Abstract
A prominent feature of transcription in Giardia lamblia is the abundant production of sterile antisense transcripts (Elmendorf et al. The abundance of sterile transcripts in Giardia lamblia. Nucleic Acids., 29, 4674–4683). Here, we use a computational biology analysis of SAGE data to assess the abundance and distribution of sense and antisense messages in the parasite genome. Sterile antisense transcripts are produced at ∼50% of loci with detectable transcription, yet their abundance at a given locus does not correlate to the abundance of the complementary sense transcripts at that locus or to transcription levels at neighboring loci. These data suggest that sterile antisense transcripts are not simply a local effect of open chromatin structure. Using 5′RACE, we demonstrate that Giardia promoters are a source of antisense transcripts through bidirectional transcription, producing both downstream coding sense and upstream sterile antisense transcripts. We use a dual reporter system to explore roles of specific promoter elements in this bidirectional initiation of transcription and suggest that the degenerate AT-rich nature of TATA and Inr elements in Giardia permits them to function interchangeably. The phenomenon of bidirectional transcription in G. lamblia gives us insight into the interaction between transcriptional machinery and promoter elements, and may be the prominent source of the abundant antisense transcription in this parasite.
INTRODUCTION
A critical stage of gene expression is the assembly of transcriptional machinery in the correct orientation at promoters, such that transcription proceeds in a single direction. Directional transcription is ensured by proper interaction between the core promoter, general transcription factors (TFs) and RNA polymerase II to form the pre-initiation complex (PIC). Components involved in the process are largely conserved (though often differently named) between eukaryotes and archaea. Transcription initiation begins with the recognition of the TATA box (Box A in archaea) by the TATA-binding protein (TBP), a component of TFIID (TFD in archaea). However, the ability of TBP to interact with the TATA box in both orientation (1–3), due to the 2-fold symmetry of their interaction (4–9), raises an important problem for the polar orientation of the PIC (10). It has been determined that the slight asymmetry of the TBP–TATA complex could only minimally account for the correct orientation of transcriptional machinery, as in solution TBP has only 60:40 preference toward binding the TATA box in the appropriate orientation (3). On the other hand, a TFIIB (TFB)-recognition-element (BRE), found immediately upstream of the TATA box in archaea (11) and eukaryotes (12), is specifically recognized by TFIIB (TFB), in a highly asymmetric fashion (12,13). This interaction is thought to ensure the correct assembly of the PIC and consequently, unidirectional transcription.
Giardia lamblia is a binucleated parasitic protozoan that is one of the most common intestinal pathogens of humans and animals worldwide and a significant cause of diarrheal disease. The parasite's haploid genome is ∼12 Mb (14) and is exceedingly tightly organized, as demonstrated by the presence of only a few introns (15,16), and extremely short intergenic regions (17) and 5′ and 3′ untranslated regions (UTRs) [reviewed in (18)]. Additionally G. lamblia has short and simple core promoters (17,19–22). Two AT-rich regions appear to be crucial for transcription initiation: one at the transcription start site (Initiator region (Inr)-like element) and the other ∼30 bp upstream (TATA-like element) [reviewed in (18,23)]. One study has identified an additional element resembling the CAAT box, ∼50 bp upstream (22). Importantly, the Inr and TATA are not highly conserved with respect to their sequence, length or the exact position relative to the transcription start site (19–22). Rather, it appears that the overall AT richness in the most proximal ∼50-bp region of the promoter is crucial for the recognition by the parasite's transcriptional machinery and determination of transcriptional efficiency. Upstream or downstream distal regulatory elements, activators or repressors have not been reported in G. lamblia, although specific regulatory elements have been reported for developmentally regulated genes (17,24).
The distinctive nature of G. lamblia genome may have direct consequences on gene expression regulation in the parasite. One of the most unusual features of transcription in G. lamblia is the abundance of sterile antisense transcripts (over 20% of total polyadenylated RNA) that do not have a usable open reading frame (ORF) and, hence, cannot code for a protein (25). These messages have been documented at developmentally regulated, as well as constitutively expressed genes (17,25,26), but it remains unclear whether they have regulatory functions in controlling gene expression and/or are consequences of a loosely regulated transcriptional process.
A genome-wide comparative analysis of G. lamblia transcription initiation machinery further serves to raise important questions regarding the control of gene expression (27). While a fairly typical set of eukaryotic RNA polymerase II subunits is present in the parasite (28), the absence of a significant portion of the general eukaryotic TFs has been reported (27). Giardia apparently has TBP (although the sequence is unexpectedly divergent from both archaeal and eukaryotic TBPs), Rrn3 (RNA polymerase I TF) and TFIIH components. Interestingly, a single protein with similarity to both TFIIB and TFIIIB domains was identified (27) (S.T. and H.G.E., unpublished data). It remains unclear whether this single protein is serving a dual role for both RNAII and RNAIII polymerases or whether one of the two TFs (II or III) is absent in Giardia. Given that the interaction of TFIIB (TFB) with the BRE is thought to play a crucial role in ensuring that transcription proceeds in the correct orientation, the uncertainty surrounding the activity of this TF in G. lamblia calls into question parasite's ability to achieve transcriptional directionality.
There is precedence for a single short intergenic region acting to direct bidirectional transcription in G. lamblia. Work by Tai and colleagues has shown that divergent transcription of G. lamblia's GTP-binding Ran gene (ran) and pHD zinc-finger protein gene (phd) occurs from a 153-bp intergenic region between these head-to-head arranged genes (29). This pattern of transcription is not unique to G. lamblia, and shared bidirectional regulatory units between head-to-head-oriented genes have also been shown to exist in yeast (30) and mammals (31–33).
The significance of the new research presented here is in our demonstration that bidirectional transcription is not limited to head-to-head gene arrangements, but is rather an inherent feature of G. lamblia promoters. Because of the exceptionally tight packing of the G. lamblia genome, bidirectional transcription produces not only the appropriate downstream sense transcript, but leads to the production of either an upstream sense transcript (for promoters between genes in a head-to-head arrangement) or an upstream sterile antisense transcript (for promoters between genes in a head-to-tail arrangement). Bidirectional transcription from promoters across the genome could therefore be a leading contributor to the abundance of sterile antisense transcripts observed in G. lamblia.
MATERIALS AND METHODS
Assessment of sense and antisense transcript ratios at individual ORFs within the genome
Files of all ORFs and their coordinates with respect to the assembled contigs, as well as trophozoite serial analysis of gene expression (SAGE) tags with respect to the ORFs were downloaded from the G. lamblia genome project database (www.mbl.edu/Giardia). The Giardia SAGE database is annotated to be able to determine a primary assignment for each tag. An algorithm was created to generate a list of all ORFs and their forward (primary and alternate sense) and reverse (primary and alternate antisense) SAGE tags. We chose to limit our analysis of ORFs to those between 200 and 10 000 bp to improve the likelihood of ORF validity; this decreased the sample size from 9646 to 7313 ORFs. Each ORF was first matched to SAGE tags corresponding to the sense transcript from that ORF (including both primary and alternative sense tags), and total sense tag frequency was tabulated for each ORF. Similarly, each ORF was matched to SAGE tags corresponding to any antisense transcripts from that ORF (again including primary and alternative antisense tags), and total antisense tag frequency was tabulated for each ORF. In SAGE libraries, tag frequency reflects the abundance of the transcripts in the initial mRNA population. The ratio of all sense tags to all antisense tags was then calculated for each ORF.
