Abstract
Expression of short hairpin RNAs via the use of PolIII-based transcription systems has proven to be an effective mechanism for triggering RNAi in mammalian cells. The most popular promoters for this purpose are the U6 and H1 promoters since they are easily manipulated for expression of shRNAs with defined start and stop signals. Multiplexing (the use of siRNAs against multiple targets) is one strategy that is being developed by a number of laboratories for the treatment of HIV infection since it increases the likelihood of suppressing the emergence of resistant virus in applications. In this context, the development of alternative small PolIII promoters other than U6 and H1 would be useful. We describe tRNALys3-shRNA chimeric expression cassettes which produce siRNAs with comparable efficacy and strand selectivity to U6-expressed shRNAs, and show that their activity is consistent with processing by endogenous 3′ tRNAse. In addition, our observations suggest general guidelines for expressing effective tRNA-shRNAs with the potential for graded response, to minimize toxicities associated with competition for components of the endogenous RNAi pathway in cells.
INTRODUCTION
Suppression of HIV-1 replication has been achieved through siRNAs directed against numerous targets including the TAR element, the 3′ UTR, vif, gag, tat, rev and reverse transcriptase (RT) transcripts, as well as the HIV-1 cellular co-receptor CCR5 [recently reviewed in (1–7)]. While these results are encouraging, many challenges still remain in developing effective HIV-1 gene therapeutics. Of particular concern is the ability of HIV-1 to rapidly evolve resistance to RNAi, especially as siRNAs are sensitive to single base-pair mismatches. Careful design and optimization of anti-viral siRNAs increase the probability of circumventing resistance in therapeutic applications. Potential RNAi-related problems include off-target effects, immunostimulation and interference with cellular microRNA pathways by competition for transport, processing or RISC loading. Choosing highly conserved HIV-1 regions as targets increases the likelihood that RNAi-resistant variants will be less fit. Multiplexing (the use of siRNAs against multiple conserved targets) further increases the likelihood of suppressing the emergence of viral variants, but must be balanced against the possibility of compromising cellular metabolism.
Expressing shRNAs from tRNA promoters has several potential advantages in this context, compared to the more commonly used U6 and H1 promoters: tRNA promoters are smaller, provide a variety of options and are typically expressed at lower levels. Smaller promoters ease multiplexing in delivery vectors with size constraints; expanding the number of promoter options eases difficulties associated with repeated use of the same promoter within a multiplex construct, and possible recombination. In addition, lower expression levels may allow multiplexing anti-HIV RNAi constructs with a lower probability of interference with endogenous RNAi pathways. Controlling the levels of shRNA expression appears to be an increasingly important consideration for therapeutic applications since sustained high-level expression of shRNAs can lead to toxicity via competition with components of the endogenous RNAi machinery and perhaps increased off-targeting (8,9).
To address the challenge of creating a simplified PolIII expression system that can be used in combination with, or in place of the existing U6 and H1 promoters, we designed a tRNALys3-shRNA chimera expression construct targeting a highly conserved sequence in an exon common to both tat and rev, previously shown to be an effective target when the same shRNA was expressed by the U6 promoter (10,11).
The first step in developing this vector system was to optimize the effectiveness of tRNALys3-shRNA chimeras while minimizing off-target effects. We have developed a series of tRNALys3-shRNA chimeric constructs which exhibit a graded set of functional activities that should prove useful for a variety of shRNA expression applications. Our studies also define critical features of the tRNA sequence that result in effective processing of the shRNAs for further processing into siRNAs.
MATERIALS AND METHODS
Plasmid construction
tRNALys3-tat/rev shRNA expression vectors
Cloning of the U6-tat/rev shRNA was previously described (11). The details of cloning and all DNA oligomers used in this study are listed in the Supplementary Data, but a brief description is included here.
tRNALys3-tat/rev shRNA BsrG1 loop constructs were cloned as follows. A PCR product encoding the tRNA and tat/rev hairpin sense strand and loop was generated using a (1) 5′ primer and template specific to either the S4 or wild-type tRNA; the 5′ primer adds a SalI site immediately upstream of the 5′ end of the mature tRNALys3 sequence and (2) a series of 3′ primers that introduce a given acceptor stem/DNt combination as well as the hairpin sense/BsrG1 loop sequences. The PCR products were digested with SalI and BsrG1 and gel purified as previously described (11). Two additional complementary oligomers having BsrG1 and PstI overhangs at the 5′ and 3′ ends (after annealing) encode the remainder of the hairpin loop, anti-sense stem and PolIII termination sequences (which is identical in all of the BsrG1 loop constructs). The PCR products and annealed oligos were ligated into the SalI and PstI sites of pBluescript (Stratagene, USA).
tRNALys3-tat/rev shRNA DC loop constructs were cloned as follows. tRNALys3 vectors were generated by PCR as above, only using a series of 3′ primers that add either a BssHII or NruI site followed by a HindIII site in the tRNALys3 acceptor stem sequence. The PCR product was digested with SalI and HindIII and cloned into the same sites of pBluescript. Annealed oligos with BssHII or NruI 5′ termini and PstI 3′ termini that code for the remainder of the tRNALys3 acceptor stem, entire tat/rev shRNA hairpin and RNA pol3 termination signal were ligated into the cognate sites of the appropriate tRNALys3 vector.
Luciferase reporter vectors
Complementary DNA oligomers designed to have 5′ XhoI and 3′ XbaI overhangs after annealing were synthesized for either the tat/rev sense or anti-sense target regions, and inserted into the XhoI and XbaI sites in the 3′ UTR of Renilla luciferase (R-luc) of the psiCHECK-2 dual reporter plasmid (Promega, USA). The correct sequence of all constructs was confirmed by DNA sequencing at the City of Hope Sequencing Core.
Tissue culture, transfections and dual luciferase assays
HCT116 colon carcinoma cells (CCL-247) or HEK-293 embryonic kidney fibroblast cells (CCL-1573) were obtained from the American Type Culture Collection (USA), and grown in DMEM high glucose (Irvine Scientific, USA) supplemented with 10% fetal bovine serum (FBS) and 1.5 mM l-glutamine (Irvine Scientific, USA) and 10 mM pyruvate (Irvine Scientific, USA). Cells were cultured in a humidified 5% CO2 incubator at 37°C.
For dual luciferase assays, HCT116 or 293 cells were seeded in 48-well culture dishes and transfected the next day at ∼90% confluency using Lipofectamine 2000 (Invitrogen, USA) according to the manufacturer's instructions. For each well, a total of 160 ng DNA (see below) in 20 μl OptiMEM was combined with an equal amount of diluted (2.0 μl/100 μl OptiMEM) Lipofectamine 2000. The incubation medium was removed from the cells and the DNA–lipofectamine mixture was added immediately, followed by 160 μl of complete medium. At 18 h post-transfection, the cell medium was replaced with 200 μl fresh medium and culture continued for another 6 h. Dual luciferase assays were performed according to the manufacturer's instructions (Promega, USA) 24 h post-transfection. All samples were transfected in duplicate or triplicate and the experiment was performed a minimum of three times.
