Abstract
There is considerable interest in the use of bacteriophage vectors for mammalian cell gene transfer applications, due to their stability, excellent safety profile and inexpensive mass production. However, to date, phage vectors have been plagued by mediocre performance as gene transfer agents. This may reflect the complexity of the viral infection process in mammalian cells and the need to refine each step of this process in order to arrive at an optimal, phage-based gene transfer system. Therefore, a flexible system was designed that alowed for the introduction of multiple modifications on the surface of bacteriophage lambda. Using this novel method, multiple peptides were displayed simultaneously from both the phage head and tail. Surface head display of an ubiquitinylation motif greatly increased the efficiency of phage-mediated gene transfer in a murine macrophage cell line. Gene transfer was further increased when this peptide was displayed in combination with a tail-displayed CD40-binding motif. Overall, this work provides a novel system that can be used to rationally improve bacteriophage gene transfer vectors and shows it may be possible to enhance the efficiency of phage-mediated gene transfer by targeting and optimizing multiple steps within the viral infection pathway.
INTRODUCTION
Bacteriophage lambda has appealing characteristics as a gene and vaccine delivery vector, which include a high degree of physical stability, compatibility with rapid and inexpensive production/purification methods, genetic tractability and inherent biological safety in mammalian cells (1–3). In addition, the dimensions of the lambda phage particles are broadly similar to those of many mammalian viruses and recent structural evidence points to a shared ancestry between tailed bacteriophages and mammalian DNA viruses (4).
Lambda phage vectors have been successfully used to transfer exogenous genes to mammalian cells, following surface modification of either the phage coat protein (gpD) or the major tail protein (gpV) (5–8). Phage heads contain between 405 and 420 copies of gpD (9), while the phage tail consists of 32 rings, each containing six subunits of gpV (10–12). Thus, both of these proteins can be used to display foreign proteins or peptides at high copy numbers on the surface of lambda phage particles.
Most attempts to enhance lambda phage-mediated gene transfer to mammalian cells have concentrated on optimizing the binding of phage particles to mammalian cells (7,8,13,14). However, the eukaryotic cell poses numerous barriers to phage-mediated gene transfer. After receptor binding and internalization, phage must gain access to the cytoplasm, uncoat and deliver their DNA payload to the nucleus. Thus, the ultimate success of phage-mediated gene transfer depends on the ability to overcome multiple intracellular barriers. This is likely to require the use of combination strategies that increase the efficiency of each step involved in phage-mediated gene transfer, including cell attachment, cytoplasmic entry, endosomal escape, uncoating and nuclear import (7,15,16).
This article reports the design and development of a novel and tractable lambda-based vector that allows for the facile generation of phage particles that display multiple peptides or proteins of interest on their surface. We hypothesized that this system could surmount current obstacles to efficient phage-mediated gene transfer, by generating phage vectors that display a combination of exogenous peptides each intended to circumvent a separate barrier to efficient phage gene delivery. This article presents the first example of single lambda phage constructs incorporating multiple surface modifications that, collectively, enhanced in vitro gene transfer to mammalian cells.
METHODS
Lysogens
The λD1180(luc) lysogen was a gift from Dr Mahito Nakanishi and DNAVEC Corporation (6); λD1180(luc) contains a firefly luciferase gene under the transcriptional control of the major human cytomegalovirus (CMV) immediate-early promoter. Lysogen λD1180 (Dam15 del EcoRI-SacI cIts857 nin5 Sam100) is deficient in gpD, the S lysis gene, and contains a temperature-sensitive mutation in the cI repressor. The E. coli lysogen host (Top10, Invitrogen) is sup0, ensuring that neither Sam100 nor Dam15 are expressed.