Pearson's correlation coefficient (r) was calculated to test the significance level of correlation between sense and antisense SAGE tags at individual ORFs. Pearson's correlation coefficient (r) values can range from −1 (indicating absolute negative association) to 1 (indicating absolute positive relationship), where under null hypothesis (r = 0) no relationship exists between the two characteristics.
Assessment of antisense and sense transcript ratios at neighboring ORFs within the genome
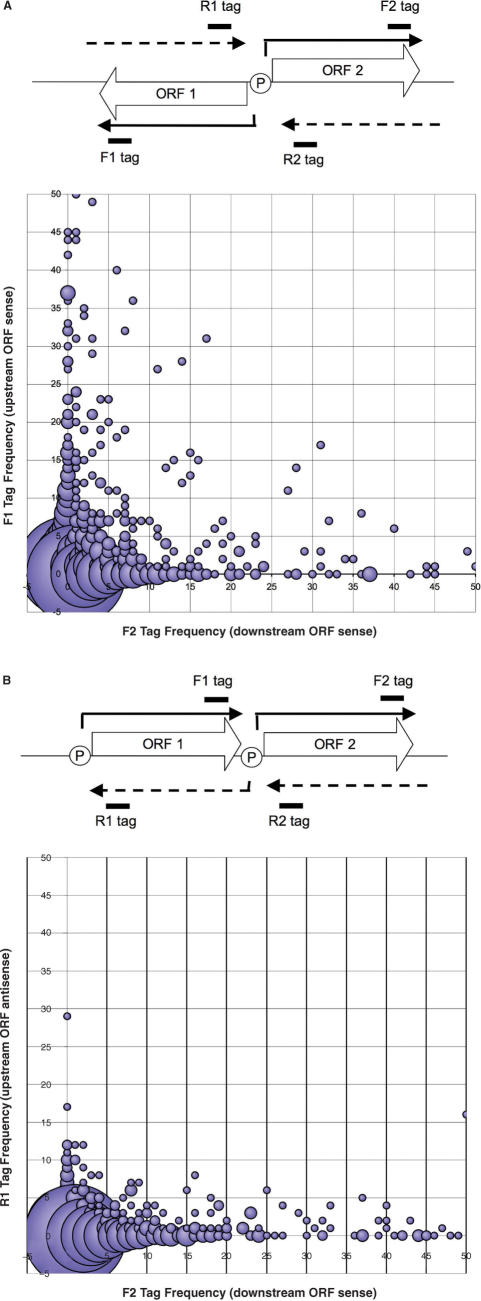
A computer algorithm mentioned in the previous section was expanded to detect neighboring ORFs and their SAGE tag frequencies (a detailed explanation of the protocol can be found in the Supplementary Data). The protocol described above was initially performed for all ORFs to correctly assign SAGE tags to each ORF. Next, ORFs with proximal neighbors were identified: two ORFs were considered neighbors if the distance between them was ≤250 bp, further decreasing the number of considered ORFs from 7313 to 4458. Two groups of ORF pairs were identified (Figure 2A and B): (1) Both adjacent ORFs are in the same orientation on the contig (head-to-tail); (2) An ORF and its upstream ORF are in the opposite orientations on the contig (head-to-head). Ratios of sense and antisense tags from neighboring ORFs were compared in appropriate combinations as described in the text.
Figure 2.
Examining the role of chromatin structure in antisense transcription. The total pool of ORFs used in Figure 1 was restricted to investigate only the adjacent ORFs in the genome; ORFs were considered neighbors if the distance between them was ≤250 bp. Two categories of neighboring ORFs were identified: head-to-head ORFs that are oriented in opposite directions on the contig [divergently transcribed ORFs, as shown on the top panel of (A)], and head-to-tail ORFs that are oriented in the same direction on the contig (B, top panel); SAGE tags indicative of divergent transcription on the chromosomal segment were compared: (A) For head-to-head-oriented neighboring genes, frequency of forward (sense) tags of both genes was evaluated. The graph represents the relationship between the frequency of forward (sense) tags of ORF2 (F2 tag) and forward (sense) tags of ORF1 (F1 tag). (B) For head-to-tail-oriented neighboring genes, frequency of forward (sense) tags of one gene and reverse (antisense) tags of another was evaluated. The graph represents the relationship between the frequency of forward (sense) tags of ORF2 (F2 tag) and reverse (antisense) tags of ORF1 (R1 tag).
Mapping transcriptional start sites
Transcriptional start sites for sense and antisense transcripts from ORF 40817 (actin), ORF 112079 (alpha-2 tubulin), ORF 14369 (hypothetical protein) ORF 10868 (hypothetical protein) and an intergenic region immediately upstream of the alpha-2 tubulin gene were determined using Rapid Amplification of cDNA Ends (5′RACE). Similarly, firefly and Renilla luciferase transcription start sites were ascertained from trophozoites stably transfected with each of the five bidirectional dual reporter plasmids described below.
The 5′RACE protocol instructions from the manufacturer (5′ RACE System for Rapid Amplification of cDNA Ends, Invitrogen) were followed. Briefly, 3 µg of total trophozoite RNA (RNA-STAT, Tel-Test) was converted into cDNA using reverse transcriptase and gene-specific oligonucleotides (Table S2) and dC-tailed with terminal deoxytransferase. PCR products (both primary and secondary) were generated from the cDNA template in the following reaction mix: 20 mM Tris-HCl, 50 mM KCl, 1.5 mM MgCl2, 200 µM each dNTP, 2.5 U Taq DNA polymerase, and either 400 nM oligonucleotide primers (for the primary PCR) or 200 nM nested oligonucleotide primers (for the secondary PCR) (Table S2). Cycling conditions for amplifications were as follows: 94°C for 2 min; 35 cycles of 94°C for 30 s, 55°C for 30 s, 72°C for 1 min; 72°C for 5 min. Nested PCR products were cloned into a TOPO-TA pCR 4 vector (TOPO TA Cloning Kit for Sequencing, Invitrogen). Ligated products were transfected into TOP10 E. coli chemically competent cells and grown on LB agar containing ampicillin. DNA was extracted from picked clones (Wizard Plus Minipreps DNA Purification System, Promega) and used as a template in the sequencing reactions, along with either M13 forward or M13 reverse oligonucleotides (Invitrogen). Sequencing was preformed in a 3100 Genetic Analyzer (Applied Biosystems). Transcription start sites were determined based on chromatograms (Sequencher 4.1.1, Gene Codes).
