 5 µM and to increase the ligand concentration to 50 or 100 µM (Figure 6). Assuming a maximum of 2 binding sites per quadruplex (Supplementary Figure S5), complete quadruplex formation would decrease free ligand concentration by no more than 2.5 µM, a negligible amount relative to the ligand concentration. Figure 6A demonstrates that, unsurprisingly, no quadruplex formation was obtained after one week at 5 µM strand concentration in the absence of 360A. In contrast, 50 or 100 µM ligand lead to near-complete quadruplex formation over the same period. Results were quantitated and are presented in Figure 6B. At 100 µM ligand, t1/2 was 140 min. It was possible to fit these curves using the same mathematical model we and others used for tetramolecular quadruplex formation (34). Apparent kon values of 3.4 × 1012 M−3 s−1 and 1.4 × 1013 M−3 s−1 were found for 50 and 100 µM ligand concentrations, respectively. The association rate constant determined in the absence of any ligand for the same 5′ phosphate oligonucleotide at a much higher strand concentration (200 or 300 µM) was 1.4–2.3 × 106 M−3 s−1; at 5 µM, in the absence of ligand, no quadruplex formation was observed. The comparison of these rate constants allows us to conclude that the presence of 50 or 100 µM ligand increases the rate constant 2 or 21 million-fold, respectively. Due to the order of 4 chosen for the fits, this impressive—but somehow misleading—difference corresponds to a 125–275-fold apparent increase in strand concentration.
Figure 6.
Induction of the tetramolecular G-quadruplex [PO4-TG3T]4 by a large excess of 360A. (A) Association kinetics of PO4-TG3T incubated at 4°C at a final strand concentration of 5 µM in absence or presence of 360A at 50 and 100 µM; incubations are performed in 10 mM lithium cacodylate (pH 7.2), 110 mM KCl. Products were resolved on a 20% acrylamide gel; electrophoresis was performed at 4°C in buffer containing 1× TBE and 20 mM KCl. (B) Quantification of the fraction of 32P-TG3T single-strand (black squares: without ligand; blue triangles: +360A 50 µM; red squares: +360A 100 µM). The fourth-order model for tetramolecular G-quadruplexes association was applied and kon values calculated for complex formation in the presence of large excess of ligand are given (34).
360A affects the kinetics of other quadruplexes
We analyzed in detail the effect of 360A on a single sequence, TG3T. To prove that the effect of this ligand was not restricted to this peculiar sequence, we analyzed its impact on the association kinetics of three other sequences, T2AG3, T2AG3T and T2G5T2 (Supplementary Figure S6). In all cases, a significant increase in the amount of quadruplex formed was observed in the presence of 360A compared to the same conditions for oligonucleotide alone, as revealed by UV-shadow. This indicates that this ligand was able to increase the kinetics of association of various tetramolecular quadruplexes.
DISCUSSION AND CONCLUSION
The number of identified G4 ligands has grown rapidly over the past few years. A range of molecules recognize the telomeric quadruplex (18,23,24,31,45,50–63). The compound 360A belongs to a family of pyridine dicarboxamide compounds that exhibit high selectivity toward quadruplex DNA relative to other DNA structures (26,47,48). Thanks to the relatively slow kinetics of tetramolecular quadruplex formation and dissociation, it was possible to perform fluorescence titration experiments with the same oligonucleotide in quadruplex or single-stranded form (Supplementary Figure S5). Little or no variation of 360A fluorescence emission was found when an excess of the single-strand was added, whereas a near-complete quenching was obtained in the presence of the tetrameric quadruplex. These results were not unexpected, considering the equilibrium dialysis profile previously found for this molecule (48).
It is a general assumption that small molecules bind to preformed nucleic acid structures. In this article, we demonstrate that 360A may actively induce the formation of a tetramolecular quadruplex, acting as a chaperone for the association of the four strands. Furthermore, this effect was not restricted to a unique sequence; quadruplex formation by several different sequences was accelerated by the compound. The presence of a 3′ terminal guanine or a TpG was not required, as evidenced by data generated on the TTAGGG sequence. During this study, we also demonstrated that introduction of a 5′-terminal phosphate through radioactive labeling affected the kinetics, presumably due to the additional negative charges. The 5′ phosphate group slowed the association rate with little effect on thermal stability (Table 1). In principle, the presence of these extra charges could also influence the binding affinity of the ligand; however, 360A bound to both 5′-OH and 5′-phosphate quadruplexes, and affected favorably the kinetics of both quadruplexes.