For psiCHECK dual luciferase transfections, DNA mixtures contained 40 ng of psiCHECK target derivative, 40 ng of test or control plasmid and 80 ng pBluescript (carrier plasmid) in 20 μl OptiMEM. Negative controls, used for normalization, contained a plasmid with the U6 or tRNA promoter for each psiCHECK target tested. The U6-tat/rev shRNA construct was used as a positive control. For each replicate, the R-luc target reading was normalized internally to the F-luc value, and an average calculated from the replicates. The average for each hairpin construct was then normalized to the value calculated from the appropriate empty promoter (S4tRNALys3, (+)tRNALys3 or U6) and target (sense or anti-sense) combination in the same experiment. The averages from independent experiments were then used to calculate the values and standard deviations shown in the figures.
pNL4-3.Luc.R−.E− (NIH AIDS Research and Reference Reagent Program, Germantown, MD) is a Env− Vpr− non-infectious clone containing the firefly luciferase (F-luc) gene inserted into the nef gene. Transfections contained a final combination of 40 ng of pNL4-3.Luc.R−.E−, 0.2 ng of pRSV-Renilla, 40 ng of test or control plasmid and 80 ng pBluescript (carrier plasmid) in 20 μl OptiMEM. For each replicate, the F-luc target reading was normalized to the R-luc internal control, and an average calculated from the replicates. The average for each hairpin construct was then normalized to the value calculated using the appropriate empty promoter (S4tRNALys3, (+)tRNALys3 or U6) in the same experiment. The averages from independent experiments were then used to calculate the values and standard deviations shown in the figures.
Northern analysis
HCT116 and HEK 293 cells were cultured and transfected as above for dual luciferase assays with the following modifications. (1) Transfections were scaled up to 60 mm dishes. (2) To enhance detection of siRNA products, all the transfected DNA consisted of the indicated construct (as opposed to a 1:1:2 mixture of shRNA construct:target construct:carrier pBluescript DNAs); consequently, 4-fold more shRNA construct DNA was transfected per cell for northern analysis, although the total amount of transfected DNA per cell remained constant in both assays. (3) Cells were incubated for 48 h post-transfection before isolating RNA, replacing the media at 18 and 42 h post-transfection. RNA was isolated from transiently transfected cells using RNA STAT60 (Tel-Test, Inc., USA) following the manufacturer's instructions. Twenty-five micrograms of total RNA was electrophoresed on a 10% polyacrylamide–8 M urea gel. RNA was transferred to Hybond-N+ membrane (Amersham Pharmacia Biotech, USA) by electroblotting. Prehybridization and hybridization were carried out using PerfectHyb Plus Hybridization buffer (Sigma, USA) at 37°C with 5 pmol of oligonucleotide probe end-labeled with T4 polynucleotide kinase and γ-32P-ATP. Filters were washed twice with 100–150 μl/cm2 with 5 × SSPE/1% SDS at 37°C for 15 min, then sequentially with 0.5–1.0 ml/cm2 with 2 × SSPE/0.5% SDS and 1 × SSPE/0.5% SDS at 45°C for 30–60 min each, prior to autoradiography or PhosphorImager quantitation using ImageQuant software. Filters were then re-hybridized with an end-labeled probe against U2 snRNA as a loading control, and autoradiography and PhosphorImager quantitation repeated.
RESULTS
Previous studies in our laboratory demonstrated U6-tat/rev shRNA (Figure 1A) to be a highly effective inhibitor of HIV replication in pNL 4–3 transient transfection assays (10,11); consequently, we chose this same tat/rev shRNA for the initial or ‘parental’ tRNALys3-promoter construct, S4tRNALys3-tat/rev (Figure 1B and C), using a design modeled on previously published tRNALys3-anti-HIV ribozyme chimeras [(12) and see below]. We initially assayed these constructs using the psiCHECK reporter system, which readily allows screening of the potencies of candidate sh/siRNAs. The psiCHECK reporter system has the advantage that knockdown of both the sense target (corresponding to the mRNA) and the anti-sense target can be assayed independently, and used as a measure of the relative incorporation of each strand into the RNA-induced silencing complex (RISC). This strand selectivity is an important factor in evaluating shRNAs; for instance, efficacy against the desired target may suffer, due to competition by the sense (passenger) strand with the anti-sense (guide) strand for RISC entry. Of equal concern, off-target effects are greater with shRNAs that efficiently incorporate the passenger strand due to unfavorable thermodynamics or structure. Off-target effects confuse interpretation of downstream events in biological or genetic applications of RNAi and potentially compromise patient health in therapeutic applications of RNAi. In practice, it is informative to measure both the total efficacy (knockdown of the sense target corresponding to the mRNA) and the strand selectivity (the ratio of sense target:anti-sense target knockdown) when evaluating shRNAs.
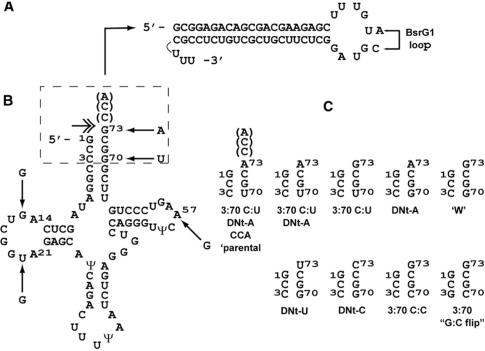
Figure 1.
(A) tat/rev shRNA, as transcribed from the U6 promoter. The hairpin orientation is sense–loop–anti-sense. In some derivatives, additional nucleotides were changed in the loop as shown to create a BsrG1 site for cloning convenience. (B) tRNALys3-tat/rev shRNAs. tRNALys3 wild-type sequence is shown starting with the mature tRNALys3 5′ end; arrows indicate SELEX-derived mutations in the S4tRNALys3 variant; link to base of tat-rev hairpin in (A) is shown schematically. The double headed arrow indicates the location of cleavage required to release the tat/rev hairpin with a sequence identical to that in (A). The dashed box indicates the acceptor stem/DNt region which was varied and shown in (C). (C) Acceptor stem/DNt variants. Only mutated bases relative to wild-type are indicated; all constructs lack the CCA linker between the discriminator nucleotide and the first base of the hairpin unless otherwise indicated. DNt-X indicates a change of the predominant (G) discriminator nucleotide to the indicated base. Third acceptor stem base-pair mutations relative to wild-type are also shown. ‘W’: wild-type acceptor stem. The acceptor stem/DNt nucluotide variants were constructed in both the SELEX4 (St4tRNALys3) and wild-type (+) tRNALys3 backgrounds.