Bidirectional plasmid constructs
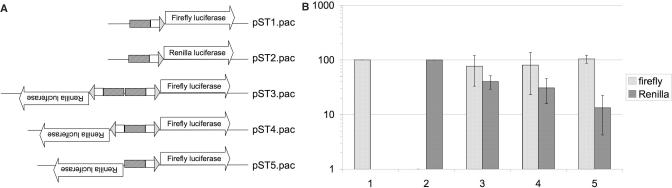
We generated five plasmids to enable us to test the bidirectionality of the alpha-2 tubulin core promoter in stably transfected G. lamblia cell lines (Figure 4A and Table S3). (1) pST1pac: The puromycin resistance cassette (pac) was inserted into a previously described plasmid pTubLucFF (22); this plasmid used the alpha-2 tubulin core promoter to drive expression of firefly luciferase and is selected for and maintained in stable transfectants using puromycin. (2) pST2pac: The firefly luciferase gene in the pST1 plasmid was replaced by the Renilla luciferase gene. (3) pST3pac: A promoter unit, consisting of two complete alpha-2 tubulin core promoters oriented adjacently and facing in opposite directions, was positioned between the divergently (head-to-head) oriented Renilla (upstream) and firefly (downstream) luciferase genes. (4) pST4pac: A promoter unit consisting of a second Inr positioned in the opposite orientation adjacent and upstream of the core alpha-2 tubulin promoter was positioned between the divergently oriented (head-to-head) Renilla (upstream) and firefly (downstream) luciferase genes (5) pST5pac: A promoter unit containing only the single core alpha-2 tubulin promoter was positioned between the divergently oriented (head-to-head) Renilla (upstream) and firefly (downstream) luciferase genes.
Figure 4.
Examination of the role of promoter elements in bidirectional transcription initiation. (A) Construction of dual reporter plasmids utilizing a shared promoter region in Giardia. Bidirectional plasmid constructs were generated to create a dual reporter system that permits us to quantify bidirectional transcription and to more closely examine the roles of specific elements within Giardia promoters. We generated several reporter constructs that contain firefly and Renilla luciferase reporter genes in a head-to-head arrangement, separated by different promoter regulatory units of the alpha-2 tubulin promoter. (B) Quantitative measurements of bidirectional promoter strength for elements of the alpha-2 tubulin promoter. Luciferase assays were conducted in triplicate on parasites stably transfected with the dual reporter constructs. Firefly and Renilla luciferase activities were established at 100% in the control (S1 and S2) plasmids, and other measurements were calculated relative to these levels.
pST1pac
A puromycin resistance gene was inserted into a previously described plasmid pTubLucFF (22).
pST2pac
The Renilla luciferase reporter gene was amplified from pRL-SV40 (Promega) using primer pair ST1F and ST1R, and the resulting amplicon was digested with SacI/NsiI, and ligated into a SacI/NsiI-digested pTubLucFF vector to generate pST2. The Renilla luciferase gene was PCR amplified with ST2F and ST2R from pST2 plasmid, digested with NsiI/BamHI, and inserted into NsiI/BamHI-digested pST1pac vector to produce pST2pac.
pST3pac
The Renilla luciferase reporter gene and alpha-2 tubulin promoter were amplified from pST2 using primer pair ST3F and ST3R, and the resulting amplicon was digested with BsaBI/HindIII, and ligated into a BsaBI/HindIII-digested pTubLucFF vector to create pST3. The pac cassette was excised from pST1pac using NdeI/EcoRV and ligated into an NdeI/EcoRV-digested pST3 to produce pST3pac.
pST4pac
The Renilla luciferase reporter gene and Inr element of the alpha-2 tubulin promoter from pST2 were excised with BsaBI/HindIII and the insert was ligated to a BsaBI/HindIII-digested pTubLucFF, creating pST4. The pac cassette was then inserted as described above, to produce pST4pac.
pST5pac
The Renilla luciferase reporter gene was amplified from pST2 using the primer pair ST4F and ST4R, and the resulting amplicon was digested with BsaBI/HindIII, and ligated into a BsaBI/HindIII-digested pST1pac, creating pST5pac.
Cell cultures
Giardia lamblia trophozoites, isolate WB 1267, were maintained anaerobically in borosilicate tubes or polystyrene flasks. Cultures were grown in a modified TYI-S-33 medium at 37°C (34), with a replacement of the traditional phosphate buffer solution with 0.024 M sodium bicarbonate.
Stable transfections and luciferase assays
Giardia lamblia trophozoites were stably transfected by electroporation, as described previously (35,36), using 20 µg of plasmid DNA (Wizard Maxiprep Kit, Promega). Episomes were maintained in stably transfected trophozoites by the addition of 10 mM puromycin to the culture medium. Transfected cultures were iced for 30 min, the parasites were counted in a hemacytometer, and pelleted by centrifugation at 1200 × g for 7 min. Pellets were washed once in phosphate buffered saline (PBS) and lysed in 40 µl of 1× reporter lysis buffer (Luciferase Assay Kit, Promega) supplemented with 5 μg/ml leupeptin. Cells were placed at −80°C for a minimum of 30 min, ensuring successful cell lysis. Lysates were briefly thawed on ice and centrifuged at 10 000 × g for 5 min to pellet cell debris. Here, 20 µl of supernatant was added to 100 µl of firefly luciferase substrate (Luciferase Assay Kit, Promega), while the remaining 20 µl of supernatant was added to 100 µl of Renilla luciferase assay reagent (Renilla Luciferase Assay System, Promega), and assayed for firefly and Renilla luciferase activities in a luminometer (Turner TD-20, Promega). Luminometer readings were normalized for the number of cells assayed. Baseline firefly and Renilla luciferase activities were set to a 100% and all the subsequent readings are displayed as relative luciferase values. Six independent experiments were performed, and results are reported as mean ± S.D.
RESULTS
Ubiquitous expression of antisense sterile transcripts across the G. lamblia genome
We began our studies by assessing levels of sense and antisense transcription at individual loci throughout the Giardia genome to document their abundance and distribution. We compared the levels of sense and antisense transcripts at each ORF in the genome, using the tag frequency detected by SAGE as a measure of gene expression (Figure 1). We eliminated from consideration ORFs without any SAGE tags, which limited the analyzed dataset to 3379 ORFs (out of 7313 ORFs), represented by at least one SAGE tag (for either sense or antisense transcripts). It is worth noting that an absence of tags does not necessarily signify that the ORF is not transcribed, but simply that levels of transcription or transcript stability are sufficiently low such that no transcripts were incorporated into the SAGE dataset.