Under the conditions of our studies, 360A did not exert a catalytic effect: one 360A molecule accelerates the formation of a single quadruplex complex and remains bound. The ligand remains attached to the newly formed quadruplex, due to its high affinity. This conclusion was reached using a molar quadruplex:ligand ratio above one (Figure 3C). Another interesting bit of information is that, contrary to the case of bimolecular quadruplexes, a single complex is formed; this is supported by the fact that (i) we observed a single band on the gel, (ii) the CD T1/2 values (Supplementary Figure S2B) and (iii) the isothermal difference spectra (Supplementary Figure S3B) are similar. Furthermore, the overall architecture of the quadruplex induced by the presence of the ligand is probably similar to the structure of the quadruplex formed alone, as the CD signatures of both quadruplexes were similar (Supplementary Figure S2A) and the migration of quadruplexes were not very different (with the exception of PIPER, and taking into account the fact that the ligand somewhat retards the complex due to its positive charges and molecular weight) (Supplementary Figure S6).
Of course, the observation that a ligand increases the association rate constant does not imply that this molecule has no effect on the dissociation rate constant. In fact, 360A did affect the apparent melting temperature of the quadruplex (Supplementary Figure S2B), indicating that 360A also leads to a decrease in koff (i.e. an increase in the lifetime and thermal stability of the complex). This is in agreement with other studies demonstrating that a quadruplex ligand affects the dissociation of a quadruplex (64,65). Several questions remain unanswered, and two main questions are the purpose of our ongoing studies: (i) Do these observations hold for different ligands? The gels shown in Figure 2 suggest that other molecules such as a trisubstituted acridine also affect the association process, although to a lesser extent than the bis quinolinium pyridine dicarboxamide derivatives 360A and 307A. We will determine how general this process is and to which parameters it could be related (charge, shape, affinity). (ii) Are other types of quadruplexes also affected, such as bi- and intramolecular complexes? These complexes, which might be more biologically relevant than the parallel-stranded, tetramolecular complexes studied here, will be more complicated to analyze because of their faster and more complicated kinetics of association and dissociation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Patrick Mailliet and Eliane Mandine (Sanofi-Aventis, Vitry, France), Jean-François Riou (Université de Reims), Laurent Lacroix, Barbara Saccà, Patrizia Alberti, Anne Bourdoncle, Julien Gros and Samir Amrane (Museum, Paris) for helpful discussions. Telomestatin was a kind gift from Prof. K. Shin-ya (U. Tokyo). This work was supported in part by an ARC grant (n°3365) and an EU FP6 ‘MolCancerMed’ grant (LSHC-CT-2004-502943 to J-L.M.). Funding to pay the Open Access publication charges was provided by INSERM.
Conflict of interest statement. None declared.
REFERENCES
- 1.Williamson JR. G-quartet structures in telomeric DNA. Annu. Rev. Biophys. Biomol. Struc. 1994;23:703–730. doi: 10.1146/annurev.bb.23.060194.003415. [DOI] [PubMed] [Google Scholar]
- 2.Davis JT. G-quartets 40 years later: from 5′-GMP to molecular biology and supramolecular chemistry. Angew. Chem. Int. Ed. 2004;43:668–698. doi: 10.1002/anie.200300589. [DOI] [PubMed] [Google Scholar]
- 3.Henderson E, Hardin CC, Walk SK, Tinoco I, Jr, Blackburn EH. Telomeric DNA oligonucleotides form novel intramolecular structures containing guanine-guanine base pairs. Cell. 1987;51:899–908. doi: 10.1016/0092-8674(87)90577-0. [DOI] [PubMed] [Google Scholar]
