Abstract
Insulators play important roles in controlling gene activity and maintaining regulatory independence between neighbouring genes. In this article, we show that the enhancer-blocking activity of the insulator present within the LTR retrotransposon Idefix can be abolished if two copies of the region containing the insulator—specifically, the long terminal repeat (LTR)—are fused to the retrotransposon's 5′ untranslated region (5′ UTR). The presence of this combination of two [LTR-5′ UTR] modules is a prerequisite for the loss of enhancer-blocking activity. We further show that the 5′ UTR causes flanking genomic sequences to be displaced to the nuclear periphery, which is not observed when two insulators are present by themselves. This study thus provides a functional link between insulators and independent genomic modules, which may cooperate to allow the specific regulation of defined genomic loci via nuclear repositioning. It further illustrates the complexity of genomic regulation within a chromatic environment with multiple functional elements.
INTRODUCTION
Eukaryotes contain thousands of genes that are expressed in unique patterns to ensure the correct establishment of cellular identities. This process requires the coordinate transcriptional regulation of hundreds of genes. Gene expression is controlled by promoter sequences located immediately upstream of the transcriptional start sites of genes as well as additional regulatory sequences present close to or within the genes themselves. These cis-regulatory elements, which can act as enhancers or silencers of gene expression, are stretches of DNA that usually span a few hundred base pairs, even while being able to exert long-range effects regardless of their position or orientation (1–3). At the same time, neighbouring genes which should theoretically be influenced by the same enhancer often display independent transcription profiles. This raises the fundamental question of how the range of enhancer action can be restricted.
Evidence suggests that the control of specific expression patterns may be correlated with the spatial positioning of genes within the nucleus. Indeed, recent data have shown that enhancers and their target promoters are in close proximity to each other within the nuclear space. These interactions persist during transcription, suggesting that the direct interactions of enhancers with their target genes may be important for activation (3–5). These specific interactions are at least partly mediated by enhancers and by specific promoter-bound proteins.
Distant enhancer and promoter interactions can also be limited by DNA elements called insulators. Insulators possess two functional properties. First, insulators positioned between enhancers and promoters can block their interaction. Second, insulators (also called boundaries) prevent the advance of nearby condensed chromatin and thereby protect gene expression from positive or negative chromatin effects (6,7). Insulators have been identified in most eukaryotic genomes, suggesting that they have a conserved role in defining domains of gene function. They have also been shown to play critical roles in many developmental processes, such as imprinting and mammalian dosage compensation.
The Drosophila genome contains many sequences with insulator function (8). The first insulators to be identified were scs and scs′, which correspond to regions of unusual chromatin structure flanking the decondensed domain produced by the transcription of two heat-shock (hs) genes. Another Drosophila insulator is the gypsy insulator. This element was identified as the region within the gypsy retrotransposon that is responsible for the induction of tissue-specific mutations in many genes. Although the gypsy insulator acts as an enhancer blocker when present as a single copy, when two gypsy insulators are present between an enhancer and a promoter they lose their enhancer-blocking activity (9,10); in other words, the presence of two gypsy insulators results in an insulator bypass. Gypsy insulators have also been shown to stabilize trans activation between distantly located enhancers and promoters (11). While the proteins bound to the gypsy insulator are uniformly distributed along polytene chromosomes, in interphase nuclei of diploid cells they coalesce into large foci called insulator bodies (12).
The enhancer-blocking effects of insulators are accomplished without affecting the intrinsic properties of any of the regulatory elements, implying that insulators somehow disrupt the signalling between enhancers, silencers and promoters (13,14). While the mechanism whereby insulators establish independent functional domains is unclear, it might involve interactions between insulators and/or nuclear substructures to form loop domains that limit the action of transcriptional regulatory elements (12).
Some years ago, we reported the identification of alleles of the Drosophila melanogaster white gene that were caused by the successive insertion of two LTR retrotransposons called ZAM and Idefix (15). Subsequent analysis of the molecular mechanisms by which ZAM and Idefix interfere with white gene regulation revealed that the 5′ untranslated region (5′ UTR) of ZAM bears cis-acting regulatory sequences that can enhance the transcription of white in the eyes of a line called RevI. In another line, called RevII, this activation can be counteracted by the long terminal repeat (LTR) of Idefix, which acts as an insulator in the flies’ eyes to isolate the white gene from the upstream ZAM enhancer (16). We also reported the surprising discovery that an additional copy of Idefix inserted between the ZAM enhancer and the white gene in a line called RevIV leads to the full reversion of the orange eye colour phenotype that is caused by the presence of a single Idefix element: in the presence of both copies of Idefix, the eyes became brick red. This reversion occurred through a mechanism that remained unelucidated (15).
In this study, we sought to advance our understanding of the insulator properties of Idefix and of its potential impact on nearby genes. We combined transgenic experiments with 3D fluorescent in situ hybridization (3D FISH)-immunoassays to further dissect its enhancer-blocker properties. We demonstrate that the ability of the Idefix insulator to protect genes from enhancer effects in cis is increased when two copies of the insulator are present. This is similar to data reported for tandem repeats of scs/scs′ elements. We also show that when an additional DNA fragment from the 5′ UTR of Idefix is fused to the insulator domain, the enhancer-blocking activity of the Idefix LTR is neutralized. This novel functional module promotes insulator bypass, as has been observed with pairs of gypsy insulators. We further show that this loss of insulator function is correlated with a displacement of the tested sequence from the interior of the nucleus towards the nuclear periphery.
RESULTS
Two insulators of Idefix are more efficient than one
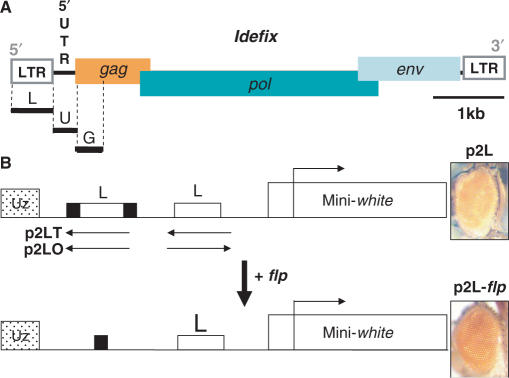
In view of the observations that pairs of gypsy insulators nullify the effects of a single element and allow insulator bypass, while pairs of scs/scs′ insulators are more efficient than one and increase enhancer-blocking activity, we decided to examine the effects of having two Idefix insulators. For this purpose, we constructed P transformation vectors in which two Idefix LTRs—which provide the insulator function—were placed between the ZAM enhancer and the mini-white reporter gene (Figure 1A and B). The two LTRs were inserted either in tail to tail or head-to-tail in vectors called p2LT and p2LO, respectively (Figure 1B). In each construct, one of the two Idefix sequences was flanked by FRT sites, the targets of the flp recombinase. The constructs could then be crossed with flies expressing flp recombinase under the control of a heat-shock promoter so that expression of flp in their descendants would lead to recombination between the two FRTs and deletion of the intervening LTR copy. These constructs were microinjected into flies carrying a null mutation in the white gene (w1118), and transgenic flies were identified by virtue of their red eye colour resulting from mini-white gene expression. Crosses with flies bearing the flp recombinase yielded flies, called p2L-flp, that carried only one insulator within the transgene. This strategy enabled us to compare the levels of mini-white gene expression when one or two insulators were present within a transgene at a defined locus. A total of six p2L transgenic lines were obtained for each construct.