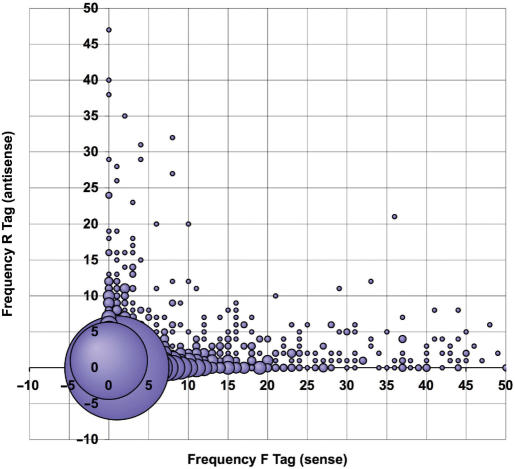
Figure 1.
The abundance of antisense transcripts in Giardia. To assess the abundance and distribution of sense and antisense messages throughout the Giardia genome, all open reading frames (ORFs) and their location with respect to the assembled contigs were downloaded from the genome project database (www.mbl.edu/Giardia), along with trophozoite SAGE tags with respect to the ORFs. A computer algorithm was created to generate a list of all ORFs and the frequency of their forward (sense) and reverse (antisense) SAGE tags for both primary and alternative transcripts. For each ORF, primary and alternative SAGE tags were added, yielding a total forward (sense) and reverse (antisense) tag frequency. The coordinate of each ORF on the graph is depicted by its forward (sense) tag frequency on the x-axis and its reverse (antisense) tag frequency on the y-axis. The size of the bubble at each coordinate is reflective of the number of ORFs with a given forward (sense):reverse (antisense) tag ratio.
Taking into consideration the frequency of sense and antisense tags, out of a total of 29 421 tags, 23 895 were sense and 5526 were antisense tags. Thus, we find that ∼19% of all messages produced in the genome are antisense, which corroborates the previous estimates (25). Here, 2691 ORFs had one or more sense tags, while 1760 ORFs had one or more antisense tags, indicating that antisense transcripts are synthesized at over half of the loci in the genome. Further examination of patterns of gene expression revealed that 1618 ORFs had only sense tags detected, 687 ORFs had only antisense tags detected and 1073 ORFs had both sense and antisense tags detected. We identified all ORFs that had 15 or more antisense SAGE tags and noted that they represent a wide spectrum of gene families, although ORFs that have exceptionally high ratios of antisense to sense tags are disproportionately annotated as hypothetical proteins (Table S1).
A role for antisense messages in down-regulating their sense counterparts would be depicted by a best-fitting regression line with a negative slope in Figure 1, which would represent an inverse relationship between sense and corresponding antisense transcripts. On the contrary, we observe a distribution of antisense tags from numerous loci throughout the genome without an apparent pattern and a lack of correlation between antisense transcripts and their sense counterparts globally in the genome (correlation coefficient r = 0.09).
A genome-wide analysis fails to provide evidence for the production of antisense transcripts as a consequence solely of local euchromatin organization
The prevalence of antisense transcripts at such a diverse array of loci led us to investigate their origins. The tight gene packing in G. lamblia initially suggested that patterns of transcription at neighboring loci might be related because of sequence availability in the euchromatin—that is, the local opening of the chromosome for transcription at one locus would also render the neighboring locus available for recruitment of transcriptional machinery. We therefore explored whether transcription from one locus had any relationship to transcription at an adjacent locus by comparing levels of gene expression, predicted by SAGE, for sense and antisense transcripts between adjacent ORFs (Figure 2A and B). Given that G. lamblia promoters are typically extremely short (∼60nt), two ORFs were considered to be adjacent only when the distance between them was ≤250 bp. Additionally, we considered head-to-head- and head-to-tail-oriented genes separately, since promoters between genes arranged in a head-to-head orientation could be purposefully bidirectional in order to drive transcription of sense transcripts for the two divergently arrayed ORFs [as in the ran/phd example previously described (29)], while promoters situated between genes arranged in a head-to-tail orientation should, in theory, be unidirectional to drive transcription of sense transcripts for the downstream ORF only.
For each pair of head-to-head-oriented genes, we calculated the ratio of sense tag frequencies between the downstream and upstream ORF, while for each pair of head-to-tail-oriented genes, we calculated the ratio of sense tag frequencies for the downstream ORF and the antisense tag frequencies for the upstream ORF (Figure 2A and B). For both datasets, instances where the frequencies of both sense tags for a pair of ORFs were zero (i.e. there was no evidence in the SAGE database for expression of either ORF) were eliminated from the analysis. In both graphs, there is a widely scattered distribution of tag ratios, indicating a lack of relationship between the expression levels of the two ORFs (Figure 2A and B); in fact, the same absence of correlation in tag ratios held true regardless of which pair of tags (sense, antisense, or sense and antisense) was being compared for any two neighboring ORFs (data not shown). There are also numerous instances in which the expression of antisense tags is higher, relative to sense tags, as demonstrated by the presence of numerous data points above the equal ratio line imposed on the graph. The most notable difference between Figure 2A and B, is the bias towards higher expression of sense tags relative to antisense tags seen in the head-to-tail graph (Figure 2B), indicating that promoters are typically composed to favor transcription into coding regions of DNA. Overall, these data indicate that there is little to no relationship between transcription at neighboring loci, unless the two ORFs are in a head-to-head arrangement and share a common promoter region.
Giardia lamblia promoters act bidirectionally to produce both sterile antisense and sense transcripts
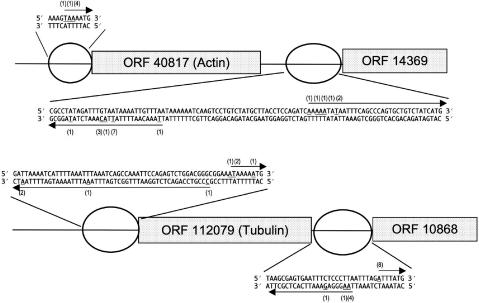
We next questioned whether antisense transcripts might be a result of bidirectional transcription occurring at promoters in Giardia. We conducted a detailed investigation of two pairs of adjacent head-to-tail-oriented loci, using 5′RACE to map their transcription start sites: (1) ORF 40817 (actin) and ORF 14369 (hypothetical protein) (Figure 3A) and (2) ORF 112079 (alpha-2 tubulin) and ORF 10868 (hypothetical protein) (Figure 3B). Intergenic regions for these two ORF pairs were 177 and 30 bp, respectively, representing a typical range of intergenic spacing in Giardia.
Figure 3.