Our initial comparison demonstrated that, as expected, the U6-tat/rev shRNA is highly effective, mediating ∼95% knockdown of the sense target; however, knockdown of the anti-sense target is much less, indicative of good strand selection (Figure 2, lane 1). In contrast, the parental S4tRNALys3-tat/rev shRNA has poorer efficacy (<80%) and strand selectivity, targeting the anti-sense strand nearly equally (Figure 2, lanes 1 and 4). If S4tRNALys3-tat/rev were less efficient at targeting the sense strand, but had a comparable anti-sense:sense target ratio as U6-tRNALys3-tat/rev the simplest interpretation would be that the S4tRNALys3 promoter produces the same tat/rev shRNA as the U6 counterpart, but at lower levels. This is not what we observe: moreover, previous experiments suggest that the reduced efficacy of tRNALys3-tat/rev shRNAs of this general design stemmed in part from poor processing, although 21-mer guide strand products were observed by Northern analysis (data not shown). Since the U6 promoter transcribes a single shRNA resulting in superior RNAi efficacy and strand selectivity, we considered the possibility that processing of the parental S4tRNALys3-shRNA construct was producing related, but poorer RNAi effectors than the U6 hairpin, as opposed to the identical hairpin at lower levels. This seemed reasonable since it is known that minor changes, such as hairpin leader sequences and single nucleotide shifts in target sequences between related siRNAs, can have a large impact on RNAi-mediated target knockdown. If so, then strand selectivity and possibly efficacy might improve if the same shRNA could be processed from the S4tRNALys3-tat/rev shRNA chimera as produced by the U6 promoter (Figure 1A).
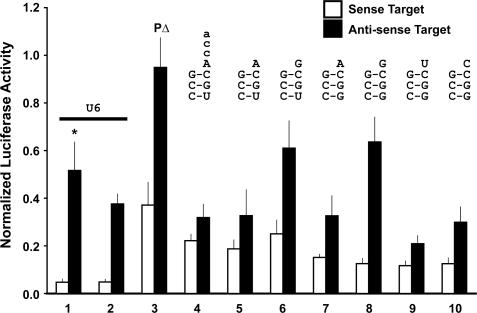
Figure 2.
tat/rev shRNA activity and strand selectivity of acceptor stem/DNt variants on the S4tRNALys3 background. Dual luciferase assays of psiCHECK sense (open bars) and anti-sense (filled bars) targets are shown. All constructs are normalized to the value of the corresponding empty promoter construct (S4tRNALys3 or the U6) in combination with the relevant target (sense or anti-sense). All tat/rev hairpins contain the BsrG1 loop except where an asterix (*) denotes the DC loop. PΔ (Lane 3) indicates a mutant tat/rev shRNA carrying a deletion (Δ) of nucleotide 10 of the tat/rev hairpin passenger (P) strand, which has no activity on the anti-sense substrate. The sequence of the tRNA acceptor stem and discriminator nucleotide (Figure 1B and C) is shown above each construct. (Lane 1) U6 tat/rev shRNA (DC loop). (Lane 2) U6 tat/rev shRNA (BsrG1 loop). (Lane 3) S4tRNALys3-3:70 C:U/DNt-A/CCA tat/rev shRNA (BsrG1 loop) ΔP. (Lane 4) S4tRNALys3-3:70 C:U/DNt-A/CCA tat/rev shRNA (BsrG1 loop). (Lane 5) S4tRNALys3-3:70 C:U/DNt-A tat/rev shRNA (BsrG1 loop). (Lane 6) S4tRNALys3-3:70 C:U tat/rev shRNA (BsrG1 loop). (Lane 7) S4tRNALys3-DNt-A tat/rev shRNA (BsrG1 loop). (Lane 8) S4tRNALys3-W tat/rev shRNA (BsrG1 loop). (Lane 9) S4tRNALys3-DNt-U tat/rev shRNA (BsrG1 loop). (Lane 10) S4tRNALys3-DNt-C tat/rev shRNA (BsrG1 loop).
We next considered the possibility that if these conjectures were correct, re-examination of the parental S4tRNALys3-tat/rev shRNA chimera characteristics might be useful. Briefly, the parental S4tRNALys3-tat/rev shRNA hybrid design was based upon the tRNALys3-PBS ribozyme chimeras (12) consisting of: a SELEX-derived tRNALys3 variant (S4tRNALys3); a CCA linker (mimicking the sequence added posttranscriptionally during normal tRNA biogenesis prior to aminoacylation); and an anti-HIV-1ribozyme directed against the sequence immediately adjacent to the HIV-1 primer binding sequence (PBS) recognized by the 3′ end of cellular tRNALys3, the obligate primer for HIV-1 reverse transcription. The S4tRNALys3 variant contains a total of five mutations (Figure 1B). The first two mutations, A14G and A21G, affect tRNA-invariant D-loop nucleotides; the third A57G is a conservative (purine–purine) change of a semi-invariant position in the TΨC-loop. The S4tRNALys3-PBS ribozyme (S4tRNALys3-Rz) maintains 4–6-fold higher steady-state levels of expression in transient transfections than the corresponding wild-type tRNALys3-Rz (data not shown). We have not determined if this is due to higher transcription levels, since A14 and A57 occur in promoter A and B boxes, respectively, or to a longer half-life since the invariant bases A14 and A21 are involved in tertiary interactions. The fourth mutation G70U changes the third 3:70 base pair (bp) of the acceptor stem from C:G to C:U. The fifth mutation changes the discriminator nucleotide (DNt), which serves as an identity element for recognition by the corresponding aminoacyl-tRNA synthetase from G to A (G73A); only 3/15 of the known human nuclear tRNALys3 genes have A as the DNt (http://lowelab.ucsc.edu/GtRNAdb) (13).
The processing mechanism producing 21-mer tat/rev effector sequences from the S4tRNALys3-tat/rev shRNA hybrid was unclear; however, one possible candidate was the cellular machinery for processing endogenous precursor tRNAs, particularly the enzyme involved in the removal of 3′ trailers. We reevaluated our initial design in the light of what is known about this enzyme, tRNAse ZL. tRNAse ZL is a 3′tRNAse that cleaves pre-tRNA 3′ trailers immediately after the DNt (in most cases), which is the first unpaired base after the acceptor stem, prior to addition of the CCA sequence [reviewed in (14); Figure 1B]. In vitro assays demonstrate that inclusion of the CCA sequence in pre-tRNA-like substrates inhibits the 3′ tRNAse processing enzyme tRNAse ZL in higher eukaryotes (15–17). The tRNALys3 moiety of the parental chimera contains a CCA linker after the DNt, which would potentially inhibit tRNAse ZL activity. The SELEX mutations affect the base pairing in the acceptor stem and potentially alter the tertiary structure of the pre-tRNA; similar changes in this stem can inhibit tRNAse ZL processing (see the Discussion section). Therefore, we decided to test the hypothesis that redesigning the tRNALys3-tat/rev shRNA chimeras to be better substrates of tRNAse ZL would improve efficacy and strand selectivity of the shRNA, as measured by the psiCHECK reporter assays already described. We expected that more efficient processing of the chimera would increase sense target knockdown, which we refer to as efficacy (provided strand selectivity were not highly compromised). Good strand selectivity, i.e. a high ratio of sense:anti-sense target knockdown, would be consistent with release of the shRNA by specific cleavage after the DNt, since it would produce the same hairpin structure as the U6 promoter. Poor strand selectivity would be indicative of the production of other, less efficacious products, either by tRNAseZL or some other cellular process.