Figure 1.
Two Idefix insulators are more efficient than one. (A) Molecular structure of Idefix. The two LTRs are symbolized by white boxes at each end; Idefix open reading frames (gag, pol and env) are symbolized by coloured rectangles. Black lines under the Idefix structure indicate the different cloned fragments used in this study. (B) The mini-white reporter gene is depicted by a white rectangle and its transcription start site by an arrow. Two Idefix LTRs (L) are inserted between the ZAM enhancer (Uz) and the mini-white gene. One of them is flanked by FRT sites (black boxes). The tested Idefix orientations are shown as arrows placed above the rectangles (p2LO and p2LT). Eye phenotypes of the resulting transgenic lines before (p2L) and after (p2L-flp) flp action are presented on the right.
All of the transgenes subjected to flp recombination, i.e. the p2L-flp transgenes, showed increased mini-white expression regardless of the LTR orientation. In the six transgenic lines, eye pigmentation was always weaker in the p2L flies carrying two copies of the Idefix insulator than in the p2L-flp flies carrying a single copy. This is illustrated in Figure 1B, showing clearly the yellow eye pigmentation of the p2L line before the LTR was flipped out, and the orange colour after. This result indicates that the insulator present in the Idefix LTR only partially blocks enhancer–promoter communication, and that two Idefix insulators reinforce the enhancer–blocking activity. This result is similar to data reported for the scs/scs′ elements in D. melanogaster (17).
The presence of nearby Idefix 5′ UTR sequences modifies Idefix insulator properties
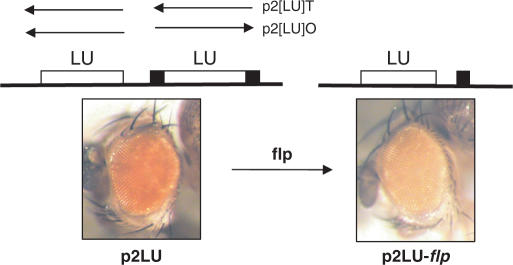
A second series of transgenes was constructed using a longer fragment that spans nucleotides 1–1010 of the Idefix sequence, encompassing the LTR and the 5′ UTR regions of Idefix. Two copies of this 1010-bp fragment were similarly inserted between the ZAM enhancer and the mini-white reporter gene. The LTR–UTR pairs were inserted either in tail to tail or head-to-tail in vectors called p2[LU]T and p2[LU]O, respectively (Figure 2). The 1010-bp fragment that was more proximal to the transcription start site of the mini-white gene was flanked by FRT sites. Crosses with flies bearing the flp recombinase yielded flies, called p2[LU]-flp, that therefore had only one LTR–UTR fragment within the transgene. A total of eight p2[LU] transgenic lines were obtained for each construct.
Figure 2.
The presence of two Idefix 5′ UTR sequences modifies Idefix insulator properties. Eye phenotypes of transgenic lines before and after flp action are presented on the left and on the right, respectively. The p2[LU] transgene structure is indicated above the eye phenotypes. The tested fragments L and U (LTR and UTR, respectively) are located between the mini-white gene and the ZAM enhancer (not indicated in the scheme). The FRT sites are shown as black rectangles. The arrows indicate the orientation of the tested sequences in the p2LUO or p2LUT transgenes.
Surprisingly, when the eye phenotypes were examined, each of the eight transgenic p2[LU]-flp lines, which had only one LTR–UTR fragment, showed a lighter eye colour than the p2[LU] lines before the flp-recombinase action. An example is presented in Figure 2B. The p2LU fly displays red eyes, while the p2LU-flp fly has pale orange eyes. This was observed regardless of the relative orientation of the pair. This result indicates that the untranslated region of Idefix, when present in two copies, introduces a novel mode of transgene regulation that reduces the insulation conferred by two Idefix LTRs. The eye pigment levels in the transgenic lines were quantified before and after flp-recombinase action and are reported in Table 1. The pigmentation levels were also compared to those measured in a series of transgenic lines called pUzW (Table 1); these lines, which were established by Conte et al. (16) to study the effect of the ZAM enhancer on white, contain transgenes in which the ZAM enhancer is located directly upstream of the mini-white gene (16). As shown in Table 1, the eye pigmentation levels were very similar between the p2[LU] and pUzW lines before flp action, suggesting that the presence of two [LU] modules brings about a complete insulator bypass. This result is similar to data reported for the gypsy insulator: when two gypsy insulators are placed between an enhancer and a reporter gene, the insulator effect is nullified and the upstream enhancer can activate the downstream gene.
Table 1.