Plasmid design
pTrc:gpD-Fusion and pTrc:gpD
The plasmids pTrc:gpD and pTrc:gpD-Fusion were derived from pTrcHis (Invitrogen) with the addition of a synthetic, codon-optimized D gene as described (17). The D gene was inserted as an NcoI to BamHI fragment, with subsequent loss of the NcoI site. A linker sequence [G(SGGG)2SGGT] was then added (BamHI to KpnI), to permit insertion of DNA sequences encoding exogenous peptides of interest between the sites KpnI and HindIII. The plasmid pTrc:gpD was constructed with the addition of a stop codon immediately after the codon optimized D gene.
pTrc:gpD-UBHA
A ubiquitinylation motif derived from the Hepatitis A Virus (HAV) 3 protease [LGVKDDWLLV; (18)] was constructed by overlapping PCR using the primers: UBHAfor (5′-AACCTGGGTACCTTAGGCGTTAAAGATGACTGGTTGCTG-3′) and UBHArev (5′-CAGGCTAAGCTTCTACACCAGCAACCAGTCATCTTTAAC-3′; underlining denotes the translational stop codon). The PCR product was digested with KpnI and HindIII and cloned into pTrc:gpD-Fusion to create pTrc:gpD-UBHA.
pTrcRSF:gpV-Fusion
The gpV expression plasmid was constructed from pTrcHis. First, the truncated gpV gene was PCR amplified from the phage genome using the primers pVForA (5′-AGCTCCATGGCGCCTGTACCAAATCCTACAATG-3′) and pV’RevLink (5′-AGCTGGATCCCCCTTTCACCACCGAGGTGC-3′). The PCR product was digested with BamHI and NcoI and cloned into pTrcHis at the corresponding sites. A linker with the sequence G(SGGG)8T was synthetically constructed by GeneART (Regensburg, Germany) and inserted as a BamHI to KpnI fragment. DNA sequences encoding the exogenous fusion peptides of interest were then inserted as KpnI to HindIII fragments, with the addition of a translational stop codon at the end of each fusion sequence.
The RSF plasmid origin of DNA replication and a linked kanamycin resistance cassette from pRSF-1b (Novagen) were inserted into the pTrc-based plasmid by replacement of the pBR-based origin and flanking antibiotic cassette. This was achieved by PCR amplification of the corresponding DNA fragment from pRSF-1b using the primers CDFDUETFOR (5′-AGCTCCATGGGAAGCACACGGTCACACTGCT-3′) and CDFDUETREV (5′-AGCTGCATGCAAGTTAGCTCACTCATTAGGGA-3′). The PCR product was partially digested with SphI and NcoI and then ligated with the promoter region from the original pTrc-based, gpV expression plasmid (which was isolated following digestion of the original pTrc:gpV-Fusion plasmid with SphI and BspHI).
pTrcRSF:gpV-CD40
A DNA sequence encoding the CD40 receptor-binding peptide, ATYSEFPGNLKP (19), was subcloned from the parent plasmid pAT-043-CD40 using KpnI and HindIII digests and then cloned into pTrcRSF:gpV-Fusion at the corresponding restriction sites to create pTrcRSF:gpV-CD40.
Production of polyclonal antiserum
The production of the polyclonal anti-gpD antiserum has been described (17). Polyclonal anti-gpV antiserum was produced by transforming the plasmid pBAD:gpVtrunc into Top10 E. coli (Invitrogen). Transformed bacteria were grown to mid-log phase and then induced for 3 h with 0.002% of L-arabinose. After expression, bacteria were lysed with BugBuster (Novagen), treated with benzonase and lysozyme and cleared by centrifugation. The lysate was purified on a Ni–NTA matrix and fractions containing the purified gpV protein were identified on immunoblot with an anti-His antibody. SDS-PAGE purified fractions containing the truncated gpV protein were submitted to Sigma Genosys for the production of rabbit anti-gpV antiserum. Antiserum reactivity was verified by immunoblot and aliquots were stored at −80°C.