In order to test this concept, we evaluated the effect of systematically reverting the SELEX mutations in the tRNALys3 moiety of the shRNA ‘parental’ chimera, using the psiCHECK reporter system as an endpoint assay. We began by first addressing the mutations in the acceptor stem and DNts, as well as the effect of the CCA sequence. Specifically (Figure 1C), we constructed a series of S4tRNALys3-based-tat/rev shRNAs where we eliminated the CCA, known to inhibit tRNAse ZL cleavage of endogenous pre-tRNA, in all constructs and (1) reverted the mutant U70 base back to G, restoring the wild-type 3:70 C:G pairing of third acceptor stem base-pair from the mutant C:U or (2) reverted the A73 base DNt-A, back to the original DNt-G or (3) reverted both. In addition, we constructed versions DNt-C and DNt-U which have the remaining possible DNts (with a restored acceptor stem 3:70 C:G pair). The first three SELEX mutations, A14G, A21G and A57G were retained; this is referred to as the ‘SELEX background’ indicated as S4tRNALys3. We tested this first set of constructs in parallel with the ‘parental’ S4tRNALys3 tat/rev shRNA and U6-tat/rev shRNAs (Figure 2).
As shown in the reporter assays (Figure 2), mere removal of the CCA did not increase efficacy (knockdown of the sense target) or enhance strand selectivity (Figure 2, lanes 4 and 5). Efficacy was most improved by restoration of the third acceptor stem Watson–Crick base-pairing (Figure 2, lanes 7–10), although not to levels obtained by the corresponding U6 constructs (Figure 2, lanes 1 and 2). However, any DNt besides G compromised strand selectivity; only restoration of the wild-type G DNt reduced knockdown of the anti-sense strand to approximately the same absolute value as seen in the U6 constructs (Figure 2, lanes 1, 6 and 8), although the efficacy was still somewhat compromised relative to U6 tat/rev shRNA.
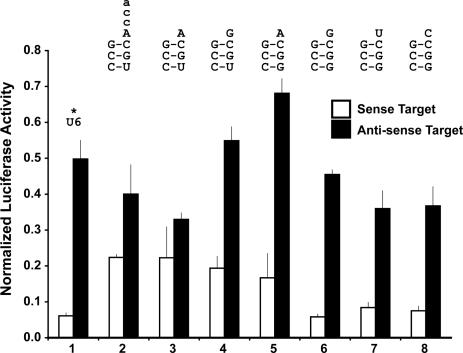
Next, we tested the effects of the remaining SELEX mutations by reverting the first three A14G, A21G and A57G SELEX mutations back to wild-type (referred to as the wild-type or (+)tRNALys3 background) in combination with the same series of acceptor stem/DNt variants (Figure 3). As with the previous series, efficacy and strand selectivity were not improved by removal of the CCA alone (Figure 3, lanes 2 and 3). Efficacy was again improved by restoration of the third Watson–Crick acceptor stem base-pairing in most cases (Figure 3, lanes 4, 6–8); however, the A DNt compromised efficacy but not strand selectivity in this background (Figure 3, lanes 5–8). The most striking result was that the completely wild-type tRNALys3-tat/rev shRNA construct had identical efficacy and strand selectivity as the corresponding U6 version in this assay (Figure 3, lane 6), and the DNt-U and DNt-C variants were nearly as good (Figure 3, lanes 7 and 8).
Figure 3.
tat/rev shRNA activity and strand selectivity of acceptor stem/DNt variants in the wild-type (+)tRNALys3 background. Dual luciferase assays of psiCHECK sense (open bars) and anti-sense (filled bars) targets. All constructs are normalized to the value of the corresponding empty promoter construct (S4tRNALys3 or the U6) in combination with the relevant target (sense or anti-sense). All tat/rev hairpins contain the BsrG1 loop except where an asterix (*) denotes the DC loop. (Lane 1) U6 tat/rev shRNA (DC loop). (Lane 2) (+)tRNALys3-3:70 C:U/DNt-A/CCA tat/rev shRNA (BsrG1 loop). (Lane 3) (+)tRNALys3-3:70 C:U/DNt-A tat/rev shRNA (BsrG1 loop). (Lane 4) (+)tRNALys3-3:70 C:U tat/rev shRNA (BsrG1 loop). (Lane 5) (+)tRNALys3-DNt-A tat/rev shRNA (BsrG1 loop). (Lane 6) (+)tRNALys3-W tat/rev shRNA (BsrG1 loop). (Lane 7) (+)tRNALys3-DNt-U tat/rev shRNA (BsrG1 loop). (Lane 8) (+)tRNALys3-DNt-C tat/rev shRNA (BsrG1 loop).
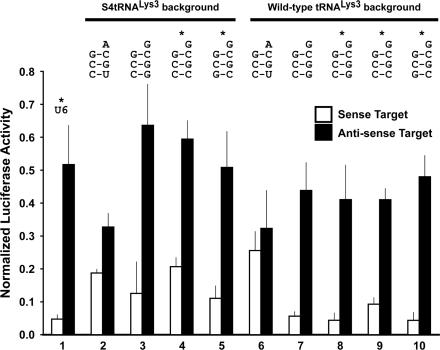
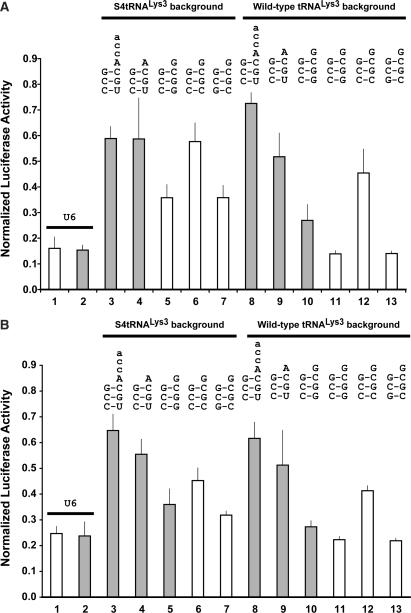
We compared additional acceptor stem variant tRNALys3 tat/rev shRNAs on both backgrounds in psiCHECK dual luciferase assays; specifically, the wild-type third 3:70 C:G acceptor stem base-pair was altered to C:C or G:C (G:C flip), We also tested tat/rev shRNAs with two slightly different hairpin loops, referred to as the ‘DC’ and ‘BsrG1’ loops (Figure 1A). On the S4tRNALys3 background, the acceptor stem third base-pair C:C mismatch (Figure 4, lane 4) had the same knockdown profile as the C:U mismatch (Figure 2, lane 6), compromising sense strand knockdown, not strand selectivity. However, restoration of the canonical Watson–Crick base-pairing, even in the flipped orientation, restored efficacy (Figure 4, lane 5). The same general pattern was observed in the wild-type tRNALys3 background (Figure 4, lanes 8–10).
Figure 4.