Quantification of the amount of eye pigment in the transgenic lines
| Transgenes | Line | No flp | flp | flp/no flp |
|---|---|---|---|---|
| p2LU | 1 | 0.16 | 0.06 | 0.4 |
| 2 | 0.10 | 0.03 | 0.3 | |
| 3 | 0.05 | 0.03 | 0.6 | |
| 4 | 0.12 | 0.07 | 0.5 | |
| 5 | 0.09 | 0.03 | 0.4 | |
| 6 | 0.16 | 0.05 | 0.3 | |
| 7 | 0.14 | 0.05 | 0.4 | |
| 8 | 0.05 | 0.02 | 0.3 | |
| PUzW | 1 | 0.16 | 0.06 | 0.4 |
| 2 | 0.17 | 0.09 | 0.5 | |
| 3 | 0.14 | 0.13 | 0.7 | |
| 4 | 0.08 | 0.04 | 0.5 | |
| 5 | 0.08 | 0.03 | 0.4 | |
| 6 | 0.15 | 0.08 | 0.5 | |
| 7 | 0.10 | 0.04 | 0.4 | |
| 8 | 0.12 | 0.09 | 0.5 | |
| 9 | 0.12 | 0.06 | 0.9 | |
| 10 | 0.12 | 0.08 | 0.7 |
A pair of Idefix 5′ UTRs coupled to a pair of Idefix LTRs is necessary to nullify the enhancer-blocker function
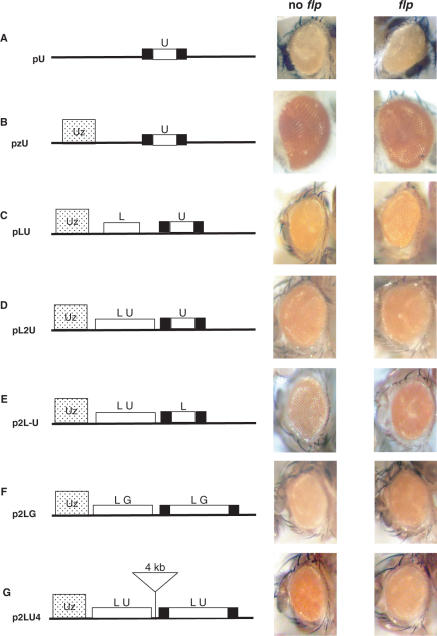
To further characterize the interaction between the 5′ UTR and LTR of Idefix in the p2[LU] lines, we first asked whether the 5′ UTR could alter the regulation of the mini-white gene by itself. We cloned a copy of the 5′ UTR flanked by FRT sites upstream of the mini-white gene. Five transgenic lines were established with this vector, called pU, which lacks the ZAM enhancer. All the lines had yellow eyes, as shown in Figure 3A, and no variation in eye colour was observed in five transgenic lines analysed before and after flp treatment. This indicates that the 5′ UTR by itself has neither enhancer nor silencer function that affects the downstream mini-white gene.
Figure 3.
Two [Insulator–UTR] pairs are necessary to bypass the insulator effect. The eye phenotypes of various transgenic lines before and after flp action are presented on the right. The structure of the transgenes is depicted on the left. The tested fragments are located upstream of the mini-white gene (not indicated in the scheme). The ZAM enhancer is represented as a dotted rectangle labelled Uz, and the FRT sites are shown as black rectangles. (A) pU transgenes: a copy of the 5′ UTR flanked by FRT sites upstream of the mini-white gene. (B) pzU transgenes: a copy of the 5′ UTR flanked by FRT sites between the ZAM enhancer and the mini-white gene. (C) pLU transgenes: a single LTR fused to a 5′ UTR flanked by FRT sequences sites between the ZAM enhancer and the mini-white gene. (D) pL2U transgenes: two 5′ UTR sequences and a single LTR were placed between the enhancer and the mini-white reporter gene. One of the two 5′ UTRs was flanked by FRT sites. (E) p2L-U transgenes: two LTRs and a single 5′ UTR were placed between the enhancer and the mini-white reporter gene. One of the two LTR was flanked by FTR sites. (F) p2LG transgenes: A 450-bp fragment from the gag gene of Idefix was fused to the LTR fragment. FRT sites are indicated as black boxes. (G) p2LU4 transgenes: Same configuration as Figure 2A, but the two [LTR–UTR] units are separated by 4 kb (triangle).
The effect of the two 5′ UTR regions in the p2[LU] lines could also result from a modification in the interaction between the ZAM enhancer and its mini-white gene target. To visualize any such alterations, we cloned a 5′ UTR fragment without the LTR region between the ZAM enhancer and the mini-white gene (pzU transgenes) (Figure 3B). When the 5′ UTR was flipped out via flp recombinase action, no variation in eye colour was observed in five pzU transgenic lines analysed (Figure 3B). This result indicates that the 5′ UTR by itself does not modify the communication between ZAM and mini-white. Further, the eye phenotypes of the five transgenic pzU lines before and after flp treatment ranged from orange to red (Figure 3B), a colour spectrum very similar to that observed in the pUzW transgenic lines (data not shown). It thus appears that the 5′ UTR of Idefix does not interfere with the enhancer effect of ZAM.
We next attempted to identify any potential interactions between the 5′ UTR and the LTR of Idefix. We first tested whether a single 5′ UTR could affect the insulator effect of a single LTR. The enhancer-blocker function was tested in a transgene, called pLU, that contains an LTR fused to a 5′ UTR that is itself flanked by FRT sequences. None of the transgenic lines tested displayed any change in eye colour in response to 5′ UTR excision, indicating that single 5′ UTRs do not modify the enhancer-blocker property of individual Idefix insulators. Figure 3C shows the pLU flies having the same orange eye pigmentation before and after flp recombinase action.
We next investigated whether two 5′ UTRs associated with a single Idefix LTR could similarly abolish the LTR's enhancer-blocking function. This was tested using a pL2U construct in which two 5′ UTR sequences and a single LTR were placed between the enhancer and the mini-white reporter gene. One of the two 5′ UTRs was flanked by FRT sites. We observed no detectable differences in the eye colour phenotypes of transgenic flies following flp gene expression compared to in unflipped flies (see the orange phenotype of the pL2U and pL2U-flp flies in Figure 3D). This result shows that two 5′ UTR sequences by themselves are unable to modify the insulator effect of a single Idefix LTR, and that pairing 5′ UTR does not alter the downstream mini-white gene regulation.
We also tested the combination of two LTRs and one 5′ UTR in lines called p2L-U. As observed with the p2L transgenic lines (Figure 1), eye pigmentation was always weaker in the p2L-U flies carrying two copies of the Idefix LTR than in the p2L-U-flp flies carrying a single copy (Figure 3E). This result indicates that the insulator function present in the LTR is not modified by the presence of a single 5′ UTR. A single 5′ UTR thus has no effect on the enhancer-blocking activity of two LTRs.