Phage production
Chemically competent lysogens of λD1180 were transformed with purified DNA plasmids of interest, and transformants were then selected using media containing the appropriate antibiotic. Single colonies were then grown overnight at 32°C with antibiotic selection, and the resulting cultures were then used to inoculate 1 l of fresh antibiotic-containing medium the next day (at a dilution of 1:100). Cultures were grown at 32°C with vigorous shaking (300 r.p.m.) until an OD600 of between 0.3 and 0.4 was reached. Lysogens were induced by transferring the bacteria to a water bath set between 51 and 53°C, followed by incubation with gentle shaking for 20 min. After thermal induction, the cultures were vigorously shaken for an additional 3 h at 38°C. Bacteria were then pelleted and resuspended in phage suspension media (SM: 50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 10 mM MgSO4, 0.01% gelatin) and lysed with the addition of 10% chloroform. After chloroform treatment, bacterial DNA was lysed with DNaseI at a final concentration of 10 µg/ml. The lysate was then cleared of cellular debris by low-speed centrifugation and phages were pelleted by ultracentrifugation at 110 000 g. Pelleted phage particles were subsequently purified by cesium chloride equilibrium density gradient centrifugation and phage were dialyzed against dialysis buffer (10 mM NaCl, 50 mM Tris-HCl, pH 8.0, 10 mM MgCl2), prior to storage at 4°C. Typical titers were 1 × 1012 p.f.u./ml.
Immunoblot analysis of purified phage
Phage was denatured and structural proteins separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). Gels for the detection of gpD contained between 15 and 20% polyacrylamide, while gels for gpV contained between 12 and 15% polyacrylamide. Phage proteins were detected by western blotting with the appropriate primary antibody (rabbit polyclonal antisera specific for either gpD or gpV, which were used at dilutions of 1:1000 or 1:2000, respectively). Detection of bound rabbit antibody was achieved by incubation with a 1:3000 dilution of anti-rabbit HRP (Sigma) for 1 h, followed by detection with ECL-Plus Substrate (Amersham Biosciences).
Detection of CD40 expression on RAW 264.7 cells
RAW 264.7 cells were purchased from the American Type Culture Collection (ATCC) and maintained according to ATCC recommendations. For detection of the CD40 receptor 1 × 106 cells were stained for 15 min on ice with 1:200 of PE-CD40 or an appropriate isotype control antibody (BD Pharmingen). After staining, cells were washed with PBS, resuspended in FACS buffer, and analyzed by fluorescence-activated cell sorting using a FACSCalibur instrument.
Luciferase assay for phage-mediated gene expression in RAW 264.7 cells
RAW 264.7 cells were plated at 1 × 105 cells/well in 12-well plates, 12 h before the planned phage transduction. After the 12-h incubation, the media was removed and replaced with lambda phage (1 × 1011 p.f.u.) diluted in 1 ml of serum-containing medium. Infection of cells was centrifugally enhanced [1200 g for 1 h at 37°C; ‘spinoculation’ (20–22)] and cells were subsequently incubated with the phage for 48 h at 37°C. Cells were then either used for quantitative DNA PCR analysis (see below) or used for luciferase assays. For the latter, cells were washed with phosphate buffered saline (PBS) and then lysed in 30 µl of Promega's passive lysis buffer (PBL) for luciferase analysis. Protein concentration in the lysates was determined by Bradford assay, and equal protein quantities were used in luciferase assays, utilizing a luciferase assay kit (Promega).
Quantitative DNA PCR assay for phage genome copies in RAW 264.7 cells
RAW 264.7 cells were transduced with lambda phage particles as described above. At the end of the 48-h incubation, cells were washed four times with PBS and total genomic DNA was extracted using the Wizard Genomic DNA Extraction Kit (Promega). Total genomic DNA was quantitated spectrophotometrically and 10 ng was then used for QPCR analysis using a luciferase-specific Taqman probe (5′-CATTTCGCAGCCTACCGTGGTGTT-3′) together with the reverse primer TaqLucR (5′-TTGCAACCCCTTTTTGGAAA-3′) and the forward primer TaqLucF (5′-AACGTGAATTGCTCAACAGTATGG-3′). The amplification conditions comprised an initial denaturation step at 95°C for 10 min followed by 40 cycles at 95°C for 15 s and 60°C for 1 min. A standard curve for lambda DNA quantitation was generated by using serially diluted phage DNA and samples were analyzed with a Bio-Rad iCycler. All standards and samples were assayed in triplicate.