Evidence for three bidirectional promoters. Transcriptional start sites for sense and antisense transcripts originating from three promoter regions (regions upstream of ORFs 14369, 112079 and 10868) were determined by 5′RACE and depicted by a schematic here. Additionally, sense transcriptional start sites originating from the fourth promoter (region upstream of ORF 40817) are shown. Actual sequences for promoter domains in intergenic regions are given. Transcripts originating from these regions are indicated by arrows; for all ORFs, arrows pointing right indicate sense transcripts and arrows pointing left indicate sterile/antisense transcripts. Exact nucleotide positions representing transcription start sites are underlined. Relative usage of each nucleotide is indicated in parentheses above and below the underlined nucleotide. Top panel: ORF 40817 (actin) and ORF 14369 (hypothetical protein). Bottom panel: ORF 112079 (alpha-2 tubulin), ORF 10868 (hypothetical protein), and an intergenic region without an ORF immediately upstream of the alpha-2 tubulin gene.
We first analyzed the promoters and mapped transcription start sites for the downstream ORFs in these two pairs of loci. Within ∼90 bp upstream of the start codon for ORF 14369, we identified two AT-rich regions. ORF 14369 sense transcripts originated from the AT-rich region proximal to the start codon (analogous to the Inr element) (Figure 3A), producing a 5′ UTR of ∼25 bp. The intergenic region upstream of ORF 10868 likewise has two AT-rich regions, with transcription for ORF 10868 sense transcripts again initiating in the AT-rich region closest to the start codon, creating an ∼3 bp 5′ UTR (Figure 3B). The size and placement of the AT-rich sequences resemble the previously defined elements of core promoters in G. lamblia, and the mapping of transcription start sites to nucleotides within the more proximal of the two AT-rich elements again corresponds with previous studies (17,19–22).
Strikingly, both of these promoters also gave rise to sterile antisense transcripts that were complementary to coding sequences of the two upstream ORFs (ORF 40817 and ORF 112079) (Figure 3A and B). These sterile antisense messages originated within the more distal (TATA) element of the promoter regions (as defined by their placement with respect to the downstream ORFs). Thus, both core promoters initiated bidirectional transcription with an apparent reversal of TATA and Inr functionality between the two directions. We further investigated the roles of the promoter elements in the bidirectional promoters through dual reporter constructs described below.
Not all loci in Giardia are closely spaced, given that longer intergenic regions exist in the parasite's genome, and we were curious to determine whether promoters were bidirectional in these instances as well. We therefore investigated the directionality of transcription at a third promoter (the alpha-2 tubulin promoter upstream of ORF 112079), for which the closest upstream ORF (ORF 16920—annotated as a WD-repeat protein) was in a head-to-head arrangement at ∼308 bp upstream of ORF 112079 (Figure 3B). We mapped sense transcripts for the alpha-2 tubulin locus (ORF 112079) originating at sites previously determined to be the dominant transcription start sites for this gene (21), and we also detected transcripts divergently transcribed in the upstream direction from this promoter (Figure 3B). Given the recent work that extremely short 5′UTRs were not only common in the parasite but also essential for efficient translation (37), any upstream transcripts originating in the alpha-2 tubulin promoter would give rise to non-translatable transcripts; the presence of a canonical polyadenylation signal only 163 bp into the upstream ORF (ORF 16920) further confirmed that such transcripts would be sterile.
Thus, the analysis of transcription initiation sites from promoters of housekeeping genes shows that G. lamblia promoters are bidirectional, directing transcription both downstream, as well as upstream, irrespective of the surrounding sequences. The phenomenon of bidirectional transcription is thus an inherent feature of the promoters in the parasite and is unrelated to transcript function.
Lastly, we note that for both the downstream (sense) and the upstream (antisense) transcripts, multiple start sites were discovered within any AT-rich region (Figure 3A and B), as previously reported at numerous loci (17,25). Additional sterile transcripts also originated from AT-rich regions within actin and tubulin genes (data not shown). This was not surprising to us, as previous work (25) and unpublished results from our laboratory (C.D.W. and H.G.E., unpublished data) have consistently detected transcription to initiate from loosely defined AT-rich patches, ‘cryptic promoters’, throughout the genome.
Defining the functionality of sequences within bidirectional promoters
We constructed a dual reporter system to permit us to quantify bidirectional transcription and to more closely examine the roles of specific elements within G. lamblia promoters. We generated several reporter constructs that contain firefly and Renilla luciferase reporter genes in a head-to-head arrangement, separated by different promoter regulatory units of the alpha-2 tubulin promoter (Figure 4A). Previous studies have shown that the G. lamblia alpha-2 tubulin promoter is a very strong promoter, and ∼60 bp of sequence upstream of the translation start site, consisting of a TATA-box-like element (TATA) and an Inr separated by ∼27 bp, was determined to function as efficiently as a full (∼350 bp of upstream sequence) promoter (21); we hereafter refer to this 60 bp segment as the ‘complete promoter’.
The complete alpha-2 tubulin promoter was used to drive the transcription of either firefly (pST1.pac) or Renilla (pST2.pac) luciferase reporter genes (Figure 4A) to determine baseline firefly and Renilla luciferase activities within this system. Firefly and Renilla luciferase readouts from pST1.pac and pST2.pac, respectively, were defined as 100% transcription (Figure 4B). The demands of transcription for two head-to-head arrayed genes on a small plasmid—i.e. the supercoiling induced by opening the DNA at each locus—posed an additional concern. Therefore, as an additional control, we constructed a plasmid (pST3.pac) in which two complete alpha-2 tubulin promoters were oriented in opposite directions, driving expression of firefly and Renilla luciferase genes (Figure 4B). Although firefly luciferase was expressed at ∼80% of the baseline value (pST1.pac), Renilla luciferase values were at only ∼40% of pST2.pac (Figure 4B). This finding confirmed our concerns about the possible steric hindrances to transcription, and we therefore refer to transcriptional levels from the subsequent test plasmids relative to transcriptional levels observed in pST3.pac.
To investigate the ability of various promoter elements to initiate bidirectional transcription, a full promoter was used to direct the expression of the downstream firefly luciferase gene, while the Renilla luciferase gene was positioned immediately upstream and in the opposite orientation (pST5.pac) (Figure 4A). Both firefly and Renilla luciferase were expressed from this construct, with reduction of Renilla luciferase expression levels to 33%, compared to pST3.pac (Figure 4B).
To further determine the contribution of different elements of the complete promoter to its bidirectionality, an inverted Inr element was cloned upstream of the complete promoter in pST5.pac to make pST4.pac (Figure 4A). Again, both firefly and Renilla luciferase were expressed from this construct, this time with only a slight decrease in Renilla activity compared to the baseline values (Figure 4B). Taken together, these results further demonstrate that the alpha-2 tubulin promoter in G. lamblia is capable of driving transcription bidirectionally. It remains unclear, however, whether the increased transcriptional efficiency of pST4.pac relative to pST5.pac is due specifically to the presence of the additional Inr element (suggesting that the TATA box is functioning bidirectionally) or whether it results from a mere increase in AT-richness in the promoter region (previously indicated to be the determining factor in promoter strength (21)).