To verify that the expression of the transgene was (or not) specifically affected by the 5′ UTR in the p2[LU] lines, we substituted the 5′ UTR sequence with an equivalently sized fragment taken from another portion of the Idefix retrotransposon. A 450-bp fragment from the gag gene was used (Figures 1A and 3F). Constructs referred to as p2[LG], which contain two [LTR–Gag] fragments inserted between the ZAM enhancer and the mini-white gene, were tested for their enhancer-blocking activity. One of the two LTR–Gag sequences, specifically that is closer to the transcription start site of the mini-white gene, was flanked by FRT sites. Four transgenic lines were tested. The four p2[LG] lines displayed a light eye colour phenotype (pale orange in Figure 3F) that became darker after the flp treatment (dark orange in Figure 3F), i.e. when only one copy of the LTR–Gag pair remained in the transgene. This result, which is similar to what was observed in the p2L lines (Figure 1B), indicates that the Gag fragment does not modify the insulator property of the Idefix LTR. Further, these data indicate that the 5′ UTR carries specific features, not present in the Gag fragment, that allow it to modify the insulator properties of the LTR.
Finally, we asked whether the modified enhancer-blocking activity observed in the p2[LU] lines was related to the short distance (150 bp) separating the two LU fragments in the p2[LU] transgene. Specifically, we tested whether a longer sequence of several kilobases (kb) between the two [LU] pairs would affect the expression of the transgene (Figure 3G). We found that the resulting transgenes, called p2LU4, were not sensitive to the length separating the two [LU] elements: the eyes were darker when a single [LU] was present, and lighter in the presence of two copies (see orange eyes before flp treatment versus yellow eyes after flp treatment in Figure 3G). Thus, the enhancer-blocking function is lost when two LUs are present, whether they are close to each other (150 bp) (Figure 2A) or are separated by 4 kb (Figure 3G).
Taken together, these results show that the enhancer-blocking property of two Idefix insulators can be modified if the insulators are associated with two copies of the 5′ UTR region. This novel form of regulation is likely to be due to specific properties of the 5′ UTR itself and to occur through a mechanism that directly affects neither the expression of the mini-white gene, nor its interactions with the ZAM enhancer, nor the insulator function associated with the LTR sequence.
Subnuclear localization of transgenes
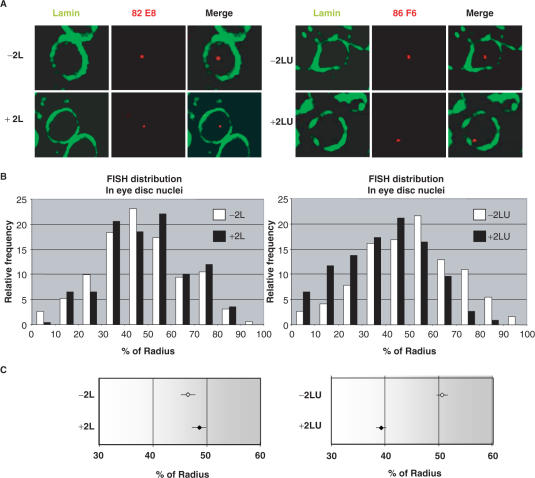
As chromatin loops have been suggested to play a role in the organization of eukaryotic chromatin domains and in insulator function (12), we hypothesized that the presence of the 5′ UTRs in the transgenes might be altering the subnuclear localization and consequently the expression of the transgenes. If this were the case, then the presence of the p2[LU] transgenes might affect the nuclear position of neighbouring genomic DNA. Thus, we performed 3D FISH-immunostaining experiments to track loci in the nuclear volume and to identify any changes in their nuclear localization in the presence or absence of the transgenes described above. In particular, we determined the effects of the p2L and p2[LU] transgenes on their flanking DNA. The 3D FISH-immunostaining experiments were performed in the differentiated cells of larval imaginal eye discs, where the mini-white gene is active.
The genomic insertion sites of p2L and p2[LU] constructs were mapped in detail by inverse PCR. Two independent transgenic lines were selected for each vector, with p2L1 and p2L2 localizing to cytological sites 82E8 and 33E7, respectively, and p2[LU]1 and p2[LU]2 to cytological sites 93B9 and 86F6, respectively. Eye imaginal discs were dissected from these four transgenic lines as well as from a w1118 control line that lacked the transgenes. FISH was performed on larval discs using probes specific for the genomic sequences surrounding the construct insertion sites. Following FISH, immunostaining was performed using a rabbit polyclonal antibody against nuclear lamin. Images were acquired along the z-axis using confocal microscopy. In most nuclei, the FISH signal representing the position of the locus of interest appeared as a single spot (red) surrounded by the lamin signal (green) (Figure 4A). In each line, the shortest distance between the FISH signal and the lamin ring was measured in a total of 100 nuclei. These distances, expressed as the percentage of the nuclear radius (% radius), were plotted as a function of their relative frequencies (see Materials and methods section). The histograms of the 3D foci distribution values are presented in Figure 4B. The relative frequencies of the foci with or without transgenes are presented as black and white bars, respectively. For the p2L and p2[LU] transgenic lines, the distribution patterns obtained for the two independent loci (cytological sites 82E8 and 33E7 for p2L, and 93B9 and 86F6 for p2[LU]) were pooled and directly compared in a single histogram with the distribution pattern of the corresponding wild-type loci (−2L and −2[LU], respectively). The distribution patterns of the p2L lines were no different from those in wild-type flies with respect to the relative nuclear positioning of the two independent loci (Figure 4B, left). Foci were centered around 50% of the radius in both the transgenic lines and the control. By contrast, a clear difference was observed when the p2[LU] lines were analysed. Indeed, DNA flanking the p2[LU] transgenes assumed a more peripheral distribution than the corresponding loci lacking the p2[LU] transgene (Figure 4B, right).
Figure 4.
Effect of the Idefix 5′ UTR and/or LTR on the nuclear localization of neighbouring chromosomal DNA. (A) FISH-immunostaining in interphase nuclei of larval eye imaginal discs. Single slices of individual nuclei showing characteristic examples of data obtained in different lines are presented. The FISH signals of the 82E8 and 86F6 loci containing the p2L and p2LU transgenes, respectively (red), and nuclear lamin (green), are shown. Arrows indicate cases of localization of the locus at the nuclear lamin. Genotypes are indicated by −2L or −2LU for the absence of the corresponding transgene at the indicated locus, or by +2L or +2LU for the presence of it. (B) Histograms showing the 3D position of each locus hybridization signal with (black bars) or without (white bars) the respective transgenes. The relative sample frequency is plotted against the FISH spot-to-lamin distance (expressed as a% of the nuclear radius). Three larval discs were analysed for each locus. For each histogram, data from two independent loci were pooled together. (C) Position (means ± SEM) of each locus, either with (black squares) or without (white squares) the respective transgene, relative to the nuclear radius.
