Abstract
RecA plays a central role in the nonmutagenic repair of stalled replication forks in bacteria. RdgC, a recombination-associated DNA-binding protein, is a potential negative regulator of RecA function. Here, we have determined the crystal structure of RdgC from Pseudomonas aeruginosa. The J-shaped monomer has a unique fold and can be divided into three structural domains: tip domain, center domain and base domain. Two such monomers dimerize to form a ring-shaped molecule of approximate 2-fold symmetry. Of the two inter-subunit interfaces within the dimer, one interface (‘interface A’) between tip/center domains is more nonpolar than the other (‘interface B’) between base domains. The structure allows us to propose that the RdgC dimer binds dsDNA through the central hole of ∼30 Å diameter. The proposed model is supported by our DNA-binding assays coupled with mutagenesis, which indicate that the conserved positively charged residues on the protein surface around the central hole play important roles in DNA binding. The novel ring-shaped architecture of the RdgC dimer has significant implications for its role in homologous recombination.
INTRODUCTION
Homologous recombination systems contribute to the maintenance of genome integrity and are essential for the repair of stalled replication forks (1). Genetic studies of the recombination-dependent growth C (rdgC) gene in bacterial systems have suggested that it is involved in DNA replication and recombination (2,3). In Neisseria gonorrhoeae (the gonococcus), RdgC is required for efficient pilin antigenic variation and plays some role in cell growth (4,5). The Escherichia coli rdgC gene encodes a DNA-binding protein of 34 kDa, whose expression level was at its highest during exponential phase, reaching its maximum at ∼1000 dimers per cell (2). Its level decreased sharply to ∼50 dimers per cell in stationary phase. This profile suggests that RdgC might function during the period of DNA replication (2).
The RecA proteins and their homologs play a key role in recombinational DNA repair in bacteria and eukaryotes. Their aberrant reactions could result in gross chromosomal rearrangements that lead to human diseases, including cancer. Therefore, their functions must be tightly regulated, and understanding how the RecA family of recombinases is regulated is of utmost importance (1). The E. coli RdgC protein is a potential negative regulator of the function of RecA (1). It inhibits RecA-promoted DNA strand exchange, RecA-mediated ATPase activity and RecA-dependent LexA cleavage (1). Sedimentation equilibrium data indicated that E. coli RdgC exists in solution as a mixture of oligomeric states in equilibrium, most likely monomers, dimers and tetramers (1). This concentration-dependent change in the oligomeric state appears to affect its mode of binding to DNA and its capacity to inhibit RecA (1). The primary mechanism of RdgC inhibition appears to involve a simple competition for DNA-binding sites, especially on double-stranded DNA (dsDNA) (1).
Deletion of the E. coli rdgC gene alone causes no obvious phenotype but is highly deleterious in strains lacking certain enzymes involved in recombination and replication restart (2,3). RdgC is essential for the growth of an E. coli strain lacking PriA, indicating that it might affect replication fork progression or fork rescue (2). PriA provides a means to load the DnaB replicative helicase at DNA replication fork and D loop structures (2). RdgC is, therefore, a key factor in the rescue of stalled or broken forks and subsequent replication restart. dnaC suppressors of PriA can overcome this inviability, especially when RecF, RecO or RecR is inactivated, which indicates that RdgC avoids or counters the toxic effect of these proteins by limiting inappropriate RecA loading on SSB-coated ssDNA (2).
The DNA-binding ability of the RdgC protein has been demonstrated clearly using electrophoretic mobility shift assays (2). Escherichia coli RdgC binds non-specifically to dsDNA and with higher affinity than ssDNA (2). RdgC from N. meningitidis also binds DNA with little specificity for sequence or structure, like the E. coli protein (4). RdgC exhibits no preference for DNA replication or recombination intermediates (2). However, no detectable domains or nuclease activity could be identified for the E. coli RdgC protein (2,6). The complexes of E. coli RdgC with both linear and supercoiled circular plasmid DNA were imaged using atomic force microscopy (7). RdgC was found to have an increased affinity to DNA ends and to promote bending of DNA (7). RdgC has the effect on DNA superstructure; the promotion of DNA condensation was observed at high protein concentrations (7). Recombination is largely enhanced by close contacts of distant regions along the DNA strands through condensation (7).
Pseudomonas aeruginosa is a ubiquitous environmental Gram-negative bacterium that belongs to the γ subdivision of the Proteobacteria. Orthologs of the rdgC gene are found only in β and γ subdivisions of the Proteobacteria (2). The amino acid sequence of P. aeruginosa RdgC, a 306-residue protein, shows 46 and 36% identities against those of E. coli and N. meningitidis, respectively. Despite the importance of DNA-binding ability of RdgC in its biological roles, the structural details of RdgC and the mechanism of its action remain unclear. Here, we report the crystal structure of P. aeruginosa RdgC as determined by the multiwavelength anomalous diffraction (MAD) method of X-ray crystallography. It reveals that RdgC dimer is ring-shaped, with a central hole of ∼30 Å diameter. The inside surface around the central hole is rich in conserved, positively charged residues, which are implicated in DNA-binding by mutagenesis. We propose that dsDNA binds to RdgC through this central hole. Our structure provides a solid structural framework for a better understanding of the role of RdgC in homologous recombination.
MATERIALS AND METHODS
Protein expression and purification
The rdgC gene (PA3263) was amplified by the polymerase chain reaction (PCR) using the genomic DNA of P. aeruginosa strain PAO1 as a template. The oligonucleotide primers designed using the published genome sequence (8) were 5′-G GAA TTC CAT ATG TGG TTC CGC AAT CTG CTC G-3′ (forward) and 5′-CCG CCG CTC GAG GAC GCC CTG GGG GAT TTC TTC-3′ (reverse). The bases in bold denote the NdeI and XhoI cleavage sites, respectively. The PCR product was digested with NdeI and XhoI, and was then inserted into the NdeI/XhoI-digested expression vector pET-21a(+) (Novagen). This vector construction adds an eight-residue tag (LEHHHHHH) to the carboxyl terminus of the gene product to facilitate protein purification.