Effect of the acceptor stem third base-pair variants on tat/rev shRNA functional activity and strand-selectivity in both the S4tRNALys3 and (+)tRNALys3 backgrounds. Dual luciferase assays of psiCHECK sense (open bars) and antisense (filled bars) targets. All constructs are normalized to the value of the corresponding empty promoter construct (S4tRNALys3 or the U6) in combination with the relevant target (sense or anti-sense). All tat/rev hairpins contain the BsrG1 loop except where an asterix (*) denotes the DC loop. (Lane 1) U6 tat/rev shRNA (DC loop). (Lane 2) S4tRNALys3-3:70 C:U/DNt-A tat/rev shRNA (BsrG1 loop). (Lane 3) S4tRNALys3-W tat/rev shRNA (BsrG1 loop). (Lane 4) S4tRNALys3-3:70 C:C tat/rev shRNA (DC loop). (Lane 5) S4tRNALys3-3:70 G:C flip/DNt-A tat/rev shRNA (DC loop). (Lane 6) (+)tRNALys3-3:70 C:U/DNt-A tat/rev shRNA (BsrG1 loop). (Lane 7) (+)tRNALys3-W tat/rev shRNA (BsrG1 loop). (Lane 8) (+)tRNALys3-W tat/rev shRNA (DC loop). (Lane 9) (+)tRNALys3-3:70 C:C tat/rev shRNA (DC loop). (Lane 10) (+)tRNALys3-3:70 G:C flip/DNt-A tat/rev shRNA (DC loop).
These trends were even more obvious in a different dual luciferase assay system. In our hands, knockdown of psiCHECK targets is sometimes artificially high, thereby minimizing potential differences in sh/siRNA efficacy that reflect more accurately their performance in more biologically relevant applications. We therefore replicated these assays using a non-infectious HIV pNL4-3 luc reporter, containing a firefly luciferase-coding sequence inserted into the viral 3′ UTR (18,19). Unlike the psiCHECK reporter, which produces a single target transcript, pNL4-3 luc produces multiple HIV-1 transcripts which share a common 3′ UTR containing the luciferase-coding sequences. Any tat or rev transcripts that escape RNAi-mediated degradation allow expression of downstream viral messages, which all express the reporter but not necessarily the tat/rev target sequence. In practice, differences in tat and rev knockdown that are difficult to detect in the psiCHECK system are more obvious in the pNL4-3 luc reporter system. As seen in the psiCHECK assays, the acceptor stem third base-pair C:C mismatch compromised efficacy against the pNL4–3 luc target as well, which was fully restored by the G:C flip within both tRNALys3 backgrounds (Figure 5A; lanes 6, 7, 12 and 13). However, the differences between the S4tRNALys3 and +S4tRNALys3 backgrounds were much more evident (e.g. Figure 5A; lane 8 versus 13). In addition, the sequence of the tat/rev loop influenced efficacy on the wild-type (+)tRNALys3 background (Figure 5A; lanes 10 and 11). Most importantly, two tRNALys3-tat/rev shRNA constructs in the wild-type background achieved knockdown levels comparable to the U6-tat/rev shRNA constructs (Figure 5A; lanes 1, 2, 12 and 13).
Figure 5.
Effect of the acceptor stem third base-pair variants on tat/rev shRNA functional activity and strand selectivity in both the S4tRNALys3 and (+) tRNALys3 backgrounds in the pNL4-3 luc reporter assay in (A) HCT116 cells and (B) HEK293 cells. Open bars: constructs with tat/rev shRNA DC loop. Gray filled bars: constructs with tat/rev shRNA BsrG1 loop. All values are normalized to the values obtained with the corresponding (+) tRNALys3, S4tRNALys3 or U6 empty promoter constructs. (Lane 1) U6 tat/rev shRNA (DC loop). (Lane 2) U6 tat/rev shRNA (BsrG1 loop). (Lane 3) S4tRNALys3-3:70 C:U/DNt-A/CCA tat/rev shRNA (BsrG1 loop). (Lane 4) S4tRNALys3-3:70 C:U/DNt-A tat/rev shRNA (BsrG1 loop). (Lane 5) S4tRNALys3 -W tat/rev shRNA (BsrG1 loop). (Lane 6) S4tRNALys3-3:70 C:C tat/rev shRNA (DC loop). (Lane 7) S4tRNALys3-3:70 G:C flip/DNt-A tat/rev shRNA (DC loop). (Lane 8) (+)tRNALys3-3:70 C:U/DNt-A/CCA tat/rev shRNA (BsrG1 loop). (Lane 9) (+)tRNALys3-3:70 C:U/DNt-A tat/rev shRNA (BsrG1 loop). (Lane 10) (+)tRNALys3 -W tat/rev shRNA (BsrG1 loop). (Lane 11) (+)tRNALys3 -W tat/rev shRNA (DC loop). (Lane 12) (+)tRNALys3-3:70 C:C tat/rev shRNA (DC loop). (Lane 13) (+)tRNALys3-3:70 G:C flip/DNt-A tat/rev shRNA (DC loop).
To be generally applicable, these observations should be reproducible in different cell types. We repeated the pNL4-3-luc reporter assays shown in Figure 5A in HEK 293 cells (Figure 5B) and observed the same general trends.
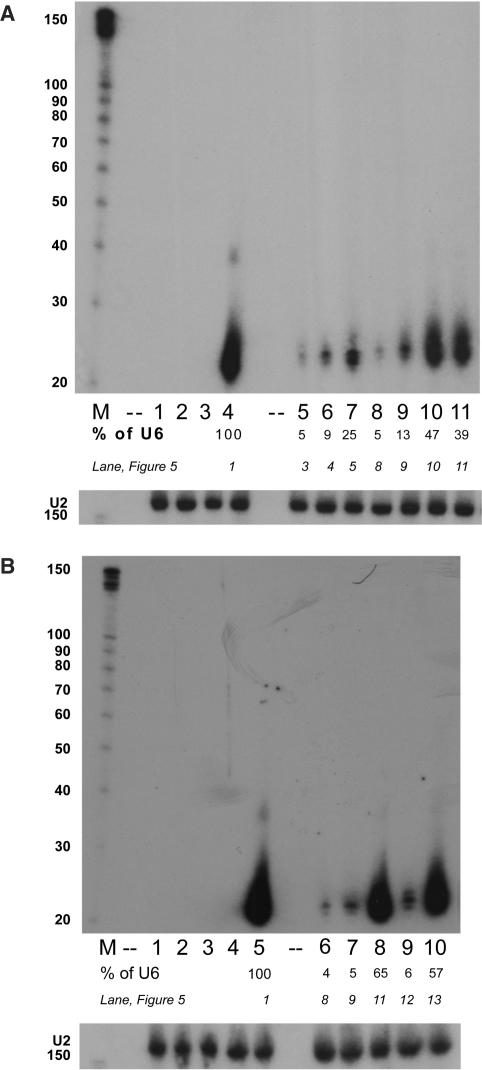
RNAi is mediated by ∼21-nt effector complexes; therefore, the tRNALys3-shRNA constructs should generate products derived from the hairpin guide strand of the same size. We carried out northern gel analyses on total RNA extracted from both HCT116 and 293 cells transiently transfected with subsets of our constructs representing the best and worst knockdown profiles, and hybridized the blots with probes against the guide strand of the hairpin (Figure 6). In both cell types, the relative levels of guide strand siRNA in the northern analysis reflected the efficacy of the construct in the dual luciferase assays.
Figure 6.