These results are further illustrated in Figure 4C, which presents the mean relative distances between the loci of interest and the nuclear lamin (means ± SEM) for the p2L and p2[LU] transgenic lines, along with data for the corresponding loci without transgenes. An analysis of the subnuclear positions of the genomic sequences flanking the p2L transgenes showed that the mean relative distance between the loci and the nuclear lamin was 47.7% (±SEM) and 49.4% (±SEM) of the nuclear radius in the p2L1 and p2L2 lines, respectively. These mean relative distances were not statistically different from the values of 46.7% (±SEM) and 46.5% (±SEM) found for the wild-type loci in the w1118 control line (P > 0.01.) The pooled values obtained for the two independent loci with or without the p2L transgenes are shown in Figure 4C.
In contrast, the mean relative distances between the loci and the nuclear lamin in the p2[LU]1 and p2[LU]2 lines were 39.2% (±SEM) and 39.3% (±SEM) of the nuclear radius, respectively. In the control line, the mean distances were 51.5% (±SEM) and 50.4% (±SEM), respectively. These differences in the relative positions of the genomic sequences in the LU transgenic lines compared to the positions in the w1118 control line were significant (P > 0.01). The pooled values obtained for the two independent loci with or without the 2LU transgenes are presented in Figure 4C. These data indicate that the presence of the 5′ UTR leads to the specific displacement of the LTR sequences and surrounding genomic sequences within the nucleus. This movement is not random within the whole nuclear space, but rather preferentially shifts genomic loci located near the two [LU] elements to the nuclear periphery.
DISCUSSION
In this study, we show that the enhancer-blocker activity mediated by the chromatin insulator found within the Idefix retrotransposon is stronger when two copies of the insulator are present than when only one is present. The activity can be abolished, however, when the 5′ UTR region of Idefix is associated with the insulator. We further show that the 5′ UTR causes flanking genomic sequences to be displaced to the nuclear periphery,
The Idefix insulator acts as an enhancer blocker regardless of its copy number or its nuclear localization
Insulators are a class of DNA sequences that restrain regulatory interactions within eukaryotic genomes (18–20). In the p2L line, we have shown that the insulator identified within the Idefix retrotransposon acts as an enhancer blocker on the eye enhancer of the ZAM retrotransposon ((16) and this study). However, a single copy of the insulator is not sufficient to completely block ZAM enhancer activity. Two copies of the Idefix insulator, on the other hand, inserted between the enhancer and the transcription start site of the target gene, can more efficiently block the enhancer. Several previously described insulators have also been shown to have greater enhancer-blocking ability when present in several copies. For example, enhancer-blocking activity is increased when two copies of the extensively described D. melanogaster insulators, scs and scs′, are associated (17). This flexibility might help modulate the function of Idefix insulators within the genome. We have further demonstrated that the function of the Idefix insulator not only depends on its copy number, but also that it is enhancer specific. For instance, while it is able to block the ZAM enhancer, it can block neither white gene enhancers, even though they are active in the same eye tissue as ZAM, nor ftz gene enhancers, which are specific to embryonic tissues (unpublished data). These results indicate that the function of the Idefix insulator also depends on specific enhancer characteristics.
Our results also indicate that the Idefix insulator is active within the whole space of the nucleus and does not localize to any particular nuclear regions. Similar results have recently been reported concerning the enhancer-blocker activity of the gypsy insulator, which is also functional from the nuclear periphery to the nuclear interior (21). Anchorage to certain specific nuclear structures has been suggested as a mechanism for the insulation effect of some insulators (12,22).
The genomic environment may interfere with the enhancer-blocker function of the Idefix insulator
Although two Idefix insulators are more effective than one, we further demonstrated that this function can be lost if each LTR, which carries the insulator function, is associated with a second fragment of Idefix. Indeed, when two Idefix LTRs are both flanked by the Idefix 5′ UTR, their enhancer-blocking activity disappears and the downstream mini-white gene is highly expressed. This loss of function is restricted to the specific combination of two pairs of [LTR–5′UTR]. The combinations of one insulator with two 5′ UTRs, or two insulators with one 5′UTR, had no effect on enhancer-blocker activity. When the two pairs of [LTR–5′UTR] were present, they abolished the enhancer-blocker function, regardless of their respective orientations. Further, we found that this loss of function is due to specific properties of the 5′ UTR sequence, because its replacement by another segment of Idefix, namely a fragment of the gag gene, had no effect on enhancer-blocker function.
In a complementary analysis, we have found that p2[LU]-tagged loci are perinuclear regardless of whether the transgenes are transcriptionally active or inactive. Indeed, loci tagged with the p2[LU] transgenes assumed similar distributions along the radius when scored in two types of cells: (i) eye disc cells, in which the mini-white gene is known to be activated by the ZAM enhancer (Figure 4); and (ii) cells from the peripodial membrane enveloping the eye disc, where the white gene is not activated by ZAM (Supplementary Data, Figure 1). A clear displacement of the foci towards the nuclear periphery was observed in both cases, indicating that the expression of transgenes does not dictate their perinuclear position. Thus, the altered localisation of the loci is not linked to transcriptional activation.
The fact that two Idefix LTRs are necessary for bypassing enhancer-blocking activity when they are associated with two 5′ UTRs, together with the fact that a single insulator cannot bypass the activity even if fused to two 5′ UTRs, suggests that the interaction between two Idefix insulator motifs is an absolute requirement for the bypass. This kind of pairing suggests that loops are created that could organize chromatin within the nucleus, as has been described for other insulators (12,23–25). However, this result also indicates that the pairing of two Idefix insulator copies is not sufficient to explain the loss of enhancer-blocking activity. Indeed, when these two copies were fused without the 5′ UTR sequences, they conserved their enhancer-blocker activity. Therefore, we postulate that the loss of enhancer-blocking activity cannot be achieved simply via higher-order ‘looped’ domains generated by specific interactions between insulators. Consistent with this, sequences other than the Su(Hw) insulator have been suggested to be responsible for the localization of gypsy retrotransposons at the nuclear periphery (21). Rather, the combination of two separable phenomena, such as the formation of loops and specific positioning within the nucleus (established, e.g. by the two 5′ UTRs), would be required to bypass enhancer-blocker activity.
Interestingly, when the 5′ UTR sequence was analysed in silico, we identified a sequence that shares characteristics with sequences described in the literature as matrix attachment regions or MARs (MAR-Wiz software: www.futursoft.org) (Supplementary Data, Figure 2A). This predicted MAR in the 5′ UTR was also detected through in vitro binding assays with high-salt-extracted nuclei, as described (26) (Supplementary Data, Figure 2B).