The protein was expressed in E. coli C41 (DE3) cells (9). The cells were grown at 37°C up to OD600 of 0.6 in Luria–Bertani medium that contained 50 μg ml−1 ampicillin, and the protein expression was induced by 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG). The cells were allowed to grow at 20°C for 22 h after IPTG induction and were harvested by centrifugation at 4200 g (6000 rev min−1; Sorvall GSA rotor) for 10 min at 4°C. The cell pellet was resuspended in an ice-cold lysis buffer [20 mM Tris-HCl (pH 7.9), 500 mM sodium chloride, 5 mM imidazole] and was then homogenized with an ultrasonic processor. The crude cell extract was centrifuged at 36 000 g (18 000 rev min−1; Hanil Supra 21 K rotor) for 60 min at 4°C, and the recombinant protein in the supernatant fraction was purified in three chromatographic steps.
The first purification step utilized the C-terminal hexahistidine-tag by Ni2+-chelated HiTrap chelating column (GE Healthcare). The eluent was diluted 3-fold with 50 mM Tris-HCl at pH 7.4. The diluted solution was applied to a 20-ml heparin-Sepharose column (GE Healthcare), which was previously equilibrated with a buffer of 50 mM Tris-HCl (pH 7.4), 1 mM dithiothreitol and 1 mM EDTA. The protein was eluted with a linear gradient of 0–2.0 M sodium chloride in the same buffer. The first peak corresponded to high molecular aggregates of RdgC and the second peak to RdgC dimers, which was also consistent with our dynamic light scattering measurements. When the aggregates were not separated from the dimers, the protein readily aggregated within a day. The dimer fractions were concentrated to ∼6 mg ml−1 concentration using an YM10 ultrafiltration membrane (Millipore-Amicon) and further purified by gel filtration on a HiLoad XK 16 Superdex 200 prep-grade column (GE Healthcare), which was previously equilibrated with a buffer of 50 mM Tris-HCl (pH 7.4), 200 mM sodium chloride, 1 mM dithiothreitol and 1 mM EDTA. The first peak corresponding to the high molecular aggregates of RdgC was only a minor component, and the second peak corresponding to RdgC dimers was stable for several weeks when frozen at −70°C. The procedure for purifying the SeMet-substituted protein was the same, except for the presence of 10 mM dithiothreitol in all buffers used during purification steps. Dynamic light scattering experiments were performed at 24°C, with the protein (at 2 mg ml−1 concentration) dissolved in a buffer consisting of 50 mM Tris-HCl (pH 7.4), 1 mM dithiothreitol, 1 mM EDTA and 200 mM sodium chloride on a Model DynaPro-801 instrument (Wyatt, Santa Barbara, CA, USA). The protein concentration was estimated by measuring the absorbance at 280 nm, employing the calculated extinction coefficient of 20 910 M−1 cm−1 (SWISS-PROT; http://www.expasy.ch/).
Mutagenesis and electrophoretic mobility shift assay
Point mutants were prepared using the QuikChange Site-Directed Mutagenesis kit (Stratagene), and the mutations were confirmed by DNA sequencing. The seven mutants R4A, F120A, K146A, R198A, R208A, Q212A and R252A were well expressed and soluble. The three mutants K70A, R81A and R211A were not expressed, while R122A mutant was expressed but was not soluble. Therefore, we mutated Lys70, Arg81, Arg122 and Arg211 into aspartate. The K70D, R81D, R122D and R211D mutants were well expressed and soluble. The soluble mutants were expressed and purified under the conditions identical to those for the wild type, except that the heparin-Sepharose column step was omitted. All of the soluble mutants had similar elution profiles as the wild type upon gel filtration, and the dimer fractions were used for DNA-binding assays. We mixed 5 μl of RdgC (1.1 μg μl−1) and 5 μl of a linear dsDNA with 414 bp (63 ng μl−1), corresponding to the molar ratio of 64:1 for RdgC dimer to dsDNA. The reaction mixtures were incubated for 10 min at room temperature. RdgC–DNA complexes were analyzed by 1.5% (w/v) agarose gel electrophoresis in 1× TAE buffer (45 mM Tris-acetate at pH 8.3, 1 mM EDTA). DNA bands were visualized by ethidium bromide staining. For DNA-binding assays, the protein concentration was measured with a Bio-Rad protein assay kit.
Crystallization and X-ray data collection
Crystals were grown by the hanging-drop vapor diffusion method at 24°C by mixing equal volumes (2 μl each) of the protein solution (at 19 mg ml−1 concentration in a buffer consisting of 50 mM Tris-HCl (pH 7.4), 1 mM dithiothreitol, 1 mM EDTA and 200 mM sodium chloride) and the reservoir solution. To grow the native crystals, we used a reservoir solution consisting of 0.1 M sodium HEPES (pH 7.5), 1.0 M tri-sodium citrate and 1% (w/v) Anapoe®35 as the detergent. Native crystals grew to approximate dimensions of 0.2 mm × 0.1 mm × 0.1 mm within a few days. The SeMet-substituted protein was crystallized under conditions identical to those for the native crystals except for the presence of 10 mM dithiothreitol in the protein solution.
A crystal of the SeMet-substituted protein was dipped into a cryoprotectant solution for a few seconds and was flash-cooled in the cold nitrogen gas stream at 100 K. The cryoprotectant solution consisted of 0.1 M sodium HEPES (pH 7.5), 1.0 M tri-sodium citrate, 1% (w/v) Anapoe®35 as the detergent and 10% (v/v) glycerol. A set of Se MAD data was collected at 100 K at three different wavelengths using an Area Detector System Corporation Quantum 210 charge-coupled device detector at the beamline NW12A of Photon Factory, Tsukuba, Japan. The crystal was rotated through a total of 180° with a 1° oscillation range per frame. The raw data were processed and scaled using the program suit HKL2000 (10). The SeMet-substituted crystal belongs to the space group P21212, with unit cell parameters of a = 87.04 Å, b = 115.60 Å and c = 76.07 Å.
The native crystals were soaked for 10 min in 10 mM ethyl mercury thiosalicylate (EMTS) to collect a set of Hg MAD data at four different wavelengths. X-ray diffraction data from the EMTS-derivatized crystal, as well as a native crystal, were collected on a Bruker charge-coupled device detector at the beamline BL-6B of Pohang Light Source, Pohang, Korea. The EMTS-soaked crystal belongs to the space group P21212, with unit cell parameters of a = 87.16 Å, b = 116.02 Å and c = 74.97 Å. The native crystal belongs to the space group P21212, with unit cell parameters of a = 87.49 Å, b = 116.04 Å and c = 76.03 Å. If the presence of a dimeric molecule in the asymmetric unit is assumed, the calculated crystal volume per protein mass (VM) is 2.75 Å3 Da−1 and the solvent content is 55.3%. Supplementary Table 1 summarizes the statistics of X-ray diffraction data collection.