RESULTS
Incorporation of truncated gpV into phage tails
We previously designed and evaluated a plasmid-based, trans-complementation system that allowed the incorporation of recalcitrant peptides into the head of phage lambda (17). We reasoned that a similar approach might also permit the display of exogenous peptides on the major tail protein, gpV.
Isolation of naturally occurring mutations in gpV indicate that up to one-third of the carboxyl terminus is unnecessary and can be deleted without adversely effecting tail assembly (23). Therefore, exogenous peptides have been fused to the C-terminus of either full-length or truncated gpV, and displayed on the tail of lambda particles (24–26). Pilot experiments showed that a truncated gpV protein bearing a hexahistidine tag was efficiently incorporated into our phage particles (not shown). Therefore, the truncated gpV gene was used in all subsequent experiments.
Selection of peptide motifs for display on the head and tail of bacteriophage lambda particles
Successful gene transfer by bacteriophage vectors is likely to depend on the circumvention of multiple intracellular barriers to efficient gene expression. Therefore, a vector displaying multiple modifications, each intended to address a distinct barrier to gene delivery, may result in more efficient gene transfer when compared to phage that display only a single modification on their surface.
Uncoating represents an important, but little studied, aspect of phage-mediated gene transfer that may be enhanced by the display of specific peptide motifs on the phage surface. We were therefore intrigued by reports that gene transduction by a number of mammalian viruses can be improved in the presence of proteasome inhibitors (27–29). This suggests that some viruses may use ubiquitinylation and proteasomal degradation to trigger head uncoating and genomic release. There is also evidence that ubiquitinylation, especially mono-ubiquitinylation, plays an important role in vesicular trafficking (30) and that ubiquitin can serve as a signal for localization to multi-vesicular bodies (31). With this in mind, we set about displaying an ubiquitinylation motif from the hepatitis A virus 3C protease (UBHA) on the surface of lambda phage particles. To do this, the ubiquitinylation motif (LGVKDDWLLV) was fused to the C-terminus of gpD, and then displayed on phage particles (17).
In order to test the feasibility of simultaneously co-expressing two different peptide motifs on the head and tail of lambda phage particles, we selected a second, receptor-binding peptide for display on the major tail protein, gpV. For this purpose, we elected to display a CD40-binding peptide (ATYSEFPGNLKP) that was previously identified in our laboratory (19). We therefore designed the plasmid pTrcRSF:gpV-CD40 to display this peptide on the major lambda phage tail protein, gpV.