Based on our present data, it is still uncertain whether this putative MAR domain in the 5′ UTR of Idefix is responsible for the loss of insulator activity in the Idefix LTR. However, it is interesting to note that gag fragments replacing the 5′ UTR fragments in the p2[LG] transgenes did not abolish the enhancer-blocker property of the Idefix LTRs, and that they do not display any MAR domains (see Supplementary Data, Figure 2B).
Might DNA sequences that are defined as MARs, because they precipitate in high salt solutions in vitro, have specific properties that allow them to interfere with insulator function? Investigating such potential genomic effects is all the more important because the data in the literature on the biological significance of the matrix and its associated DNA sequences are far from clear, and because various reports have attributed different and apparently opposite functions to MARs (27–29).
Whatever the functional modules involved, our present data helps us understand the phenotype of flies from the RevIV line which was identified in our laboratory some years ago (15). As mentioned above, RevIV contains an allele of the white gene that has two adjacent copies of Idefix inserted between a ZAM element and the endogenous white gene promoter. It is derived from the de novo insertion of Idefix at the white locus in a line called RevII, which already had an Idefix element inserted between ZAM and the white gene (15). The two Idefix copies abolish the action of the single Idefix element, producing flies with a brick-red instead of an orange eye colour phenotype (16). These flies containing an endogenous mutation involving successive Idefix insertions are consistent with our present results. The opposite effects of one versus two copies of the full-length Idefix retrotransposon may be accounted for by the fact that two [LTR–UTR] fragments are present in the white allele of RevIV, whereas only one [LTR–UTR] fragment is present in the white allele of RevII.
Pairing of Idefix insulators and the 5′ UTR sequences are necessary for modifying gene regulation
We have shown here that the length of the sequence separating the two [LU] pairs is not critical for bypassing the enhancer-blocking activity, since we observed an equivalent loss of activity whether the two [LU] pairs were separated by 150 bp or by 4 kb. Although longer genomic fragments remain to be tested, our data are consistent with a model in which two independent pairs of [LTR-5′UTR] sequences can interact over long distances to enable the insulator bypass, regardless of their respective genomic locations.
Consistent with the idea of long-range interactions, some studies have shown that scs and scs’ are in close proximity in Drosophila nuclei (24). Furthermore, recent studies have provided examples of eukaryotic genes that are located on separate chromosomes but that associate physically in the nucleus via interactions that may have a function in coordinating gene expression (30,31). Such examples of long-range interactions in D. melanogaster have involved elements of the Bithorax complex (32,33). Interestingly, the regulatory elements required for these latter examples of long-distance communication contain both an insulator and a Polycomb-response element (34,35). In our model, since two LTRs plus two UTRs are necessary to convert an enhancer-blocker function into an insulator bypass, it can be hypothesized that the two insulators act as an essential module that drives the interaction between the distant sites, with the two 5′ UTRs being involved in the positioning within the nucleus.
One major issue raised by this study is why the subnuclear localization established by the 5′ UTR might allow the reactivation of target genes by the enhancer. Tethering genes to the nuclear pore complex may be one way in which gene activity could be restored. Could the 5′ UTR help tether chromatin sites to the nuclear pores? Alternatively, could it modify chromatin accessibility or promote RNA export? These are the questions that are currently under investigation.
MATERIALS AND METHODS
Construction of transgenes
All P-element constructs used in the adult enhancer-blocking assays were derivatives of the pCaSPER vector. For the p2LT and p2LO transgenes, the second Idefix insulator was inserted at the EcoRI site of the Idefix insulator-containing plasmid, as previously described (16). The second Idefix Insulator was amplified by PCR using primers L1 (GTCGACGTGACATATCCATAAG) and L2 (CTTCAGTTGATCAGTACCGTAC) and cloned into the P7 vector. The EcoRI fragments containing the LTR were then inserted into the final vector to give p2LT and p2LO (Figure 1).
For the p2[LU]T and p2[LU]O transgenes, the second [LTR–UTR] fragment was inserted at the SacII site of the Idefix [LTR–UTR]-containing plasmid pL, as previously described (16). The second Idefix [LTR–UTR] fragment was amplified by PCR using primers L1 and L3 (TGATGTTTTTAGTTTTCTAGC), and cloned into pGEM-T Easy. The SacII fragment containing the Idefix [LTR–UTR] was then inserted into the final vector to give p2[LU]T and p2[LU]O (Figure 2A). Details on the other constructions used in this study can be provided upon request.
P transformation
Each construct was introduced into the Drosophila germ line by injection into w1118 embryos, as described previously (36).
Fly strains and heat-shock regimens
Fly stocks were maintained on cornmeal-glucose-yeast media at 20°C. The hsFLP flies (y[1] w[1118] P{ry[+t7.2] = 70FLP}3F), provided by the Bloomington Stock Center, express flp recombinase under the control of the heat-shock promoter. Virgin female hsFLP flies were crossed with transgenic males for 24 h at 20°C. Heat shocks of embryos <24 h old were performed as described by Ahmad and Golic (37). For each recombinant P element transformation vector injected, transgenic lines were heat shocked to compare the eye colour between heat-shocked and non-heat-shocked flies.
Quantification of the amount of eye pigment in transgenic lines
Transgenic females were crossed to hs-FLP or w1118 males. One-hour-old eggs from each cross were grown at 37°C for 1 h and then placed at 25°C. Transgenic males from the two resulting categories of crosses, i.e. hs-FLP and w1118, were compared. Pigment concentrations were determined at 485 nm, as described by Evans and Howells (38).
Inverse PCR
DNA sequences adjacent to the P-element insertion sites in the genome were cloned by inverse PCR using specific primers to the 5′ inverted P-element repeat and the 3′ region of the white gene, as described by (39). Clones were sequenced using an automated sequencing system. A BLAST search was performed using the EMBL/GenBank database to identify matching sequences.