Processed siRNAs from tRNALys3-tat/rev shRNAs. (A) Top: Northern analysis of transiently transfected tRNALys3-tat/rev shRNA constructs in HCT116 cells, probed for tat/rev shRNA guide strand. (M) Ambion decade marker, numbers indicate size in nucleotides. (Lane 1) pBluescript. (Lane 2) U6-promoter. (Lane 3) S4tRNALys3-PBS ribozyme. (Lane 4) U6-tat/rev shRNA. (Lane 5) S4tRNALys3-3:70 C:U/DNt-A/CCA tat/rev shRNA (BsrG1 loop). (Lane 6) S4tRNALys3-3:70 C:U/DNt-A tat/rev shRNA (BsrG1 loop). (Lane 7) S4tRNALys3-(W) tat/rev shRNA. (Lane 8) (+)tRNALys3-3:70 C:U/DNt-A/CCA tat/rev shRNA (BsrG1 loop). (Lane 9) (+)tRNALys3-3:70 C:U/DNt-A tat/rev shRNA (BsrG1 loop). (Lane 10) (+)tRNALys3-W tat/rev shRNA (BsrG1 loop). (Lane 11) (+)tRNALys3-W tat/rev shRNA. (B) Northern analysis of transiently transfected tRNALys3-tat/rev shRNA constructs in HEK 293 cells, probed for tat/rev shRNA guide strand. (M) Ambion decade marker. (Lane 1) pBluescript. (Lane 2) U6-irrelevant shRNA. (Lane 3) (+)tRNALys3-promoter. (Lane 4) U6(+)tRNALys3-PBS ribozyme. (Lane 5) U6-tat/rev shRNA. (Lane 6) (+)tRNALys3-3:70 C:U/DNt-A/CCA tat/rev shRNA (BsrG1 loop). (Lane 7) (+)tRNALys3-3:70 C:U/DNt-A tat/rev shRNA (BsrG1 loop). (Lane 8) (+)tRNALys3-(W) tat/rev shRNA. (Lane 9) (+)tRNALys3-3:70 C:C tat/rev shRNA (Lane 10).
DISCUSSION
Endogenous nuclear and mitochondrial pre-tRNAs undergo a multi-step maturation that includes removal of the 5′ leader sequence by RNAse P [reviewed in (20–23)]; removal of the 3′ trailer sequence by 3′ tRNAse endonucleolytic cleavage, typically immediately after the discriminator base [reviewed in (24–26)]; and addition of the CCA trinucleotide to the processed 3′ end by the CCA nucleotidyl-transferase (27) prior to aminoacylation. In addition, tRNAs undergo many posttranscriptional modifications (28–30). [See also (14,31) for overviews of tRNA processing.]
The tRNA 3′ processing enzyme, or tRNAse Z occurs in long and short forms, tRNAse ZL and tRNAse ZS, respectively; all eukaryotes have the long form, and some have both [for nomenclature, see (24)]. The human homologs HsatRNAse ZS and HsatRNAse ZL are encoded by the related but separate genes ELAC1 and ELAC2, respectively (32–34). Recombinant HsatRNAse ZL processes nuclear-encoded pre-tRNA 1600-fold more efficiently than the short form, suggesting that it is the predominant active form for processing endogenous tRNAs when both forms are present in the genome (35). Drosophila cells retain only the long form, which processes nuclear and mitochondrial pre-tRNAs in vivo and in vitro (36). Recombinant HsatRNAse ZL, but not HsatRNAse ZS, contains a mitochondrial import signal, and processes mitochondrial pre-tRNA substrates in vitro, although with lower efficiency than nuclear pre-tRNAs on the tested substrates (35,37).
We designed tRNALys3-tat/rev shRNA chimeras taking into account their potential as tRNAse ZL substrates. Accurate cleavage after the discriminator base will produce the same hairpin as the U6 promoter, which we used as the standard for optimal RNAi-mediated knockdown efficiency and strand selectivity. Our studies delineate some structural characteristics of tRNALys3 that influence the RNAi activity of the chimera, which we consider in the context of what is currently known about the activity of tRNAse ZL.
Mismatches in the third base-pair (3:70) of the tRNALys3 acceptor stem reduce overall RNAi efficacy, but not strand selectivity in both wild-type and the SELEX A14G/A21G/A57G mutant backgrounds. This is consistent with reduced efficiency, but not reduced accuracy of hairpin release from the tRNALys3 chimeras in these constructs. However, the acceptor stem need not carry the exact wild-type sequence for full activity, as constructs where the third acceptor stem base-pair is flipped from C:G to G:C are equivalent within the same tRNALys3 background. This result parallels the negative effects of acceptor stem mutations that alter Watson–Crick base-pairing in mitochondrial tRNAs (mt-tRNAs) on cleavage efficiency by HsatRNAse ZL (35,37–39). Similar effects are seen with porcine tRNAse Z activities on bipartite tRNA-like structures (40) and Drosophila tRNAse ZL on nuclear tRNAHis.
The negative effects of the acceptor stem base-pair mismatches are intensified in the SELEX A14G/A21G/A57G mutant background, although only efficiency, not strand selectivity, is compromised; this again is consistent with a decrease in activity, rather than accuracy of processing. The canonical tertiary tRNA U8-A14-A21 interaction may be adversely affected in S4tRNALys3 variant. In this context, it is interesting that the A3243G mutation in mt-tRNALeu(UUR), corresponding to the A14G mutation found in the S4tRNA variant, is processed less efficiently by tRNAse ZL in HeLa cell extracts (37). This comparison is even more striking, given that of all the mitochondrial tRNA families, tRNALeu(UUR) retain the most classical structure, including all potential tertiary interactions, unlike many mt-RNAs which often differ from the canonical nuclear tRNAs (28,41).
The CCA of mature tRNA is an anti-determinant for eukaryotic tRNAse ZL (15–17), preventing non-productive and energetically expensive recycling of addition and removal of the CCA triplet, impeding subsequent aminoacylation. Our ‘parental’ constructs contained a CCA linker between the DNt and the tat/rev hairpin. Somewhat surprisingly, removal of the CCA had no appreciable effect, but we only tested CCA removal in the context of the DNt G73A mutation and C3:U70 acceptor stem mismatches, which may mask any effect of the CCA in these assays.
As a whole, our results are consistent with a model in which cellular tRNAse ZL cleaves tRNALys3-tat/rev shRNA chimeras immediately after the discriminator base, in the same relative position as endogenous pre-tRNAs, when the chimeras resemble a normal substrate, releasing a tat/rev hairpin which functions in RNAi equivalently to the U6 promoter product. This hypothesis seems reasonable given that mammalian tRNAse ZL is capable of cleaving very minimal or unusual substrates in vitro, though with varying efficiencies (40,42). Synthetic or expressed small guide RNAs can cleave target sequences in vivo that form tRNA-like structure in trans both in vitro (43–46) and in vivo (44,47). Experiments are underway to more directly determine if tRNAse ZL is actually responsible for processing tRNA-shRNA and if the structure–function relationships observed in this work generalize to other tRNA isotypes. A possible application of our results is that tRNA-hairpin chimeras could provide a rapid endpoint assay of the effect of tRNA mutations on pre-tRNA processing.