- 4.Oka Y, Thomas C.A., Jr The cohering telomeres of Oxytricha. Nucleic Acids Res. 1987;15:8877–8898. doi: 10.1093/nar/15.21.8877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sundquist WI, Klug A. Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature. 1989;342:825–829. doi: 10.1038/342825a0. [DOI] [PubMed] [Google Scholar]
- 6.Williamson JR, Raghuraman MK, Cech TR. Monovalent cation induced structure of telomeric DNA: the G-quartet model. Cell. 1989;59:871–880. doi: 10.1016/0092-8674(89)90610-7. [DOI] [PubMed] [Google Scholar]
- 7.Zahler AM, Williamson JR, Cech TR, Prescott DM. Inhibition of telomerase by G-quartet DNA structures. Nature. 1991;350:718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- 8.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grand CL, Powell TJ, Nagle RB, Bearss DJ, Tye D, Gleason-Guzman M, Hurley LH. Mutations in the G-quadruplex silencer element and their relationship to c-MYC overexpression, NM23 repression, and therapeutic rescue. Proc. Natl. Acad. Sci. USA. 2004;101:6140–6145. doi: 10.1073/pnas.0400460101. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Schaffitzel C, Berger I, Postberg J, Hanes J, Lipps HJ, Plückthun A. In vitro generated antibodies specific for telomeric guanine quadruplex DNA react with Stylonychia lemnae macronuclei. Proc. Natl. Acad. Sci. USA. 2001;98:8572–8577. doi: 10.1073/pnas.141229498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duquette ML, Handa P, Vincent JA, Taylor AF, Maizels N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 2004;18:1618–1629. doi: 10.1101/gad.1200804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paeschke K, Simonsson T, Postberg J, Rhodes D, Lipps H. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat. Struct. Mol. Biol. 2005;12:847–854. doi: 10.1038/nsmb982. [DOI] [PubMed] [Google Scholar]
- 13.Li JJ, Tan W. A single DNA molecule nanomotor. Nano Lett. 2002;2:315–318. doi: 10.1021/nl9011694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alberti P, Mergny JL. DNA duplex-quadruplex exchange as the basis for a nanomolecular machine. Proc. Natl. Acad. Sci. USA. 2003;100:1569–1573. doi: 10.1073/pnas.0335459100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bourdoncle A, Estévez Torres A, Gosse C, Lacroix L, Vekhoff P, Le Saux T, Jullien L, Mergny JL. Quadruplex-based molecular beacons as tunable DNA probes. J. Am. Chem. Soc. 2006;128:11094–11105. doi: 10.1021/ja0608040. [DOI] [PubMed] [Google Scholar]
- 16.Maizels N. Dynamic roles for G4 DNA in the biology of eukaryotic cells. Nature Struct. Mol. Biol. 2006;13:1055–1059. doi: 10.1038/nsmb1171. [DOI] [PubMed] [Google Scholar]
- 17.Oganesian L, Bryan TM. Physiological relevance of telomeric G-quadruplex formation: a potential drug target. Bioessays. 2007;29:155–165. doi: 10.1002/bies.20523. [DOI] [PubMed] [Google Scholar]
- 18.Sun D, Thompson B, Cathers BE, Salazar M, Kerwin SM, Trent JO, Jenkins TC, Neidle S, Hurley LH. Inhibition of human telomerase by a G-quadruplex-interactive compound. J. Med. Chem. 1997;40:2113–2116. doi: 10.1021/jm970199z. [DOI] [PubMed] [Google Scholar]
- 19.Mergny JL, Hélène C. G-quadruplex DNA: A target for drug design. Nature Med. 1998;4:1366–1367. doi: 10.1038/3949. [DOI] [PubMed] [Google Scholar]
- 20.Guittat L, Alberti P, Rosu F, Van Miert S, Thetiot E, Pieters L, Gabelica V, De Pauw E, Ottaviani A, et al. Interactions of cryptolepine and neocryptolepine with unusual DNA structures. Biochimie. 2003;85:535–547. doi: 10.1016/s0300-9084(03)00035-x. [DOI] [PubMed] [Google Scholar]