DISCUSSION
Antisense transcripts were first documented in G. lamblia for the developmentally regulated gene Gln6PI-B (17), and subsequently for several other developmentally regulated and constitutively expressed genes (25,26,38). Previous research from random sampling of cDNA libraries indicated that antisense transcripts represent a surprisingly high percentage (∼20%) of total G. lamblia mRNA pool in the cell (25), but it has been less clear what the origins, fate and potential function of these messages might be. We present research here that sheds light on both the genome-wide abundance and distribution of antisense transcripts and on the unusual properties of promoters in G. lamblia that contribute to their generation.
Patterns of antisense transcription argue against a regulatory role for antisense transcripts in gene regulation in G. lamblia
Perhaps the most favored hypothesis in the literature has been that antisense transcripts play a role in RNAi or a related antisense-mediated gene regulation mechanism (17,26,38). Work by von Allmen et al., on the contrary, showed parallel, and not reciprocal, expression of variant surface-specific protein H7 (vsp H7) and cyst wall protein 1 (cwp1) sense and antisense transcripts in G. lamblia GS isolate (38), arguing against the use of antisense RNA for locus-specific regulation of gene expression. The data we present in this article further argue against a regulatory role for the antisense transcripts. We present evidence from SAGE demonstrating that antisense messages are abundantly produced throughout the genome, at low levels and from many different loci (Figures 1 and 2).
Local chromatin unwinding is likely not responsible for the generation of antisense transcripts
As an alternative hypothesis, von Allmen and colleagues speculated that antisense transcripts are simply a product of DNA unwinding during transcription (38). If unwinding of the chromatin at a chromosomal segment itself were sufficient to cause the antisense RNA production, one would expect that transcription levels from the adjacent genes should correlate with each other. However, our comparisons revealed a lack of correlation between the abundance of sense or antisense transcripts from proximal neighboring ORFs (Figure 2 and data not shown).
Although we propose that antisense transcripts might simply result from unusual loose regulation of transcription in Giardia, we recognize that the abundant antisense RNA production requires significant energy expenditure, as cautioned by von Allmen and colleagues. However, this does not necessarily imply possible biological significance of the process. It is worth remembering that tight and precise regulation of transcription, as exists in higher eukaryotes, is also an energy-demanding process—and the abundant transcription of long 5′ and 3′ UTRs and introns in higher eukaryotes also requires energy yet does not result in translated product for those non-coding sequences.
Bidirectional promoters as a source of antisense transcripts
Here, we demonstrate that G. lamblia promoters are inherently bidirectional, and that this characteristic makes them an important source of antisense sterile transcripts in the parasite (Figures 3 and 4). Additionally, we show generation of upstream and divergently oriented transcripts from the alpha-2 tubulin promoter (Figure 4B), indicating that bidirectional promoters might be significant contributors to not only antisense, but also sterile transcripts in the cell in non-coding regions. Additional source of sterile transcripts are ‘cryptic’ promoters, simple and degenerate AT-rich regions, (transcripts originating within actin and tubulin-coding regions—Figure 3A and B) and by other studies (25). Thus, it is not the unwinding of the DNA per se, but rather the bidirectionality of G. lamblia promoters, as well as promiscuous transcription from loosely defined AT-rich patches, that causes the abundance of antisense transcripts in the cell. Taken together, our findings point out that the vast majority of G. lamblia's genome is, in fact, routinely transcribed in trophozoites regardless of coding potential, given that we show that in addition to correct sense transcripts, sterile, antisense and transcripts spanning intergenic regions are continuously generated throughout the genome.
Identifying the role of promoter elements in directionality of transcription
In order to determine whether specific promoter elements are responsible for bidirectionality, we generated the first dual reporter system in Giardia (Figure 4). It is noteworthy that Renilla luciferase overall proved to be poorly expressed and is, thus, not an ideal reporter gene for studies of gene expression regulation in Giardia. The most likely explanation of this observation is that majority of transcripts mapped by 5′RACE in the baseline pST2.pac vector originated from two AT-rich regions within the Renilla gene itself (data not shown). Comparable expression levels of Renilla luciferase in pST3.pac and pST4.pac constructs (Figure 4B), suggested the involvement of the oppositely oriented, upstream TATA box in orchestrating Renilla transcription. Further, even in the complete absence of a correctly oriented promoter (as in pST5.pac) (Figure 4A), Renilla reporter gene was expressed from a bidirectional TATA (and potentially bidirectional Inr) (Figure 4B). However, a reduction in Renilla activity, in the absence of ‘correctly’ positioned Inr (pST4.pac versus pST5.pac) (Figure 4B), indicated expression from a weaker promoter, therefore supporting earlier studies (21) that additional AT-rich sequences play a notable role in determining the strength of the promoter in G. lamblia.
Thus, while our work clearly shows the ability of the TATA box to act bidirectionally, it is less clear whether this is a TATA-box-specific feature. For example, we cannot exclude the possibility that Inr also acts bidirectionally, as we did not directly ascertain the contribution of the Inr, placed in front of the firefly luciferase gene, to the expression of Renilla luciferase (Figure 4). Additionally, as transcription initiates from ‘cryptic’ promoters throughout the genome (25), it is quite possible that these degenerate AT-rich patches also have the capacity to orchestrate transcription bidirectionally, further contributing to the excessive pool of sterile and antisense mRNAs in the cell. Future work is necessary to elucidate these questions.
(Bi)directionality of transcription process in Giardia
Given that TBP has the ability to recognize and bind the TATA box in both orientations (1–9), the TBP–TATA complex alone cannot ensure the correct orientation of transcriptional machinery in eukaryotes and archaea. Specificity of this interaction might be even less defined in G. lamblia, since three of the four residues essential for TBP binding in other organisms have been substituted in the parasite (27). This is perhaps not surprising since G. lamblia's TATA-box-like elements are far more degenerate, and thus, TBP in G. lamblia must have the potential to bind an array of AT-rich sequences. Bidirectional transcription, prevented in other organisms by the formation of TFIIB (TFB in archaea)–BRE complex, might simply be a consequence of the absence of such a complex in G. lamblia. In this scenario, TBP could bind TATA box (or any AT-rich element) in either orientation, followed by an organization of the transcriptional machinery in both directions. Yet, directionality of transcription in Giardia is not wholly lacking, as documented by the following: (1) At most neighboring ORFs, the downstream sense transcript is produced more abundantly than the upstream antisense transcript (Figure 2); (2) Quantification of transcription from the alpha-2 tubulin promoter argues for preferential sense transcription (Figure 3); (3) The fact that a promoter functions bidirectionally does not seem to affect its strength in the forward direction (pST5.pac versus pST3.pac and pST5.pac versus pST1.pac). What enables this directionality?