Generation of bi-functional phage particles with modifications to both the head and tail
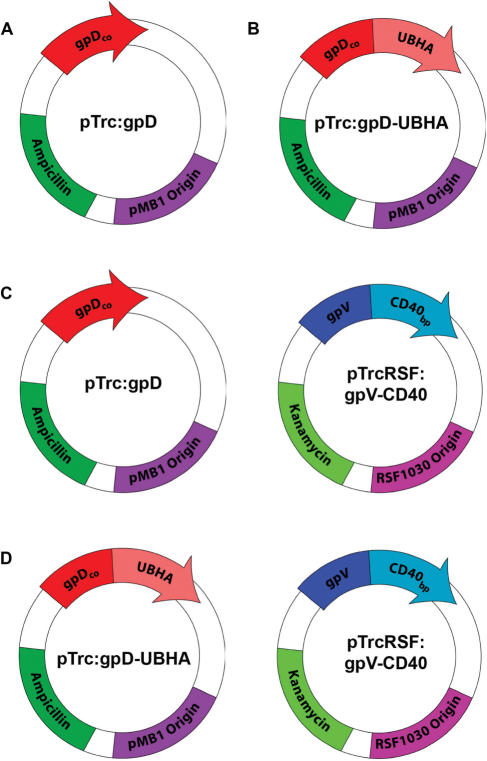
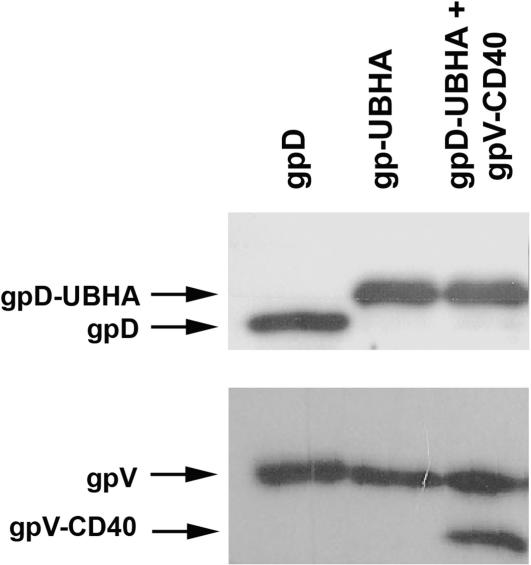
The lysogen λD1180(luc) was transformed with either pTrc:gpD or pTrc:gpD-UBHA to produce phage particles that displayed either wild-type gpD (Figure 1A) or a modified gpD head protein fused to a ubiquitinylation motif (Figure 1B). To produce phage particles that displayed a modified gpV tail protein fused to a CD40-binding peptide, the λD1180(luc) lysogen was transformed with pTrcRSF:gpV-CD40 and pTrc:gpD (Figure 1C). Finally, bifunctional phage displaying both peptide modifications were produced by dual transformation with pTrc:gpD-UBHA and pTrcRSF:gpV-CD40 into λD1180(luc) lysogens (Figure 1D). Phage particles were then produced following lytic induction of the lysogen, and CsCl density-gradient-purified phage particles were analyzed by western blot with antisera specific for gpD or gpV. As expected, introduction of pTrc:gpD-UBHA into the lysogen resulted in the generation of progeny phage particles that had full replacement of wild-type gpD head protein with gpD-UBHA (Figure 2; top panel). Similarly, introduction of the expression plasmid for gpV-CD40 into the lysogen resulted in the generation of progeny phage particles that had ∼50% replacement of wild-type gpV tail protein with the gpV-CD40 fusion protein (Figure 2; lower panel). Full replacement of wild-type gpV on the phage tail was not expected, since the lambda lysogen contains a functional wild-type gpV gene. In contrast, the lysogen contains an amber-mutated gpD gene that permits full replacement of gpD by exogenously expressed recombinant gpD, in host cells that lack the amber suppressor tRNA. In addition to the partial replacement of gpV that is evident in Figure 2 (lower panel), it can also be appreciated that the gpV-CD40 fusion protein is smaller than the endogenous gpV protein. This is because we used the truncated form of gpV to construct the gpV-CD40 fusion (see Materials and methods section).
Figure 1.
Expression plasmids for the production of bi-functional phage. Production of bi-functional phage displaying peptide modifications on both gpD and gpV was achieved by transformation of lambda lysogens with a series of compatible plasmid expression constructs. (A) pTrc:gpD for the expression of unmodified wild-type gpD. (B) pTrc:gpD-UBHA for the expression of gpD displaying a ubiquitinylation motif from the hepatitis A virus 3C protease (UBHA). (C) pTrc:gpD co-transformed with pTrcRSF:gpV-CD40 for the expression of wild-type gpD in conjunction with gpV displaying a CD40-binding peptide (CD40). (D) pTrc:gpD-UBHA co-transformed with pTrcRSF:gpV-CD40 for the simultaneous expression of gpD-UBHA and gpV-CD40.
Figure 2.