- 21.Franceschin M, Rossetti L, Dambrosio A, Schirripa S, Bianco A, Ortaggi G, Savino M, Schultes C, Neidle S. Natural and synthetic G-quadruplex interactive berberine derivatives. Bioorg. Medicinal Chem. Lett. 2006;16:1707–1711. doi: 10.1016/j.bmcl.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 22.Shin-ya K, Wierzba K, Matsuo K, Ohtani T, Yamada Y, Furihata K, Hayakawa Y, Seto H. Telomestatin, a novel telomerase inhibitor from Streptomyces anulatus. J. Am. Chem. Soc. 2001;123:1262–1263. doi: 10.1021/ja005780q. [DOI] [PubMed] [Google Scholar]
- 23.Kim MY, Vankayalapati H, Shin-ya K, Wierzba K, Hurley LH. Telomestatin, a potent telomerase inhibitor that interacts quite specifically with the human telomeric intramolecular G-quadruplex. J. Am. Chem. Soc. 2002;124:2098–2099. doi: 10.1021/ja017308q. [DOI] [PubMed] [Google Scholar]
- 24.Gowan S, Heald R, Stevens M, Kelland L. Potent inhibition of telomerase by small-molecule pentacyclic acridines capable of interacting with G-quadruplexes. Mol. Pharmacol. 2001;60:981–988. doi: 10.1124/mol.60.5.981. [DOI] [PubMed] [Google Scholar]
- 25.Han FXG, Wheelhouse RT, Hurley LH. Interactions of TMPyP4 and TMPyP2 with quadruplex DNA. Structural basis for the differential effects on telomerase inhibition. J. Am. Chem. Soc. 1999;121:3561–3570. [Google Scholar]
- 26.Pennarun G, Granotier C, Gauthier LR, Gomez D, Hoffschir F, Mandine E, Riou JF, Mergny JL, Mailliet P, et al. Apoptosis related to telomere instability and cell cycle alterations in human glioma cells treated by new highly selective G-quadruplex ligands. Oncogene. 2005;24:2917–2928. doi: 10.1038/sj.onc.1208468. [DOI] [PubMed] [Google Scholar]
- 27.De Cian A, DeLemos E, Mergny JL, Teulade-Fichou MP, Monchaud D. Highly efficient G-Quadruplex recognition by bisquinolinium compounds. J. Am. Chem. Soc. 2007;129:1856–1857. doi: 10.1021/ja067352b. [DOI] [PubMed] [Google Scholar]
- 28.Riou JF, Guittat L, Mailliet P, Laoui A, Petigenet O, Megnin-Chanet F, Hélène C, Mergny JL. Cell senescence and telomere shortening induced by a new series of specific G-quadruplex DNA ligands. Proc. Natl. Acad. Sci. USA. 2002;99:2672–2677. doi: 10.1073/pnas.052698099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han HY, Cliff CL, Hurley LH. Accelerated assembly of G-quadruplex structures by a small molecule. Biochemistry. 1999;38:6981–6986. doi: 10.1021/bi9905922. [DOI] [PubMed] [Google Scholar]
- 30.Kern JT, Kerwin SM. The aggregation and G-quadruplex DNA selectivity of charged 3,4,9,10-perylenetetracarboxylic acid diimides. Bioorg. Med. Chem. Lett. 2002;12:3395–3398. doi: 10.1016/s0960-894x(02)00763-1. [DOI] [PubMed] [Google Scholar]
- 31.Harrison RJ, Cuesta J, Chessari G, Read MA, Basra SK, Reszka AP, Morrell J, Gowan SM, Incles CM, et al. Trisubstituted acridine derivatives as potent and selective telomerase inhibitors. J. Med. Chem. 2003;46:4463–4476. doi: 10.1021/jm0308693. [DOI] [PubMed] [Google Scholar]
- 32.Burger AM, Dai FP, Schultes CM, Reszka AP, Moore MJ, Double JA, Neidle S. The G-quadruplex-interactive molecule BRACO-19 inhibits tumor growth, consistent with telomere targeting and interference with telomerase function. Cancer Res. 2005;65:1489–1496. doi: 10.1158/0008-5472.CAN-04-2910. [DOI] [PubMed] [Google Scholar]
- 33.Wyatt JR, Davis PW, Freier SM. Kinetics of G-quartet-mediated tetramer formation. Biochemistry. 1996;35:8002–8008. doi: 10.1021/bi960124h. [DOI] [PubMed] [Google Scholar]
- 34.Mergny JL, De Cian A, Ghelab A, Saccà B, Lacroix L. Kinetics of tetramolecular quadruplexes. Nucleic Acids Res. 2005;33:81–94. doi: 10.1093/nar/gki148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mergny JL, De Cian A, Amrane S, Webba da Silva M. Kinetics of double-chain reversals bridging contiguous quartets in tetramolecular quadruplexes. Nucleic Acids Res. 2006;34:2386–2397. doi: 10.1093/nar/gkl098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petraccone L, Pagano B, Esposito V, Randazzo A, Piccialli G, Barone G, Mattia CA, Giancola C. Thermodynamics and kinetics of PNA-DNA quadruplex-forming chimeras. J. Am. Chem. Soc. 2005;127:16215–16223. doi: 10.1021/ja0545923. [DOI] [PubMed] [Google Scholar]