Structure solution and refinement
Heavy atom sites were located with the program SOLVE (11). Four Se sites from the SeMet-substituted crystal and two Hg sites from the EMTS-soaked crystal were located. Two sets of the initial Se and Hg MAD phases were separately improved using the program RESOLVE (12). Two electron density maps were independently interpreted, and the fragments of the two models were combined into a single model, since the two maps were complementary to each other. Non-crystallographic symmetry (NCS) matrices were found from a partial model built from the initial electron density map, and phases were further improved by the 2-fold NCS averaging, solvent flattening and histogram matching with the program DM (13). The model was built with O (14). The model was refined with the program CNS (15), including the bulk solvent correction. 10% of the data were randomly set aside as the test data for the calculation of Rfree (16). Several rounds of model building, simulated annealing, positional refinement and individual B-factor refinement were performed. Subsequently, this model was used to refine the structure of the native protein. The model has excellent stereochemistry, as evaluated by the program PROCHECK (17). Refinement statistics are summarized in Supplementary Table 1.
RESULTS AND DISCUSSION
Model quality and monomer structure
The structure of P. aeruginosa RdgC was solved using two sets of MAD data collected from a crystal of the selenomethionine (SeMet)-substituted protein and from a mercury derivative crystal of the native protein (Supplementary Table 1). The model has subsequently been refined using the 20.0–2.50 Å data from a native crystal to crystallographic Rwork and Rfree values of 23.4 and 27.9%, respectively, with no sigma cut-off. The refined model contains 612 residues of the two monomers in the asymmetric unit of the crystal (residues 1–306 for both chains A and B) and 87 water molecules. The C-terminal eight-residue fusion tag has no electron density in both subunits and has not been modeled. Conformations of the two monomers in the asymmetric unit are similar. The root-mean-square (r.m.s.) difference between the two monomers is 0.69 Å for 306 Cα atom pairs, with a maximum deviation of 8.8 Å for the Cα atom of Gly305. The residues giving r.m.s. differences larger than 2.0 Å are 14–16, 94–96, 201–202 and 303–306, which correspond to loops connecting secondary structure elements or the C-terminus. They are located on the surface of the dimer. Subunit A has a better map quality than subunit B, and thus has a lower mean B-factor than subunit B (40.3 Å2 versus. 56.1 Å2 for all 4788 atoms) (Supplementary Figure 1). Therefore, subunit A is chosen for discussions, unless otherwise stated.
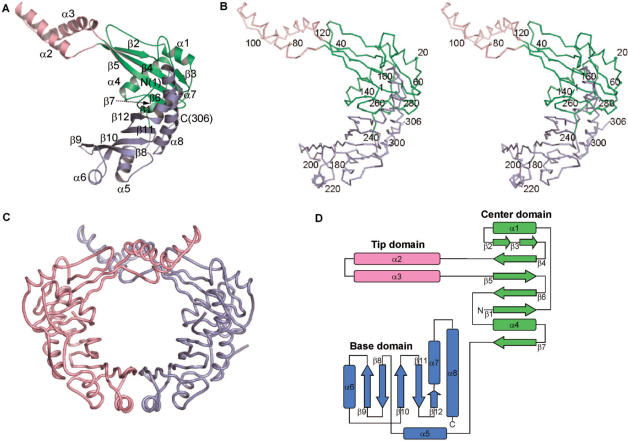
The P. aeruginosa RdgC monomer is J-shaped and has approximate dimensions of 72 Å × 60 Å × 40 Å (Figure 1). It can be divided into three structural domains: center domain, tip domain and base domain. Center domain (residues 1–73 and 121–167) includes a six-stranded anti-parallel β-sheet with two flanking α helices (α1 and α4). The helix α1 runs nearly parallel to the two outermost strands β2 and β3, which in turn lie adjacent to β4. Strands β2 and β3 are separated by a loop. The helix α4 is nearly orthogonal to the β-strands. Tip domain (residues 74–120) contains two α helices (α2 and α3) and is inserted between strands β4 and β5 of the center domain. It sticks out from the center domain. The two connections between the center domain and the tip domain are rich in conserved residues, hinting at an important functional role of these joints. They could act as a hinge for a possible conformational change. The C-terminal region of the polypeptide chain is folded into the base domain (residues 168–306), which consists of a five-stranded anti-parallel β-sheet with four α-helices, three of which (α5, α6 and α8) cover one side of the β-sheet. The long helix α8 is slightly bent.
Figure 1.
Overall structure of P. aeruginosa RdgC. (A) Ribbon diagram of a monomer. Secondary structure elements were assigned by the TOPS server (http://tops.ebi.ac.uk/tops/). (B) Stereo Cα trace of a monomer. Every tenth residue is marked by a dot and every twentieth residue is labeled. (C) Pseudomonas aeruginosa RdgC dimer. A and B chains are colored in light purple and pink, respectively. (D) Topology diagram. β-Strands are depicted as arrows and α-helices as rectangles. All figures except Figures 1D, 3 and 4D are drawn with PyMOL (DeLano, 2002, The PyMOL Molecular Graphics System, http://www.pymol.org/).
Structural similarity searches
When we searched for structural similarity against the Protein Data Bank database using the program DALI (18), the P. aeruginosa RdgC monomer showed no significant similarity with a Z score above 5. Therefore, we conclude that the overall fold of the RdgC polypeptide chain is unique. When we elaborated the structural comparisons with individual domains of P. aeruginosa RdgC, only the center domain gave a Z score above 5.