Our basic design is substantially different than that of previously reported tRNAVal-based shRNAs (48,49). In these studies, tRNAVal was used only as a promoter for the attached shRNA. The tRNAVal sequence was truncated after the B-box sequences, keeping only the first 65 nt intact; consequently, there is no acceptor stem or DNt. Instead, the shRNA is tethered by a 25 nt, non-tRNA sequence. In that study, the authors did not address issues of strand selectivity, design optimization or the potential processing mechanism.
A feature of the chimeric tRNALys3-shRNA chimeric constructs, which is significantly different from that of the U6-expressed shRNAs, is the fact that the hairpin is processed from the primary transcript in the tRNALys3 expression systems, whereas the U6 promoter results in direct expression of intact hairpins which are exported to the cytoplasm and processed by RISC. Since the shRNAs in our constructs do not fit the criteria for Drosha/DGCR8 processing (50,51), the hairpin is most probably released by tRNAseZL prior to export and RISC processing.
In summary, we describe tRNALys3-shRNAs with comparable RNAi efficiency and strand selectivity to the corresponding U6-driven shRNA, and determine some of the parameters that influence the degree of and specificity of knockdown, expanding the repertoire of promoters for shRNA expression. Furthermore, the tRNALys3 variants we describe have the potential to mediate graded RNAi knockdown (for example, Figure 5, lanes 7, 8 and 13). This may be of value in avoiding the toxicity that can arise in applications where using multiple shRNAs are advantageous, such as interfering with HIV-1 replication. Another possible application would be in modulating knockdown of an in vivo target to produce intermediate phenotypes in genetic studies.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank M. Amarzguioui and H. Soifer for helpful discussions. This work was supported by NIH grants AI29329, AI42552 and HL07470 awarded to J.J.R. Funding to pay the Open Access publication charge was provided by a National Institutes of Health grant to J.J.R.
Conflict of interest statement. None declared.
REFERENCES
- 1.Morris KV, Rossi JJ. Lentiviral-mediated delivery of siRNAs for antiviral therapy. Gene Ther. 2006;13:553–558. doi: 10.1038/sj.gt.3302688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rossi JJ. RNAi as a treatment for HIV-1 infection. Biotechniques. 2006;40:s25–s29. doi: 10.2144/000112167. [DOI] [PubMed] [Google Scholar]
- 3.Nekhai S, Jerebtsova M. Therapies for HIV with RNAi. Curr. Opin. Mol. Ther. 2006;8:52–61. [PubMed] [Google Scholar]
- 4.Cullen BR. Does RNA interference have a future as a treatment for HIV-1 induced disease? AIDS Rev. 2005;7:22–25. [PubMed] [Google Scholar]
- 5.Bagasra O. RNAi as an antiviral therapy. Expert. Opin. Biol. Ther. 2005;5:1463–1474. doi: 10.1517/14712598.5.11.1463. [DOI] [PubMed] [Google Scholar]
- 6.Nielsen MH, Pedersen FS, Kjems J. Molecular strategies to inhibit HIV-1 replication. Retrovirology. 2005;2:10. doi: 10.1186/1742-4690-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yeung ML, Bennasser Y, Le SY, Jeang KT. siRNA, miRNA and HIV: promises and challenges. Cell Res. 2005;15:935–946. doi: 10.1038/sj.cr.7290371. [DOI] [PubMed] [Google Scholar]
- 8.Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 9.An DS, Qin FX, Auyeung VC, Mao SH, Kung SK, Baltimore D, Chen IS. Optimization and functional effects of stable short hairpin RNA expression in primary human lymphocytes via lentiviral vectors. Mol. Ther. 2006;14:494–504. doi: 10.1016/j.ymthe.2006.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee NS, Dohjima T, Bauer G, Li H, Li MJ, Ehsani A, Salvaterra P, Rossi J. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat. Biotechnol. 2002;20:500–505. doi: 10.1038/nbt0502-500. [DOI] [PubMed] [Google Scholar]
- 11.Scherer LJ, Yildiz Y, Kim J, Cagnon L, Heale B, Rossi JJ. Rapid assessment of anti-HIV siRNA efficacy using PCR-derived Pol III shRNA cassettes. Mol. Ther. 2004;10:597–603. doi: 10.1016/j.ymthe.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 12.Westaway SK, Cagnon L, Chang Z, Li S, Li H, Larson GP, Zaia JA, Rossi JJ. Virion encapsidation of tRNA(3Lys)-ribozyme chimeric RNAs inhibits HIV infection. Antisense Nucleic Acid Drug. Dev. 1998;8:185–197. doi: 10.1089/oli.1.1998.8.185. [DOI] [PubMed] [Google Scholar]
- 13.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hopper AK, Phizicky EM. tRNA transfers to the limelight. Genes Dev. 2003;17:162–180. doi: 10.1101/gad.1049103. [DOI] [PubMed] [Google Scholar]
- 15.Mohan A, Whyte S, Wang X, Nashimoto M, Levinger L. The 3′ end CCA of mature tRNA is an antideterminant for eukaryotic 3′-tRNase. RNA. 1999;5:245–256. doi: 10.1017/s1355838299981256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nashimoto M. Distribution of both lengths and 5′ terminal nucleotides of mammalian pre-tRNA 3′ trailers reflects properties of 3′ processing endoribonuclease. Nucleic Acids Res. 1997;25:1148–1154. doi: 10.1093/nar/25.6.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zareen N, Hopkinson A, Levinger L. Residues in two homology blocks on the amino side of the tRNase Z His domain contribute unexpectedly to pre-tRNA 3′ end processing. RNA. 2006;12:1104–1115. doi: 10.1261/rna.4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen BK, Saksela K, Andino R, Baltimore D. Distinct modes of human immunodeficiency virus type 1 proviral latency revealed by superinfection of nonproductively infected cell lines with recombinant luciferase-encoding viruses. J. Virol. 1994;68:654–660. doi: 10.1128/jvi.68.2.654-660.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Connor RI, Chen BK, Choe S, Landau NR. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 20.Evans D, Marquez SM, Pace NR. RNase P: interface of the RNA and protein worlds. Trends Biochem. Sci. 2006;31:333–341. doi: 10.1016/j.tibs.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 21.Kirsebom LA. RNase P RNA-mediated catalysis. Biochem. Soc. Trans. 2002;30:1153–1158. doi: 10.1042/bst0301153. [DOI] [PubMed] [Google Scholar]
- 22.Jarrous N. Human ribonuclease P: subunits, function, and intranuclear localization. RNA. 2002;8:1–7. doi: 10.1017/s1355838202011184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao S, Scott F, Fierke CA, Engelke DR. Eukaryotic ribonuclease P: a plurality of ribonucleoprotein enzymes. Annu. Rev. Biochem. 2002;71:165–189. doi: 10.1146/annurev.biochem.71.110601.135352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vogel A, Schilling O, Spath B, Marchfelder A. The tRNase Z family of proteins: physiological functions, substrate specificity and structural properties. Biol. Chem. 2005;386:1253–1264. doi: 10.1515/BC.2005.142. [DOI] [PubMed] [Google Scholar]