Best et al. have shown the existence of a single protein with TFB similarity, in G. lamblia using bioinformatics approaches (27), although its expression and function have never been put to test. Given the common ancestral transcription machinery between eukaryotes and archaea (13), perhaps this single protein functions both as TFIIB and TFIIIB in G. lamblia. If so, it is interesting to speculate what its recognition sequence would be. BRE is only modestly conserved, and its sequence might be completely divergent in G. lamblia, explaining its apparent absence. However, visual inspection of several housekeeping genes failed to find any conserved promoter element, directly upstream of TATA, or anywhere else in the promoter region (data not shown), although a detailed, genome-wide search for such element is necessary. Interestingly, BRE is absent from yeast and plants [reviewed in (39)], implying the existence of BRE/TFIIB-independent ways of establishing transcriptional directionality. It has been hypothesized that even though TATA–TBP complex formation alone is not sufficient for directional PIC organization, cumulative effects of the PIC components positioning might contribute to proper unidirectional transcription (3).
Giardia lamblia's TATA-box-like and Inr-like elements are far more degenerate compared to other eukaryotes and archaea, and are mostly defined by AT-richness and their position relative to the transcription start site (21). Given the importance of AT-richness for initiating transcription in the parasite, one of the possibilities is that TATA-box-like element and Inr-like element are interchangeably used in G. lamblia's bidirectional promoters. In such an instance, bidirectionality is a feature of loosely defined promoter elements, while transcription is, in fact, perfectly directional. Observed transcription patterns would then presuppose that the two promoter elements are less effective playing the opposite role. Intriguingly, Sun and colleagues have recently identified two homologs of the ARID TF family in Giardia (Wang, Su and Sun, Molecular Parasitology Meeting, September 2006 and personal communication). ARID (AT-rich interaction domain) TFs are common among eukaryotes and are so named because of their preference for binding to AT-rich DNA sequences, raising the possibility that these proteins may be important at the initiation of bidirectional transcription in Giardia. Additional studies are needed to help us understand the extent of ‘looseness’ of transcriptional regulation at cis- and trans- levels in the parasite.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The authors thank Andrew G. McArthur and Sarah Preheim at the Giardia SAGE project, for prepublication access to the Giardia trophozoite SAGE data in the early stages of this project (data now available publicly at GiardiaDB), and also thank Andrew G. McArthur for critical reading of the manuscript. We thank Steven Moore, Paul Kennedy and Chad LaJoie at the Academic Research Computing Group, Georgetown University, for creating the algorithms used in the study for analysis of the SAGE data. We thank Sun Chin-Hung for sharing information about the recent identification of the ARID transcription factor family prior to publication. This work was supported by the NIH Grant 1R01AI/GM48922 to H.G.E. S.T. was supported in part by a Sigma Xi Grant-in-Aid of Research Award and a Georgetown Graduate School Dissertation Fellowship Award. Instrumentation for DNA sequencing was supported by an award from NSF (DBI-0100061) and Georgetown University. Funding to pay the Open Access publication charges for this article was provided by NIH Grant 1R01AI/GM48922.
Conflict of interest statement. None declared.
REFERENCES
- 1.Xu LC, Thali M, Schaffner W. Upstream box/TATA box order is the major determinant of the direction of transcription. Nucleic Acids Res. 1991;19:6699–6704. doi: 10.1093/nar/19.24.6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li JJ, Kim RH, Sodek J. An inverted TATA box directs downstream transcription of the bone sialoprotein gene. Biochem. J. 1995;310(Pt 1):33–40. doi: 10.1042/bj3100033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cox JM, Hayward MM, Sanchez JF, Gegnas LD, van der Zee S, Dennis JH, Sigler PB, Schepartz A. Bidirectional binding of the TATA box binding protein to the TATA box. Proc. Natl. Acad. Sci. USA. 1997;94:13475–13480. doi: 10.1073/pnas.94.25.13475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim JL, Nikolov DB, Burley SK. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature. 1993;365:520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- 5.Kim Y, Geiger JH, Hahn S, Sigler PB. Crystal structure of a yeast TBP/TATA-box complex. Nature. 1993;365:512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- 6.Nikolov DB, Chen H, Halay ED, Usheva AA, Hisatake K, Lee DK, Roeder RG, Burley SK. Crystal structure of a TFIIB-TBP-TATA-element ternary complex. Nature. 1995;377:119–128. doi: 10.1038/377119a0. [DOI] [PubMed] [Google Scholar]
- 7.Nikolov DB, Chen H, Halay ED, Hoffman A, Roeder RG, Burley SK. Crystal structure of a human TATA box-binding protein/TATA element complex. Proc. Natl. Acad. Sci. USA. 1996;93:4862–4867. doi: 10.1073/pnas.93.10.4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geiger JH, Hahn S, Lee S, Sigler PB. Crystal structure of the yeast TFIIA/TBP/DNA complex. Science. 1996;272:830–836. doi: 10.1126/science.272.5263.830. [DOI] [PubMed] [Google Scholar]
- 9.Tan S, Richmond TJ. Eukaryotic transcription factors. Curr. Opin. Struct. Biol. 1998;8:41–48. doi: 10.1016/s0959-440x(98)80008-0. [DOI] [PubMed] [Google Scholar]
- 10.Tsai FT, Littlefield O, Kosa PF, Cox JM, Schepartz A, Sigler PB. Polarity of transcription on Pol II and archaeal promoters: where is the ‘one-way sign’ and how is it read? Cold Spring Harb. Symp. Quant. Biol. 1998;63:53–61. doi: 10.1101/sqb.1998.63.53. [DOI] [PubMed] [Google Scholar]
- 11.Bell SD, Kosa PL, Sigler PB, Jackson SP. Orientation of the transcription preinitiation complex in archaea. Proc. Natl. Acad. Sci. USA. 1999;96:13662–13667. doi: 10.1073/pnas.96.24.13662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lagrange T, Kapanidis AN, Tang H, Reinberg D, Ebright RH. New core promoter element in RNA polymerase II-dependent transcription: sequence-specific DNA binding by transcription factor IIB. Genes Dev. 1998;12:34–44. doi: 10.1101/gad.12.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qureshi SA, Jackson SP. Sequence-specific DNA binding by the S. shibatae TFIIB homolog, TFB, and its effect on promoter strength. Mol. Cell. 1998;1:389–400. doi: 10.1016/s1097-2765(00)80039-8. [DOI] [PubMed] [Google Scholar]