Using the center domain (residues 1–73 and 121–167) alone, the highest structural similarity is found with the human β2-adaptin appendage domain (PDB code 1E42, Z score = 7.1, r.m.s. deviation = 2.5 Å for 85 structurally aligned residues, residues 1–12, 14–30, 44–47, 58–72, 122–133 and 135–160 of P. aeruginosa RdgC chain A and residues 838–841, 848–880, 885–906 and 912–967 of the human β2-adaptin appendage domain chain A) (19). There does not appear to be any functional relatedness between these two proteins. The next highest similarity is found with the yeast TATA-box-binding protein (PDB code 1YTB, Z score = 5.7, r.m.s. deviation = 2.6 Å for 79 structurally aligned residues, residues 4–12, 14–19, 21–27, 56–70, 124–132 and 134–166 of P. aeruginosa RdgC chain A and residues 69–96, 100–126 and 132–155 of TATA-box-binding protein, N-terminal half of chain A) (20). The TATA-box-binding protein is composed of a ten-stranded antiparallel β-sheet with four flanking α-helices on the convex side of the β-sheet (20). It has an internal quasi 2-fold symmetry, and its β-strands are involved in DNA binding. There is, however, no structural similarity of functional significance, because the two center domains of the RdgC dimer are well separated and do not associate into a single unit (Figure 1C and D).
Using the tip domain (residues 74–120) alone, the ATPase domain of bovine Hsc70 chaperone shows highest structural similarity (PDB code 1BA1, Z score = 4.8, r.m.s. deviation = 1.7 Å for 45 structurally aligned residues, residues 76–120 of P. aeruginosa RdgC chain A and residues 230–253 and 256–276 of the Hsc70 ATPase domain) (21). There does not seem to be any significant functional relationship between these two proteins. The α-helix-loop-α-helix structure of the tip domain is reminiscent of the helix–hairpin–helix (HhH) DNA-binding motif that is present in a number of DNA repair enzymes (22–25). The HhH motifs are characterized by the conserved sequence motif hxxhxGhGxxxAxxxhh, where h is any hydrophobic residue (VILMWFYA) and x any residue (23). Two conserved glycines allow a tight turn between two α-helices. However, the tip domain of P. aeruginosa RdgC protein does not have such a sequence motif, and the loop in the RdgC tip domain is much more extended than that of the HhH motifs. Therefore, the RdgC tip domain is uniquely folded.
With the base domain (residues 168–306) alone, the highest similarity is found with P. aeruginosa isochorismate-pyruvate lyase (unpublished deposition; PDB code 2H9C, Z score = 4.0, r.m.s. deviation = 4.8 Å for 51 structurally aligned residues, residues 186–191, 237–240, 257–272 and 274–298 of P. aeruginosa RdgC chain A and residues 36–41, 50–53, 55–66 and 68–96 of P. aeruginosa isochorismate-pyruvate lyase chain A). There does not appear to be any functional relationship between the two proteins, and the base domain of RdgC is uniquely folded. The 101-residue chain of P. aeruginosa isochorismate-pyruvate lyase is folded into three α-helices, two of which overlap with two α-helices (α2 and α3) of the base domain of RdgC.
Ring-shaped dimer structure and inter-subunit interface
Our structure reveals that P. aeruginosa RdgC forms a ring-shaped dimer of approximate 2-fold symmetry in the crystal (Figure 1). This observation is consistent with the results of previous gel filtration and sedimentation equilibrium experiments, which suggested the presence of dimeric species of E. coli RdgC in solution (1,2). Depending on the concentration, RdgC could also exist in solution as monomers, tetramers and higher oligomers (1). In the crystal lattice, no monomeric, trimeric or tetrameric species exist. Our structure unequivocally rules out the monomer–trimer model, which fits the sedimentation equilibrium data approximately, as well as the monomer–dimer–tetramer model (1). The RdgC dimer has approximate dimensions of 80 Å × 60 Å × 40 Å, with a central hole of ∼30 Å diameter.
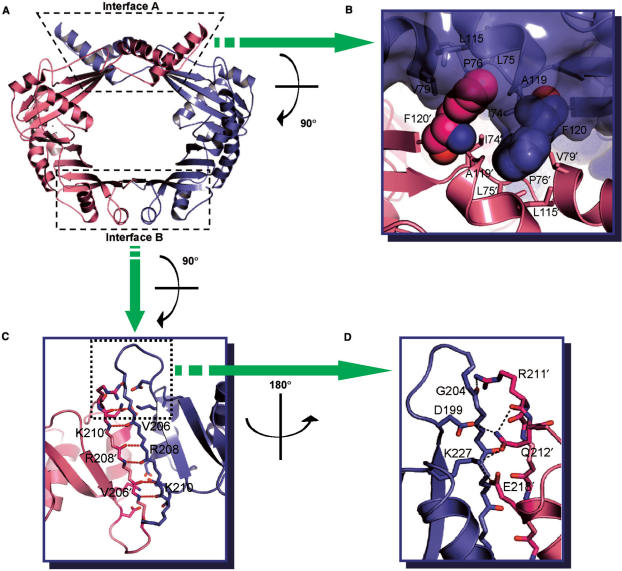
Only a moderate amount of surface area is buried upon the formation of the RdgC dimer. The monomer–monomer interface buries a solvent-accessible surface area of ∼1360 Å2 per monomer, corresponding to ∼8% of the total surface area of the monomer (Protein–Protein Interaction Server at http://www.biochem.ucl.ac.uk/bsm/PP/server/). Polar and nonpolar atoms in interface are 44 and 56%, respectively. The interface can be divided into two kinds: ‘interface A’ involving both the tip domains and center domains, and ‘interface B’ involving the base domains (Figure 2A). These two interfaces have significantly different characters.
Figure 2.
Inter-subunit interactions. (A) Ribbon diagram of RdgC dimer. The dotted boxes indicate two kinds of interfaces. (B) Interactions between tip domains. Phe120 (shown in spheres) plays a critical role in the interactions. (C), (D) Interactions between base domains. (C) Hydrogen bonding interactions involving the main chain atoms of Val206, Arg208 and Lys210. (D) A hydrogen bond network involving side chains of Asp199, Arg211, Gln212, Glu218 and Lys227.