In situ hybridizations
FISH
For each probe used in this study, eight genomic PCR fragments of 1 kb each surrounding the insertion site of the transgene on both sides were pooled for probe labelling. These PCR fragments were spaced 1 kb apart, thus covering ∼16 kb of the genomic region of interest. Probes were labelled by nick translation with digoxigenin-11-dUTP (Roche Diagnosis) according to the manufacturer's instructions. Detailed coordinates of the PCR fragments used to produce the probes can be provided upon request. FISH was performed on larval imaginal discs, as described previously (Bantignies et al., 2003; a detailed protocol is available at http://www.epigenome-noe.net/researchtools/protocol.php?protid=5). After post-hybridization washes, the larval discs were blocked in PBS, 0.3% Triton, 1% BSA, 10% Normal Goat Serum for 2 h at room temperature, and incubated overnight at 4°C in the same buffer with anti-lamin rabbit polyclonal antibody R836 at a dilution of 1:1000. Immunodetection of both FISH and lamin labels was performed after simultaneously incubating the larval discs with anti-digoxigenin-rhodamine (Roche Diagnostics) at a dilution of 1:45 and with anti-rabbit Alexa 488 (Molecular Probes) at a dilution of 1:1000 (secondary antibodies) for 1 h at room temperature. The larval eye discs were then mounted in Prolong Antifade medium (Molecular Probes).
Microscopy and image analysis
3D images were acquired on a Zeiss Meta 510 confocal microscope using a 63X objective. Optical sections were collected at 0.3 µm intervals along the z-axis for each colour channel. Distance analysis was processed using Volocity software (Improvision) and Microsoft Excel. For each nucleus, the nuclear volume as well as the centre of this volume were determined based on the lamin immunolabelling. For each FISH spot, the centre of mass (x,y,z centroid coordinates) was determined and internalized in its specific nucleus using the ‘internalized’ function ‘Volocity classification’ (http://www.improvision.com/application_center/). Data were exported to Excel for mathematical analysis: The nuclear radius (nr) was determined from the nuclear volume, the distance between the centre of mass of the FISH spot and the centre of the nuclear volume (d1) was calculated, and the distance between the FISH spot and the nuclear periphery was then calculated as d2 = nr − d1. The average nuclear radius was 3 ± 0.3 µm, indicating that the larval nuclei analysed were very homogeneous in size. Distances between the loci and the nuclear periphery were therefore expressed directly as a percentage of the nuclear radius (% of radius). A small fraction of nuclei strongly deviating from a generally circular shape was not included in this analysis. Due to almost complete somatic pairing in the larvae, all nuclei analysed contained a single FISH spot for all probes. For each data set, an analysis was performed with two-channel 3D stacks collected from 3–4 imaginal larval discs, and the FISH signals were analysed in 30 nuclei per larval disc. Statistical analyses of data from paired groups were performed via a Student's t-test and a Chi-square analysis (Kruskal–Wallis test). The level of significance was set at 0.01 for all tests.
SUPPLEMENTARY DATA
Supplementary Data is available at NAR Online.
ACKNOWLEDGEMENTS
We are grateful to Dr Giacomo Cavalli for helpful discussions and Herry Herman for critical review of the manuscript. We thank Dr D.J. Arndt-Jovin for the Lamin antibody and Raphaël Rigoard for the Excel mathematical analysis. This work was supported in part by grants from the Association pour la Recherche contre le Cancer (ARC 3441), from Ministère délégué à la Recherche (ACI/BCMS2004), and from réseau structurant EpiPro. E.B. received a grant from the Fondation de la Recherche médicale (FRM). Funding to pay the Open Access publication charge was provided by INSERM (Institut national de la santé et de la recherche médicale).
Conflict of interest statement. None declared.
REFERENCES
- 1.West AG, Fraser P. Remote control of gene transcription. Hum. Mol. Genet. 2005;14(Spec. No. 1):R101–R111. doi: 10.1093/hmg/ddi104. [DOI] [PubMed] [Google Scholar]
- 2.Dorsett D. Distant liaisons: long-range enhancer-promoter interactions in drosophila. Curr. Opin. Genet. Dev. 1999;9:505–514. doi: 10.1016/s0959-437x(99)00002-7. [DOI] [PubMed] [Google Scholar]
- 3.Carter D, Chakalova L, Osborne CS, Dai YF, Fraser P. Long-range chromatin regulatory interactions in vivo. Natl Genet. 2002;32:623–626. doi: 10.1038/ng1051. [DOI] [PubMed] [Google Scholar]
- 4.de Laat W, Grosveld F. Spatial organization of gene expression: the active chromatin hub. Chromosome Res. 2003;11:447–459. doi: 10.1023/a:1024922626726. [DOI] [PubMed] [Google Scholar]
- 5.Palstra RJ, Tolhuis B, Splinter E, Nijmeijer R, Grosveld F, de Laat W. The beta-globin nuclear compartment in development and erythroid differentiation. Nat. Genet. 2003;35:190–194. doi: 10.1038/ng1244. [DOI] [PubMed] [Google Scholar]
- 6.Kellum R, Schedl P. A position-effect assay for boundaries of higher order chromosomal domains. Cell. 1991;64:941–950. doi: 10.1016/0092-8674(91)90318-s. [DOI] [PubMed] [Google Scholar]
- 7.Kellum R, Schedl P. A group of scs elements function as domain boundaries in an enhancer-blocking assay. Mol. Cell. Biol. 1992;12:2424–2431. doi: 10.1128/mcb.12.5.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brasset E, Vaury C. Insulators are fundamental components of the eukaryotic genomes. Heredity. 2005;94:571–576. doi: 10.1038/sj.hdy.6800669. [DOI] [PubMed] [Google Scholar]
- 9.Cai HN, Shen P. Effects of cis arrangement of chromatin insulators on enhancer-blocking activity. Science. 2001;291:493–495. doi: 10.1126/science.291.5503.493. [DOI] [PubMed] [Google Scholar]
- 10.Muravyova E, Golovnin A, Gracheva E, Parshikov A, Belenkaya T, Pirrotta V, Georgiev P. Loss of insulator activity by paired Su(Hw) chromatin insulators. Science. 2001;291:495–498. doi: 10.1126/science.291.5503.495. [DOI] [PubMed] [Google Scholar]
- 11.Kravchenko E, Savitskaya E, Kravchuk O, Parshikov A, Georgiev P, Savitsky M. Pairing between gypsy insulators facilitates the enhancer action in trans throughout the Drosophila genome. Mol. Cell. Biol. 2005;25:9283–9291. doi: 10.1128/MCB.25.21.9283-9291.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerasimova TI, Byrd K, Corces VG. A chromatin insulator determines the nuclear localization of DNA. Mol. Cell. 2000;6:1025–1035. doi: 10.1016/s1097-2765(00)00101-5. [DOI] [PubMed] [Google Scholar]