- 37.Petraccone L, Erra E, Randazzo A, Giancola C. Energetic aspects of locked nucleic acids quadruplex association and dissociation. Biopolymers. 2006;83:584–594. doi: 10.1002/bip.20591. [DOI] [PubMed] [Google Scholar]
- 38.Sen D, Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its applications for meiosis. Nature. 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 39.Sen D, Gilbert W. A sodium-potassium switch in the formation of four-stranded G4-DNA. Nature. 1990;344:410–414. doi: 10.1038/344410a0. [DOI] [PubMed] [Google Scholar]
- 40.Cantor CR, Warshaw MM, Shapiro H. Oligonucleotide interactions. 3. Circular dichroism studies of the conformation of deoxyoligonucleotides. Biopolymers. 1970;9:1059–1077. doi: 10.1002/bip.1970.360090909. [DOI] [PubMed] [Google Scholar]
- 41.Kejnovska I, Kypr J, Vorlickova M. Oligo(dT) is not a correct native PAGE marker for single-stranded DNA. Biochem. Biophys. Res. Commun. 2007;353:776–779. doi: 10.1016/j.bbrc.2006.12.093. [DOI] [PubMed] [Google Scholar]
- 42.Mergny JL, Phan AT, Lacroix L. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 1998;435:74–78. doi: 10.1016/s0014-5793(98)01043-6. [DOI] [PubMed] [Google Scholar]
- 43.Petraccone L, Erra E, Esposito V, Randazzo A, Galeone A, Barone G, Giancola C. Biophysical properties of quadruple helices of modified human telomeric DNA. Biopolymers. 2005;77:75–85. doi: 10.1002/bip.20189. [DOI] [PubMed] [Google Scholar]
- 44.Guo Q, Lu M, Marky LA, Kallenbach NR. Interaction of the dye ethidium bromide with DNA containing guanine repeats. Biochemistry. 1992;31:2451–2455. doi: 10.1021/bi00124a002. [DOI] [PubMed] [Google Scholar]
- 45.Koeppel F, Riou JF, Laoui A, Mailliet P, Arimondo PB, Labit D, Petigenet O, Hélène C, Mergny JL. Ethidium derivatives bind to G-quartets, inhibit telomerase and act as fluorescent probes for quadruplexes. Nucleic Acids Res. 2001;29:1087–1096. doi: 10.1093/nar/29.5.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosu F, Pauw ED, Guittat L, Alberti P, Lacroix L, Mailliet P, Riou J-F, Mergny J-L. Selective interaction of ethidium derivatives with quadruplexes. Biochemistry. 2003;42:10361–10371. doi: 10.1021/bi034531m. [DOI] [PubMed] [Google Scholar]
- 47.Lemarteleur T, Gomez D, Paterski R, Mandine E, Mailliet P, Riou J-F. Stabilization of the c-myc gene promoter quadruplex by specific ligands inhibitors of telomerase. Biochem. Biophys. Res. Com. 2004;323:802–808. doi: 10.1016/j.bbrc.2004.08.150. [DOI] [PubMed] [Google Scholar]
- 48.Granotier C, Pennarun G, Riou L, Hoffschir F, Gauthier LR, DeCian A, Gomez D, Mandine E, Riou JF, et al. Preferential binding of a G-quadruplex ligand to human chromosome ends. Nucl Acid Res. 2005;33:4182–4190. doi: 10.1093/nar/gki722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Uddin MK, Kato Y, Takagi Y, Mikuma T, Taira K. Phosphorylation at 5′ end of guanosine stretches inhibits dimerization of G-quadruplexes and formation of a G-quadruplex interferes with the enzymatic activities of DNA enzymes. Nucleic Acids Res. 2004;32:4618–4629. doi: 10.1093/nar/gkh766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wheelhouse RT, Sun D, Han H, Han FX, Hurley LH. Cationic porphyrins as telomerase inhibitors: the interaction of tetra (N-methyl-4-pyridyl) porphyrin with quadruplex DNA. J. Am. Chem. Soc. 1998;120:3261–3262. [Google Scholar]
- 51.Fedoroff OY, Salazar M, Han H, Chemeris VV, Kerwin SM, Hurley LH. NMR-based model of a telomerase inhibiting compound bound to G-quadruplex DNA. Biochemistry. 1998;37:12367–12374. doi: 10.1021/bi981330n. [DOI] [PubMed] [Google Scholar]