The interface A is formed largely by the crossing over of the two tip domains in the dimer (Figure 1). This interface covers a surface area of ∼565 Å2 and is largely nonpolar, with the polar and nonpolar atoms in the interface being 37.5 and 2.5%, respectively. Phe120 contributes considerably to the nonpolar character of the interface between the tip domains, accounting for ∼150 Å2 of the interface area. Phe120 is part of a highly conserved sequence segment and lies at the boundary between the tip domain and the center domain. The aromatic ring of Phe120′ inserts into a hydrophobic pocket, which is formed by the hydrophobic residues Ile74, Leu75, Pro76, Val79, Leu115, Ala119 and Phe120 of the neighboring monomer (Figure 2B). The primed residue belongs to the adjacent subunit. These residues are well conserved in the RdgC family (indicated by green triangles below the aligned sequences in Figure 3). As these hydrophobic residues are located near the molecular 2-fold axis, the two clusters of hydrophobic side chains are merged into a single, large cluster in the dimer (Figure 2B). Of the 16 hydrogen bonds that exist between the two subunits, only one is made in this interface between Arg118 and Arg118′.
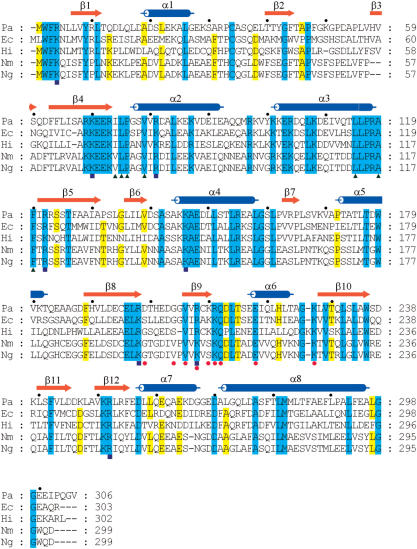
Figure 3.
Sequence alignment of the RdgC proteins. Pa, P. aeruginosa PAO1 (SWISS-PROT accession number Q9HYX7); Ec, E. coli K12 (P36767); Hi, Haemophilus influenzae (P44628); Nm, N. meningitidis serogroup B (Q9JZY2); Ng, N. gonorrhoeae strain ATCC 700825/FA 1090 (O87408). Cylinders above the sequences denote α-helices, and arrows denote β-strands. Green triangles below the sequences indicate conserved hydrophobic residues in inter-subunit interface A. Red circles indicate the residues participating in the hydrogen bond networks in inter-subunit interface B. Blue squares indicate the residues lining the central channel that are implicated for binding dsDNA through mutagenesis studies. This figure was drawn with ClustalX (36) and GeneDoc (http://www.psc.edu/biomed/genedoc/).
The interface B is contributed to exclusively by the two base domains. This interface covers a surface area of ∼795 Å2. The polar and nonpolar atoms in this interface are 50% each. The two β-sheets of the two base domains merge into a single ten-stranded antiparallel β-sheet in this interface (Figure 2A). Thus, 15 of 16 inter-subunit hydrogen bonds are clustered in this interface. The hydrogen bond networks can be divided into two types. The first type involves the main chain atoms of Val206, Arg208 and Lys210 in strand β9; this strand associates with the equivalent strand from the other subunit in an antiparallel manner (Figure 2C). The other type involves both the side chain and main chain atoms. The side chain of Arg211′ interacts with the backbone of Gly204, while the side chain of Lys227 hydrogen bonds to those of both Gln212′ and Glu218 (Figure 2D). In addition, the side chain of Gln212′ interacts with that of Asp199 and the backbone of Arg211′ (Figure 2D). Arg211, Gln212, Glu218 and Lys227 are highly conserved among the RdgC family members. The residues participating in the hydrogen bond networks are indicated by red circles below the aligned sequences in Figure 3. The observation that conserved residues play important roles in forming the dimer implies that dimerization of RdgC is critical for its function.
If one or both of interfaces A and B are to be broken for functional reasons such as for allowing dsDNA to enter into the central hole of the RdgC dimer, we can conceive of three scenarios. The first possibility would be a simultaneous breakage of the two interfaces A and B. The second would be the sequential disruption of the interfaces A and B. The third would be the disruption of only one of the two interfaces A and B. Sedimentation equilibrium indicated the presence of monomers in solution (1), which is consistent with the first two scenarios. If the third scenario holds, RdgC dimers could possibly exist in two conformations in solution, i.e. a closed form as observed in the present crystal structure, and an open form, in which one of the two interfaces A and B is broken.
A model for DNA binding by RdgC and DNA-binding assay
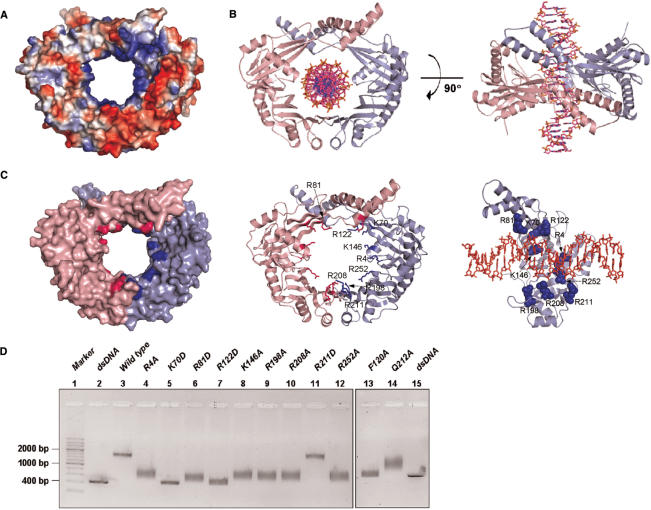
The net charge of P. aeruginosa RdgC is highly negative, since each monomer contains 12 aspartate, 13 glutamate, 10 lysine, 9 arginine and 2 histidine residues. The net charge of a dimer would be −12 (or −8), if we assume none (or all) of histidines to be positively charged. The electrostatic potential at the molecular surface of RdgC dimer shows that the charge distribution on the protein surface is highly nonuniform (Figure 4A). Since the inner surface of the RdgC dimer is rich in conserved, positively charged residues and the diameter of the central hole (∼30 Å) is approximately similar to that of other toroidal DNA-binding proteins (26), it appears likely that RdgC dimers bind dsDNA through the central hole. Despite extensive co-crystallization and soaking trials with linear dsDNAs of different lengths (14-mer, 16-mer and 18-mer), we could not locate the bound DNA experimentally. We attribute this difficulty to the nature of sequence-nonspecific DNA binding by RdgC. Therefore, we built a crude model of a DNA complex of the RdgC dimer by fitting dsDNA into the central hole (Figure 4B). This model is supported by the DNA-binding assays coupled with mutagenesis, as described below. It is also consistent with the suggested binding site size of ∼16 nt for the E. coli RdgC dimer (1) and the approximate thickness (∼40 Å) of the P. aeruginosa RdgC dimer.