- 13.Cai H, Levine M. Modulation of enhancer-promoter interactions by insulators in the Drosophila embryo [see comments] Nature. 1995;376:533–536. doi: 10.1038/376533a0. [DOI] [PubMed] [Google Scholar]
- 14.Scott KS, Geyer PK. Effects of the su(Hw) insulator protein on the expression of the divergently transcribed Drosophila yolk protein genes. EMBO J. 1995;14:6258–6267. doi: 10.1002/j.1460-2075.1995.tb00316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Desset S, Conte C, Dimitri P, Calco V, Dastugue B, Vaury C. Mobilization of two retroelements, ZAM and Idefix, in a novel unstable line of Drosophila melanogaster. Mol. Biol. Evol. 1999;16:54–66. doi: 10.1093/oxfordjournals.molbev.a026038. [DOI] [PubMed] [Google Scholar]
- 16.Conte C, Dastugue B, Vaury C. Coupling of enhancer and insulator properties identified in two retrotransposons modulates their mutagenic impact on nearby genes. Mol. Cell. Biol. 2002;22:1767–1777. doi: 10.1128/MCB.22.6.1767-1777.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Majumder P, Cai HN. The functional analysis of insulator interactions in the Drosophila embryo. Proc. Natl Acad. Sci. USA. 2003;100:5223–5228. doi: 10.1073/pnas.0830190100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerasimova TI, Corces VG. Chromatin insulators and boundaries: effects on transcription and nuclear organization. Annu. Rev. Genet. 2001;35:193–208. doi: 10.1146/annurev.genet.35.102401.090349. [DOI] [PubMed] [Google Scholar]
- 19.Kuhn EJ, Geyer PK. Genomic insulators: connecting properties to mechanism. Curr. Opin. Cell. Biol. 2003;15:259–265. doi: 10.1016/s0955-0674(03)00039-5. [DOI] [PubMed] [Google Scholar]
- 20.Oki M, Kamakaka RT. Blockers and barriers to transcription: competing activities? Curr. Opin. Cell. Biol. 2002;14:299–304. doi: 10.1016/s0955-0674(02)00327-7. [DOI] [PubMed] [Google Scholar]
- 21.Xu Q, Li M, Adams J, Cai HN. Nuclear location of a chromatin insulator in Drosophila melanogaster. J. Cell. Sci. 2004;117:1025–1032. doi: 10.1242/jcs.00964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Byrd K, Corces VG. Visualization of chromatin domains created by the gypsy insulator of Drosophila. J. Cell. Biol. 2003;162:565–574. doi: 10.1083/jcb.200305013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sigrist CJ, Pirrotta V. Chromatin insulator elements block the silencing of a target gene by the Drosophila polycomb response element (PRE) but allow trans interactions between PREs on different chromosomes. Genetics. 1997;147:209–221. doi: 10.1093/genetics/147.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blanton J, Gaszner M, Schedl P. Protein: protein interactions and the pairing of boundary elements in vivo. Genes Dev. 2003;17:664–675. doi: 10.1101/gad.1052003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishii K, Arib G, Lin C, Van Houwe G, Laemmli UK. Chromatin boundaries in budding yeast: the nuclear pore connection. Cell. 2002;109:551–562. doi: 10.1016/s0092-8674(02)00756-0. [DOI] [PubMed] [Google Scholar]
- 26.Cockerill PN, Garrard WT. Chromosomal loop anchorage of the kappa immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites. Cell. 1986;44:273–282. doi: 10.1016/0092-8674(86)90761-0. [DOI] [PubMed] [Google Scholar]
- 27.Forrester WC, Fernandez LA, Grosschedl R. Nuclear matrix attachment regions antagonize methylation-dependent repression of long-range enhancer-promoter interactions. Genes Dev. 1999;13:3003–3014. doi: 10.1101/gad.13.22.3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loc P, Strätling W. The matrix attachment regions of the chicken lysozyme gene co-map with the boundaries of the chromatin domain. EMBO J. 1988;7:655–664. doi: 10.1002/j.1460-2075.1988.tb02860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McKnight RA, Shamay A, Sankaran L, Wall RJ, Hennighausen L. Matrix-attachment regions can impart position-independent regulation of a tissue-specific gene in transgenic mice. Proc. Natl Acad. Sci. USA. 1992;89:6943–6947. doi: 10.1073/pnas.89.15.6943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435:579–580. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- 31.Brown JM, Leach J, Reittie JE, Atzberger A, Lee-Prudhoe J, Wood WG, Higgs DR, Iborra FJ, Buckle VJ. Coregulated human globin genes are frequently in spatial proximity when active. J. Cell. Biol. 2006;172:177–187. doi: 10.1083/jcb.200507073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muller M, Hagstrom K, Gyurkovics H, Pirrotta V, Schedl P. The mcp element from the Drosophila melanogaster bithorax complex mediates long-distance regulatory interactions. Genetics. 1999;153:1333–1356. doi: 10.1093/genetics/153.3.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bantignies F, Grimaud C, Lavrov S, Gabut M, Cavalli G. Inheritance of Polycomb-dependent chromosomal interactions in Drosophila. Genes Dev. 2003;17:2406–2420. doi: 10.1101/gad.269503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mihaly J, Hogga I, Gausz J, Gyurkovics H, Karch F. In situ dissection of the Fab-7 region of the bithorax complex into a chromatin domain boundary and a Polycomb-response element. Development. 1997;124:1809–1820. doi: 10.1242/dev.124.9.1809. [DOI] [PubMed] [Google Scholar]
- 35.Gruzdeva N, Kyrchanova O, Parshikov A, Kullyev A, Georgiev P. The mcp element from the bithorax complex contains an insulator that is capable of pairwise interactions and can facilitate enhancer-promoter communication. Mol. Cell. Biol. 2005;25:3682–3689. doi: 10.1128/MCB.25.9.3682-3689.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rubin GM, Spradling AC. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 37.Ahmad K, Golic KG. Somatic reversion of chromosomal position effects in Drosophila melanogaster. Genetics. 1996;144:657–670. doi: 10.1093/genetics/144.2.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Evans BA, Howells AJ. Control of drosopterin synthesis in Drosophila melanogaster: mutants showing an altered pattern of GTP cyclohydrolase activity during development. Biochem. Genet. 1978;16:13–26. doi: 10.1007/BF00484381. [DOI] [PubMed] [Google Scholar]
- 39.Cryderman DE, Cuaycong MH, Elgin SC, Wallrath LL. Characterization of sequences associated with position-effect variegation at pericentric sites in Drosophila heterochromatin. Chromosoma. 1998;107:277–285. doi: 10.1007/s004120050309. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.