- 52.Perry PJ, Read MA, Davies RT, Gowan SM, Reszka AP, Wood AA, Kelland LR, Neidle S. 2,7-disubstituted amidofluorenone derivatives as inhibitors of human telomerase. J. Med. Chem. 1999;42:2679–2684. doi: 10.1021/jm990084q. [DOI] [PubMed] [Google Scholar]
- 53.Harrison RJ, Gowan SM, Kelland LR, Neidle S. Human telomerase inhibition by substituted acridine derivatives. Bioorg. Med. Chem. Lett. 1999;9:2463–2468. doi: 10.1016/s0960-894x(99)00394-7. [DOI] [PubMed] [Google Scholar]
- 54.Neidle S, Harrison RJ, Reszka AP, Read MA. Structure-activity relationships among guanine-quadruplex telomerase inhibitors. Pharmacol. Ther. 2000;85:133–139. doi: 10.1016/s0163-7258(99)00065-0. [DOI] [PubMed] [Google Scholar]
- 55.Caprio V, Guyen B, Opoku-Boahen Y, Mann J, Gowan SM, Kelland LM, Read MA, Neidle S. A novel inhibitor of human telomerase derived from 10H-indolo[3,2-b]quinoline. Bioorg. Med. Chem. Lett. 2000;10:2063–2066. doi: 10.1016/s0960-894x(00)00378-4. [DOI] [PubMed] [Google Scholar]
- 56.Mergny JL, Lacroix L, Teulade-Fichou MP, Hounsou C, Guittat L, Hoarau M, Arimondo PB, Vigneron JP, Lehn JM, et al. Telomerase inhibitors based on quadruplex ligands selected by a fluorescent assay. Proc. Natl. Acad. Sci. USA. 2001;98:3062–3067. doi: 10.1073/pnas.051620698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alberti P, Schmidt P, Nguyen CH, Hoarau M, Grierson D, Mergny JL. Benzoindoloquinolines interact with DNA quadruplexes and inhibit telomerase. Bioorg. Med. Chem. Lett. 2002;12:1071–7074. doi: 10.1016/s0960-894x(02)00080-x. [DOI] [PubMed] [Google Scholar]
- 58.Shi DF, Wheelhouse RT, Sun DY, Hurley LH. Quadruplex-interactive agents as telomerase inhibitors: Synthesis of porphyrins and structure-activity relationship for the inhibition of telomerase. J. Med. Chem. 2001;44:4509–4523. doi: 10.1021/jm010246u. [DOI] [PubMed] [Google Scholar]
- 59.Heald RA, Modi C, Cookson JC, Hutchinson I, Laughton CA, Gowan SM, Kelland LR, Stevens MFG. Antitumor polycyclic acridines. 8. Synthesis and telomerase-inhibitory activity of methylated pentacyclic acridinium salts. J. Med. Chem. 2002;45:590–597. doi: 10.1021/jm011015q. [DOI] [PubMed] [Google Scholar]
- 60.Gowan SM, Harrison JR, Patterson L, Valenti M, Read MA, Neidle S, Kelland LR. A G-quadruplex-interactive potent small-molecule inhibitor of telomerase exhibiting in vitro and in vivo antitumor activity. Mol. Pharmacol. 2002;61:1154–1162. doi: 10.1124/mol.61.5.1154. [DOI] [PubMed] [Google Scholar]
- 61.Rossetti L, Franceschin M, Bianco A, Ortaggi G, Savino M. Perylene diimides with different side chains are selective in inducing different G-quadruplex DNA structures and in inhibiting telomerase. Bioorg. Med. Chem. Lett. 2002;12:2527–2533. doi: 10.1016/s0960-894x(02)00504-8. [DOI] [PubMed] [Google Scholar]
- 62.Read M, Harrison RJ, Romagnoli B, Tanious FA, Gowan SH, Reszka AP, Wilson WD, Kelland LR, Neidle S. Structure-based design of selective and potent G quadruplex-mediated telomerase inhibitors. Proc. Natl. Acad. Sci. USA. 2001;98:4844–4849. doi: 10.1073/pnas.081560598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maraval A, Franco S, Vialas C, Pratviel G, Blasco MA, Meunier B. Porphyrin-aminoquinoline conjugates as telomerase inhibitors. Org. Biomol. Chem. 2003;1:921–927. doi: 10.1039/b211634k. [DOI] [PubMed] [Google Scholar]
- 64.Rosu F, Gabelica V, Shin-ya K, DePauw E. Telomestatin induced stabilization of the human telomeric DNA quadruplex monitored by electrospray mass spectrometry. Chem. Commun. 2003;34:2702–2703. doi: 10.1039/b309394h. [DOI] [PubMed] [Google Scholar]
- 65.Green JJ, Ladame S, Ying L, Klenerman D, Balasubramanian S. Investigating a quadruplex-ligand interaction by unfolding kinetics. J. Am. Chem. Soc. 2006;128:9809–9812. doi: 10.1021/ja0615425. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.