- 14.Fan JB, Korman SH, Cantor CR, Smith CL. Giardia lamblia: haploid genome size determined by pulsed field gel electrophoresis is less than 12 Mb. Nucleic Acids Res. 1991;19:1905–1908. doi: 10.1093/nar/19.8.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nixon JE, Wang A, Morrison HG, McArthur AG, Sogin ML, Loftus BJ, Samuelson J. A spliceosomal intron in Giardia lamblia. Proc. Natl. Acad. Sci. USA. 2002;99:3701–3705. doi: 10.1073/pnas.042700299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Russell AG, Shutt TE, Watkins RF, Gray MW. An ancient spliceosomal intron in the ribosomal protein L7a gene (Rpl7a) of Giardia lamblia. BMC Evol. Biol. 2005;5:45. doi: 10.1186/1471-2148-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knodler LA, Svard SG, Silberman JD, Davids BJ, Gillin FD. Developmental gene regulation in Giardia lamblia: first evidence for an encystation-specific promoter and differential 5′ mRNA processing. Mol. Microbiol. 1999;34:327–340. doi: 10.1046/j.1365-2958.1999.01602.x. [DOI] [PubMed] [Google Scholar]
- 18.Adam RD. Biology of Giardia lamblia. Clin. Microbiol. Rev. 2001;14:447–475. doi: 10.1128/CMR.14.3.447-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holberton DV, Marshall J. Analysis of consensus sequence patterns in Giardia cytoskeleton gene promoters. Nucleic Acids Res. 1995;23:2945–2953. doi: 10.1093/nar/23.15.2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun CH, Tai JH. Identification and characterization of a ran gene promoter in the protozoan pathogen Giardia lamblia. J. Biol. Chem. 1999;274:19699–19706. doi: 10.1074/jbc.274.28.19699. [DOI] [PubMed] [Google Scholar]
- 21.Elmendorf HG, Singer SM, Pierce J, Cowan J, Nash TE. Initiator and upstream elements in the alpha2-tubulin promoter of Giardia lamblia. Mol. Biochem. Parasitol. 2001;113:157–169. doi: 10.1016/s0166-6851(01)00211-0. [DOI] [PubMed] [Google Scholar]
- 22.Yee J, Mowatt MR, Dennis PP, Nash TE. Transcriptional analysis of the glutamate dehydrogenase gene in the primitive eukaryote, Giardia lamblia. Identification of a primordial gene promoter. J. Biol. Chem. 2000;275:11432–11439. doi: 10.1074/jbc.275.15.11432. [DOI] [PubMed] [Google Scholar]
- 23.Vanacova S, Liston DR, Tachezy J, Johnson PJ. Molecular biology of the amitochondriate parasites, Giardia intestinalis, Entamoeba histolytica and Trichomonas vaginalis. Int. J. Parasitol. 2003;33:235–255. doi: 10.1016/s0020-7519(02)00267-9. [DOI] [PubMed] [Google Scholar]
- 24.Davis-Hayman SR, Hayman JR, Nash TE. Encystation-specific regulation of the cyst wall protein 2 gene in Giardia lamblia by multiple cis-acting elements. Int. J. Parasitol. 2003;33:1005–1012. doi: 10.1016/s0020-7519(03)00177-2. [DOI] [PubMed] [Google Scholar]
- 25.Elmendorf HG, Singer SM, Nash TE. The abundance of sterile transcripts in Giardia lamblia. Nucleic Acids Res. 2001;29:4674–4683. doi: 10.1093/nar/29.22.4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ullu E, Lujan HD, Tschudi C. Small sense and antisense RNAs derived from a telomeric retroposon family in Giardia intestinalis. Eukaryot. Cell. 2005;4:1155–1157. doi: 10.1128/EC.4.6.1155-1157.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Best AA, Morrison HG, McArthur AG, Sogin ML, Olsen GJ. Evolution of eukaryotic transcription: insights from the genome of Giardia lamblia. Genome Res. 2004;14:1537–1547. doi: 10.1101/gr.2256604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seshadri V, McArthur AG, Sogin ML, Adam RD. Giardia lamblia RNA polymerase II: amanitin-resistant transcription. J. Biol. Chem. 2003;278:27804–27810. doi: 10.1074/jbc.M303316200. [DOI] [PubMed] [Google Scholar]
- 29.Ong SJ, Huang LC, Liu HW, Chang SC, Yang YC, Bessarab I, Tai JH. Characterization of a bi-directional promoter for divergent transcription of a PHD-zinc finger protein gene and a ran gene in the protozoan pathogen Giardia lamblia. Mol. Microbiol. 2002;43:665–676. doi: 10.1046/j.1365-2958.2002.02772.x. [DOI] [PubMed] [Google Scholar]
- 30.Kruglyak S, Tang H. Regulation of adjacent yeast genes. Trends Genet. 2000;16:109–111. doi: 10.1016/s0168-9525(99)01941-1. [DOI] [PubMed] [Google Scholar]
- 31.Trinklein ND, Aldred SF, Hartman SJ, Schroeder DI, Otillar RP, Myers RM. An abundance of bidirectional promoters in the human genome. Genome Res. 2004;14:62–66. doi: 10.1101/gr.1982804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Travers MT, Cambot M, Kennedy HT, Lenoir GM, Barber MC, Joulin V. Asymmetric expression of transcripts derived from the shared promoter between the divergently oriented ACACA and TADA2L genes. Genomics. 2005;85:71–84. doi: 10.1016/j.ygeno.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 33.Koyanagi KO, Hagiwara M, Itoh T, Gojobori T, Imanishi T. Comparative genomics of bidirectional gene pairs and its implications for the evolution of a transcriptional regulation system. Gene. 2005;353:169–176. doi: 10.1016/j.gene.2005.04.027. [DOI] [PubMed] [Google Scholar]
- 34.Keister DB. Axenic culture of Giardia lamblia in TYI-S-33 medium supplemented with bile. Trans. R. Soc. Trop. Med. Hyg. 1983;77:487–488. doi: 10.1016/0035-9203(83)90120-7. [DOI] [PubMed] [Google Scholar]
- 35.Singer SM, Yee J, Nash TE. Episomal and integrated maintenance of foreign DNA in Giardia lamblia. Mol. Biochem. Parasitol. 1998;92:59–69. doi: 10.1016/s0166-6851(97)00225-9. [DOI] [PubMed] [Google Scholar]
- 36.Yee J, Nash TE. Transient transfection and expression of firefly luciferase in Giardia lamblia. Proc. Natl. Acad. Sci. USA. 1995;92:5615–5619. doi: 10.1073/pnas.92.12.5615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li L, Wang CC. Capped mRNA with a single nucleotide leader is optimally translated in a primitive eukaryote, Giardia lamblia. J. Biol. Chem. 2004;279:14656–14664. doi: 10.1074/jbc.M309879200. [DOI] [PubMed] [Google Scholar]
- 38.von Allmen N, Bienz M, Hemphill A, Muller N. Quantitative assessment of sense and antisense transcripts from genes involved in antigenic variation (vsp genes) and encystation (cwp 1 gene) of Giardia lamblia clone GS/M-83-H7. Parasitology. 2005;130:389–396. doi: 10.1017/s0031182004006742. [DOI] [PubMed] [Google Scholar]
- 39.Smale ST, Kadonaga JT. The RNA polymerase II core promoter. Annu. Rev. Biochem. 2003;72:449–479. doi: 10.1146/annurev.biochem.72.121801.161520. [DOI] [PubMed] [Google Scholar]