Figure 4.
Model and assay for dsDNA binding by P. aeruginosa RdgC dimer. (A) Electrostatic potential at the molecular surface of RdgC dimer. The molecular surface was colored according to the electrostatic potential (positive in blue and negative in red). (B) A proposed model of the complex between RdgC dimer and dsDNA (22-mer). (C) Residues that are suggested to interact with dsDNA by the complex model. Residues selected for mutagenesis are colored in blue. DNA is shown in sticks. (D) Electrophoretic mobility shift assay for dsDNA binding by P. aeruginosa RdgC. Detailed reaction conditions are described in the Materials and methods section. Lane 1, 200 bp dsDNA ladder marker; lane 2, dsDNA (414 bp); lane 3, wild type; lane 4, R4A mutant; lane 5, K70D mutant; lane 6, R81D mutant; lane 7, R122D mutant; lane 8, K146A mutant; lane 9, R198A mutant; lane 10, R208A mutant; lane 11, R211D mutant; lane 12, R252A mutant; lane 13, F120A mutant; lane 14, Q212A mutant; lane 15, dsDNA (414 bp).
When we performed electrophoretic mobility shift assay for dsDNA binding, P. aeruginosa RdgC did not require Mg2+ ions (Supplementary Figure 2), unlike Deinococcus radiodurans RecR, which required high concentrations of Mg2+ ions (27). ATP was not required for dsDNA binding by P. aeruginosa RdgC, either (Supplementary Figure 2). This is consistent with an apparent lack of an ATP-binding pocket in the RdgC dimer structure.
In order to identify the residues that are important for dsDNA binding, we carried out mutational analyses. The residues for mutagenesis were selected on the basis of the modeled complex between RdgC dimer and dsDNA, together with sequence conservation (Figure 4C). We prepared eleven point mutants (R4A, K70D, R81D, F120A, R122D, K146A, R198A, R208A, R211D, Q212A and R252A). These eleven mutated residues are well conserved among RdgC proteins. Eight mutants (R4A, K70D, R81D, R122D, K146A, R198A, R208A and R252A) had a lower affinity for dsDNA than the wild-type RdgC (Figure 4D). These eight mutated residues are indicated by blue squares below the aligned sequences in Figure 3. This result indicates that dsDNA binds to the inside surface around the central hole of the RdgC dimer, since the mutated residues are located on the inner surface of the RdgC dimer. It is worth mentioning that a single mutation introduces two alterations per RdgC dimer.
The three mutants (F120A, R211D and Q212A) were prepared to investigate whether dimerization of RdgC is crucial for dsDNA binding. Phe120 is a key residue located in the inter-subunit interface A (Figure 2B), while Arg211 makes hydrogen bonds in the interface B (Figure 2D) and Gln212 is involved in both intra- and inter-subunit interactions (Figure 2D). dsDNA binding by the F120A mutant is considerably weakened, compared to either the R211D or Q212A mutant (Figure 4D). This result suggests that destabilization of the ring-shaped architecture of the RdgC dimer is deleterious for tight dsDNA binding and that interface A is probably more important than interface B for dsDNA binding. dsDNA-binding affinity of the Q212A mutant was lower than the wild type (Figure 4D), whereas that of the R211D mutant was comparable to the wild type (Figure 4D). This result suggests that disruption of the inter-subunit interface B is also detrimental for dsDNA binding but to a smaller extent than disruption of the interface A. It further suggests that the side chain of Arg211 is not involved in recognizing dsDNA. It is interesting that the DNA complex for wild type forms a distinct band (lane 3 in Figure 4D), as opposed to a more diffuse band as one would expect if the complexes contained different numbers of RdgC dimers. Assuming a binding site size of ∼16 nt (1), the 414-bp duplex DNA would bind ∼25 RdgC dimers if it is fully saturated.
RdgC is unique among ring-shaped DNA-binding proteins
Pseudomonas aeruginosa RdgC adds a unique member to the repertoire of toroidal DNA-binding proteins (26) such as D. radiodurans RecR (27) and DNA polymerase processivity factors, including the β subunit of E. coli DNA polymerase III (28), gp45 of T4 DNA polymerase (29) and PCNA of eukaryotic DNA polymerase δ (30,31) (Figure 5). These proteins do not have an overall sequence similarity among themselves, and their quaternary structures also vary. P. aeruginosa RdgC and E. coli β clamp are homodimers, T4 gp45 and eukaryotic PCNAs are homotrimers and D. radiodurans RecR is a homotetramer.
Figure 5.
Structural comparison of P. aeruginosa RdgC with other ring-shaped DNA-binding proteins. Ribbon diagrams and electrostatic potential at the molecular surface are shown for P. aeruginosa RdgC, gp45 of T4 DNA polymerase, human PCNA, E. coli DNA polymerase III β subunit and D. radiodurans RecR. The molecular surface was colored according to the electrostatic potential (positive in blue and negative in red).
In P. aeruginosa RdgC, the inner surface of the ring-shaped dimer is largely formed by β-strands, while the outer surface of the ring is mostly formed by α-helices (Figure 5). The predicted DNA-binding site of P. aeruginosa RdgC dimer is primarily formed by β-strands and loops. In contrast, in other ring-shaped DNA-binding proteins such as the β subunit of E. coli DNA polymerase III (28), gp45 of T4 DNA polymerase (29), PCNA of eukaryotic DNA polymerases δ (30,31) and D. radiodurans RecR (27), α-helices are located toward the center of the ring and are surrounded by β-strands in the outermost side of the ring (Figure 5). Despite a superficial resemblance of the overall ring-shaped architecture of the RdgC dimer with those of other toroidal DNA-binding proteins, the internal arrangement of the secondary structure elements in the RdgC dimer is strikingly different from those of other toroidal DNA-binding proteins. Thus, RdgC is unique among ring-shaped DNA-binding proteins.