- 25.Levinger L, Morl M, Florentz C. Mitochondrial tRNA 3′ end metabolism and human disease. Nucleic Acids Res. 2004;32:5430–5441. doi: 10.1093/nar/gkh884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morl M, Marchfelder A. The final cut. The importance of tRNA 3′-processing. EMBO Rep. 2001;2:17–20. doi: 10.1093/embo-reports/kve006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weiner AM. tRNA maturation: RNA polymerization without a nucleic acid template. Curr. Biol. 2004;14:R883–R885. doi: 10.1016/j.cub.2004.09.069. [DOI] [PubMed] [Google Scholar]
- 28.Helm M. Post-transcriptional nucleotide modification and alternative folding of RNA. Nucleic Acids Res. 2006;34:721–733. doi: 10.1093/nar/gkj471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geslain R, Ribas de Pouplana L. Regulation of RNA function by aminoacylation and editing? Trends Genet. 2004;20:604–610. doi: 10.1016/j.tig.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 30.Engelke DR, Hopper AK. Modified view of tRNA: stability amid sequence diversity. Mol. Cell. 2006;21:144–145. doi: 10.1016/j.molcel.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 31.Nakanishi K, Nureki O. Recent progress of structural biology of tRNA processing and modification. Mol. Cells. 2005;19:157–166. [PubMed] [Google Scholar]
- 32.Tavtigian SV, Simard J, Teng DH, Abtin V, Baumgard M, Beck A, Camp NJ, Carillo AR, Chen Y, et al. A candidate prostate cancer susceptibility gene at chromosome 17p. Nat. Genet. 2001;27:172–180. doi: 10.1038/84808. [DOI] [PubMed] [Google Scholar]
- 33.Schiffer S, Rosch S, Marchfelder A. Assigning a function to a conserved group of proteins: the tRNA 3′-processing enzymes. EMBO J. 2002;21:2769–2777. doi: 10.1093/emboj/21.11.2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takaku H, Minagawa A, Takagi M, Nashimoto M. A candidate prostate cancer susceptibility gene encodes tRNA 3′ processing endoribonuclease. Nucleic Acids Res. 2003;31:2272–2278. doi: 10.1093/nar/gkg337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan H, Zareen N, Levinger L. Naturally occurring mutations in human mitochondrial pre-tRNASer(UCN) can affect the transfer ribonuclease Z cleavage site, processing kinetics, and substrate secondary structure. J. Biol. Chem. 2006;281:3926–3935. doi: 10.1074/jbc.M509822200. [DOI] [PubMed] [Google Scholar]
- 36.Dubrovsky EB, Dubrovskaya VA, Levinger L, Schiffer S, Marchfelder A. Drosophila RNase Z processes mitochondrial and nuclear pre-tRNA 3′ ends in vivo. Nucleic Acids Res. 2004;32:255–262. doi: 10.1093/nar/gkh182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levinger L, Oestreich I, Florentz C, Morl M. A pathogenesis-associated mutation in human mitochondrial tRNALeu(UUR) leads to reduced 3′-end processing and CCA addition. J. Mol. Biol. 2004;337:535–544. doi: 10.1016/j.jmb.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 38.Levinger L, Giege R, Florentz C. Pathology-related substitutions in human mitochondrial tRNA(Ile) reduce precursor 3′ end processing efficiency in vitro. Nucleic Acids Res. 2003;31:1904–1912. doi: 10.1093/nar/gkg282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levinger L, Jacobs O, James M. In vitro 3′-end endonucleolytic processing defect in a human mitochondrial tRNA(Ser(UCN)) precursor with the U7445C substitution, which causes non-syndromic deafness. Nucleic Acids Res. 2001;29:4334–4340. doi: 10.1093/nar/29.21.4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nashimoto M. Anomalous RNA substrates for mammalian tRNA 3′ processing endoribonuclease. FEBS Lett. 2000;472:179–186. doi: 10.1016/s0014-5793(00)01462-9. [DOI] [PubMed] [Google Scholar]
- 41.Helm M, Brule H, Friede D, Giege R, Putz D, Florentz C. Search for characteristic structural features of mammalian mitochondrial tRNAs. RNA. 2000;6:1356–1379. doi: 10.1017/s1355838200001047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nashimoto M, Geary S, Tamura M, Kaspar R. RNA heptamers that direct RNA cleavage by mammalian tRNA 3′ processing endoribonuclease. Nucleic Acids Res. 1998;26:2565–2572. doi: 10.1093/nar/26.11.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tamura M, Nashimoto C, Miyake N, Daikuhara Y, Ochi K, Nashimoto M. Intracellular mRNA cleavage by 3′ tRNase under the direction of 2′-O-methyl RNA heptamers. Nucleic Acids Res. 2003;31:4354–4360. doi: 10.1093/nar/gkg641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Habu Y, Miyano-Kurosaki N, Kitano M, Endo Y, Yukita M, Ohira S, Takaku H, Nashimoto M. Inhibition of HIV-1 gene expression by retroviral vector-mediated small-guide RNAs that direct specific RNA cleavage by tRNase ZL. Nucleic Acids Res. 2005;33:235–243. doi: 10.1093/nar/gki164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yukita M, Kitano M, Miyano-Kurosaki N, Takeuchi H, Nashimoto M, Takaku H. RNA cleavage by a mammalian tRNA 3′processing endoribonuclease (3′tRNase) reduces HIV-1 expression. Nucleic Acids Res. Suppl. 2002;(Supp 2):297–298. doi: 10.1093/nass/2.1.297. [DOI] [PubMed] [Google Scholar]
- 46.Takaku H, Minagawa A, Takagi M, Nashimoto M. A novel 4-base-recognizing RNA cutter that can remove the single 3′ terminal nucleotides from RNA molecules. Nucleic Acids Res. 2004;32:e91. doi: 10.1093/nar/gnh092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakashima A, Takaku H, Shibata HS, Negishi Y, Takagi M, Tamura M, Nashimoto M. Gene silencing by the tRNA maturase tRNase Z(L) under the direction of small-guide RNA. Gene Ther. 2007;41:78–85. doi: 10.1038/sj.gt.3302841. [DOI] [PubMed] [Google Scholar]
- 48.Kawasaki H, Taira K. Short hairpin type of dsRNAs that are controlled by tRNA(Val) promoter significantly induce RNAi-mediated gene silencing in the cytoplasm of human cells. Nucleic Acids Res. 2003;31:700–707. doi: 10.1093/nar/gkg158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Watanabe T, Onuki R, Yamashita S, Taira K, Sugiyama Y. Construction of a functional transporter analysis system using MDR1 knockdown Caco-2 cells. Pharm Res. 2005;22:1287–1293. doi: 10.1007/s11095-005-5270-z. [DOI] [PubMed] [Google Scholar]
- 50.Kim VN, Nam JW. Genomics of microRNA. Trends Genet. 2006;22:165–173. doi: 10.1016/j.tig.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 51.Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, et al. Molecular basis for the recognition of primary microRNAs by the drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]