A number of other proteins involved in DNA metabolism also adopt a ring-shaped structure despite diversity in the molecular functions (26). They include NAD+-dependent DNA ligase (25), λ exonuclease (32), hexameric helicases, topoisomerases, the trp RNA-binding attenuation protein, the bacteriophage head-to-tail connector and translin (26). There are also many examples of the proteins in DNA recombination pathways that assemble into multi-subunit toroids containing a central channel (26). Electron microscopy showed that the E. coli RuvB protein, in the presence of ATP, forms a dodecamer on double-stranded DNA in which two stacked hexameric rings encircle the DNA and are oriented in opposite directions with D6 symmetry (33). The human RAD52 protein was found to assemble into heptameric rings with a large funnel-shaped channel, 40–60 Å in diameter (34). The human DMC1 protein has so far been detected only as an octameric ring (35).
Inhibition mechanism of RecA function by RdgC
RdgC inhibits RecA-promoted DNA strand exchange, RecA-mediated ATPase activity and RecA-dependent LexA cleavage (1). It was observed that the effects of RdgC on RecA-promoted DNA strand exchange were much more pronounced than the effects of RdgC on other RecA functions, when RdgC was added to the reaction late (1). By examining the effects of the DinI protein on RdgC-mediated inhibition of RecA activities, it was concluded that the potent effect of RdgC in the inhibition of strand exchange did not primarily reflect a displacement of RecA in the nucleoprotein filament by RdgC, but instead a binding of RdgC to the dsDNA substrate so as to make it unavailable to the filaments for strand exchange (1).
Our work contributes to a better understanding of the inhibition mechanism of RecA function by RdgC. The ring-shaped architecture with a central hole that is suitable for the binding of dsDNA lends RdgC with a capacity to hold dsDNA very tightly. Therefore, we suggest that RdgC dimers probably function as a tight dsDNA gripper and physically block access to the target dsDNA by RecA filaments, because tight encircling of dsDNA by RdgC dimers, possibly as a linear aggregate, would prevent the two strands from dissociating. However, it remains unclear how the closed-ring structure of the RdgC dimer might allow binding of a large number of RdgC dimers on the duplex DNA. It may be conceivable that each RdgC dimer can open temporarily at one or both of its two inter-subunit interfaces so that dsDNA can enter the central hole.
RdgC was at its highest level during exponential phase, reaching its maximum of ∼1000 dimers per E. coli cell. Its level decreased sharply to ∼50 dimers per cell in stationary phase (2). Under normal cellular conditions, RecA is maintained at ∼1000 molecules per cell (http://www.els.net/). Following LexA repressor cleavage, the level of RecA protein in the cell increases by as much as 20-fold. RdgC inhibition of the RecA function by a simple competition mechanism at the cellular concentration of RdgC clearly requires a much higher affinity for DNA by RdgC than RecA. Possible protein–protein interactions between RdgC dimers might have a profound effect on the affinity for DNA, when a large number of RdgC dimers are bound on a duplex DNA. If only one of the two inter-subunit interfaces can open up and the 2-fold symmetry axes of the RdgC dimers are misaligned sufficiently, the escape of DNA from a linear array of interacting RdgC dimers would be extremely difficult. Alternatively, both inter-subunit interfaces of each dimer may open up simultaneously. A line of indirect evidence for the possible protein–protein interactions between RdgC dimers is provided by the cooperative binding of RdgC to DNA (7). Cooperativity of RdgC binding may be caused by local and distant protein–protein interactions (7). A kind of side-by-side interaction between two adjacent molecules of RdgC dimer is present in the crystal, burying an accessible surface area of ∼750 Å2 at the interface. The interaction involves mostly the less polar side surface of the dimer (left side of Figure 4A), with 2-fold symmetry axes of the two interacting RdgC molecules making an angle of ∼90° when viewed down the central holes and ∼15° when viewed perpendicular to the holes. This may explain the observed DNA bending by RdgC, which was speculated to be a necessary step in the working mechanism of RdgC, if one can assume that the strong protein–protein interactions in the crystal were relevant for protein–protein interactions in solution (7).
RdgC protein was shown to have a potent effect on RecA protein-promoted DNA strand exchange in E. coli (1). In a series of experiments, RdgC was added after RecA filaments had formed on the ssDNA and 5 min after the linear dsDNA was added. With 3 μM RecA, a sharp reduction in DNA strand exchange products was seen with 0.4 μM RdgC, and the generation of products was abolished at 0.8 μM RdgC (1). This suggests that the binding affinity of RdgC to dsDNA is considerably higher than that of RecA. The high affinity of RdgC could be explained by its ring shape and the suggested mode of DNA binding. Tetrameric species of E. coli RdgC, which started to form at around 2 µM concentrations, were speculated to be the species with the most potent inhibitory effect, possibly by having a higher affinity for DNA (1). Higher affinity for DNA of the tetrameric species could be understood, if the formation of a tetramer in solution involves a protein–protein interaction similar to the side-by-side interaction between two adjacent RdgC dimers in the crystal, although there is no experimental evidence. Obviously, there are still many open questions that need to be addressed in future biochemical experiments.
SUMMARY
The crystal structure of RdgC, a recombination-associated DNA-binding protein, determined in this study reveals that the polypeptide chain is uniquely folded and two monomers associate to form a ring-shaped dimer with a central hole of ∼30 Å diameter. The inner surface around the central hole is rich in positively charged residues. These structural features suggest that dsDNA runs through the central hole when it binds to RdgC dimers. This idea is supported by our mutational studies, which indicate that the conserved, positively charged residues around the central hole are important for dsDNA binding. The toroidal architecture of the RdgC dimer provides a sound framework for a better understanding of its role in homologous recombination.
Coordinates
Atomic coordinates and the structure factor data have been deposited in the Protein Data Bank (accession code 2OWY).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank the staff at beamlines BL-5A, BL-6A and NW-12A of Photon Factory, Tsukuba, Japan, and beamlines BL-4A and BL-6B of Pohang Light Source, Pohang, Korea, for assistance during X-ray diffraction experiments. This work was supported by grants from the Korean Ministry of Science and Technology (NRL-2001) and Korea Sanhak Foundation. J.Y.H., D.J.K., K.H.K., S.J.O. and H.H.L. are recipients of BK21 fellowships from the Korean Ministry of Education & Human Resources Development. Funding to pay the open Access publication charge was provided by Seoul National University.
Conflict of interest statement. None declared.
REFERENCES
- 1.Drees JC, Chitteni-Pattu S, McCaslin DR, Inman RB, Cox MM. Inhibition of RecA protein function by the RdgC protein from Escherichia coli. J. Biol. Chem. 2006;281:4708–4717. doi: 10.1074/jbc.M513592200. [DOI] [PubMed] [Google Scholar]
- 2.Moore T, McGlynn P, Ngo HP, Sharples GJ, Lloyd RG. The RdgC protein of Escherichia coli binds DNA and counters a toxic effect of RecFOR in strains lacking the replication restart protein PriA. EMBO J. 2003;22:735–745. doi: 10.1093/emboj/cdg048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ryder L, Sharples GJ, Lloyd RG. Recombination-dependent growth in exonuclease-depleted recBC sbcBC strains of Escherichia coli K-12. Genetics. 1996;143:735–745. doi: 10.1093/genetics/143.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moore T, Sharples GJ, Lloyd RG. DNA binding by the meningococcal RdgC protein, associated with pilin antigenic variation. J. Bacteriol. 2004;186:870–874. doi: 10.1128/JB.186.3.870-874.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mehr IJ, Long CD, Serkin CD, Seifert HS. A homologue of the recombination-dependent growth gene, rdgC, is involved in gonococcal pilin antigenic variation. Genetics. 2000;154:523–532. doi: 10.1093/genetics/154.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murphy LD, Rosner JL, Zimmerman SB, Esposito D. Identification of two new proteins in spermidine nucleoids isolated from Escherichia coli. J. Bacteriol. 1999;181:3842–3844. doi: 10.1128/jb.181.12.3842-3844.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tessmer I, Moore T, Lloyd RG, Wilson A, Erie DA, Allen S, Tendler SJ. AFM studies on the role of the protein RdgC in bacterial DNA recombination. J. Mol. Biol. 2005;350:254–262. doi: 10.1016/j.jmb.2005.04.043. [DOI] [PubMed] [Google Scholar]
- 8.Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 9.Miroux B, Walker JE. Over-expression of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 10.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 11.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr. 1999;D55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr. 2000;D56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 14.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. 1991;D47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 15.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 16.Brünger AT. The free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355:472–474. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 17.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 18.Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 19.Owen DJ, Vallis Y, Pearse BM, McMahon HT, Evans PR. The structure and function of the β2-adaptin appendage domain. EMBO J. 2000;19:4216–4227. doi: 10.1093/emboj/19.16.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim Y, Geiger JH, Hahn S, Sigler PB. Crystal structure of a yeast TBP/TATA-box complex. Nature. 1993;365:512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- 21.Wilbanks SM, McKay DB. Structural replacement of active site monovalent cations by the ε-amino group of lysine in the ATPase fragment of bovine Hsc70. Biochemistry. 1998;37:7456–7462. doi: 10.1021/bi973046m. [DOI] [PubMed] [Google Scholar]
- 22.Thayer MM, Ahern H, Xing D, Cunningham RP, Tainer JA. Novel DNA binding motifs in the DNA repair enzyme endonuclease III crystal structure. EMBO J. 1995;14:4108–4120. doi: 10.1002/j.1460-2075.1995.tb00083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doherty AJ, Serpell LC, Ponting CP. The helix-hairpin-helix DNA-binding motif: a structural basis for non-sequence-specific recognition of DNA. Nucleic Acids Res. 1996;24:2488–2797. doi: 10.1093/nar/24.13.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aravind L, Walker DR, Koonin EV. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 1999;27:1223–1242. doi: 10.1093/nar/27.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee JY, Chang C, Song HK, Moon J, Yang JK, Kim HK, Kwon ST, Suh SW. Crystal structure of NAD+-dependent DNA ligase: modular architecture and functional implications. EMBO J. 2000;19:1119–1129. doi: 10.1093/emboj/19.5.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hingorani MM, O’Donnell M. A tale of toroids in DNA metabolism. Nat. Rev. Mol. Cell Biol. 2000;1:22–30. doi: 10.1038/35036044. [DOI] [PubMed] [Google Scholar]
- 27.Lee BI, Kim KH, Park SJ, Eom SH, Song HK, Suh SW. Ring-shaped architecture of RecR: implications for its role in homologous recombinational DNA repair. EMBO J. 2004;23:2029–2038. doi: 10.1038/sj.emboj.7600222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kong XP, Onrust R, O’Donnell M, Kuriyan J. Three-dimensional structure of the beta subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell. 1992;69:425–437. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- 29.Moarefi I, Jeruzalmi D, Turner J, O’Donnell M, Kuriyan J. Crystal structure of the DNA polymerase processivity factor of T4 bacteriophage. J. Mol. Biol. 2000;296:1215–1223. doi: 10.1006/jmbi.1999.3511. [DOI] [PubMed] [Google Scholar]
- 30.Krishna TS, Kong XP, Gary S, Burgers PM, Kuriyan J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell. 1994;79:1233–1243. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 31.Gulbis JM, Kelman Z, Hurwitz J, O’Donnell M, Kuriyan J. Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell. 1996;87:297–306. doi: 10.1016/s0092-8674(00)81347-1. [DOI] [PubMed] [Google Scholar]
- 32.Kovall R, Matthews BW. Toroidal structure of λ-exonuclease. Science. 1997;277:1824–1827. doi: 10.1126/science.277.5333.1824. [DOI] [PubMed] [Google Scholar]
- 33.Stasiak A, Tsaneva IR, West SC, Benson CJ, Yu X, Egelman EH. The Escherichia coli RuvB branch migration protein forms double hexameric rings around DNA. Proc. Natl Acad. Sci. USA. 1994;91:7618–7622. doi: 10.1073/pnas.91.16.7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stasiak AZ, Larquet E, Stasiak A, Muller S, Engel A, Van Dyck E, West SC, Egelman EH. The human Rad52 protein exists as a heptameric ring. Curr. Biol. 2000;10:337–340. doi: 10.1016/s0960-9822(00)00385-7. [DOI] [PubMed] [Google Scholar]
- 35.Passy SI, Yu X, Li Z, Radding CM, Masson JY, West SC, Egelman EH. Human Dmc1 protein binds DNA as an octameric ring. Proc. Natl Acad. Sci. USA. 1999;96:10684–10688. doi: 10.1073/pnas.96.19.10684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.