Co-transformation of gpD-UBHA and gpV-CD40 expression plasmids into lysogens yields bi-functional phage displaying both peptide fusions. The plasmid pTrc:gpD-UBHA and the plasmid pTrcRSF:gpV-CD40 were co-transformed into lambda lysogens. After lytic induction, phage were purified by CsCl density gradient centrifugation and analyzed. (A) Phage (5 × 108 p.f.u.) were loaded on a 12% polyacrylamide gel, subjected to SDS-PAGE and transferred to nitrocellulose. Immunoblot analysis was then performed using an anti-gpD rabbit polyclonal antiserum. This revealed full replacement of gpD with gpD-UBHA in phage produced from lysogens carrying the pTrc:gpD-UBHA plasmid. The molecular weight of gpD is ∼12 kDa and of gpD-UBHA is ∼16 kDa. (B) Phage (5 × 108 p.f.u.) were loaded on a 12% polyacrylamide gel, subjected to SDS-PAGE and transferred to nitrocellulose. Immunoblot analysis was then performed with an anti-gpV polyclonal antiserum. This revealed that the phage expressing the gpV-CD40 fusion protein contained roughly a 1:1 ratio of wild-type gpV and recombinant gpV-CD40. The projected molecular weights are ∼30 kDa for wild type gpV and ∼27 kDa for recombinant gpV-CD40 (Note that the recombinant gpV fusion protein is based on a truncated form of gpV, and is therefore smaller than its wild-type counterpart).
Display of two distinct peptide modifications on the surface of lambda phage results in an enhancement of phage-mediated gene transfer
We hypothesized that phage particles displaying both a CD40-binding motif on the phage tail and an ubiquitinylation motif on the phage head would be capable of mediating more efficient gene transfer in CD40-positive mammalian cells, when compared to unmodified phage particles or phage particles that displayed only a single capsid modification.
In order to test this prediction, we performed in vitro gene transfer experiments using the RAW 264.7 cell line. This murine macrophage cell line retains key properties of primary macrophages, including (i) cell surface expression of modest levels of CD40 and (ii) the ability to take up exogenous particles and macromolecules by phagocytosis (32–35). The phagocytic competence of RAW cells may be important since phagocytosis has been proposed to represent the major mechanism by which lambda phage vectors transduce cells in vivo (36).
RAW 264.7 cells were exposed to luciferase-encoding phage particles at high, intermediate and low multiplicities of infection (MOI = 1 × 106, 3 × 105 or 1 × 105, respectively). Forty-eight hours later, the cells were washed extensively and extracts were prepared for analysis of luciferase expression levels.
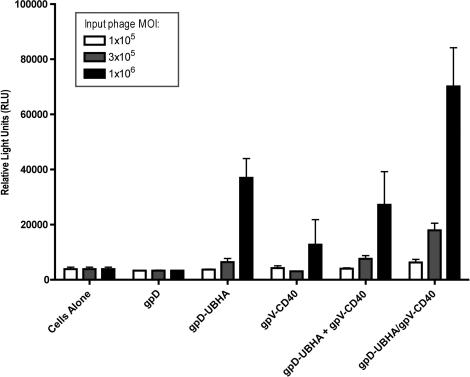
Unmodified phage particles (gpD) and particles which displayed only the CD40-binding peptide (gpV-CD40) did not mediate detectable levels of luciferase gene expression when compared to control cell lysates (Figure 3; note that there is a modest but non-statistically significant trend towards increased gene transduction in the case of the gpV-CD40 phage, at the high-dose level).
Figure 3.
Display of two distinct modifications on the phage capsid results in a dose-dependent enhancement of phage-mediated gene transfer in RAW 264.7 cells. RAW 264.7 cells were incubated with phage particles at the indicated MOIs (high, intermediate and low doses corresponding respectively to 1 × 106, 3 × 105 or 1 × 105 p.f.u./cell). After an initial centrifugal enhancement step (1200 g, 1 h), cells were incubated with the phage for 48 h at 37°C. Cells were then washed, lysed, and analyzed for luciferase expression. Luciferase assays were performed using equal total protein quantities for each sample. Unmodified phage particles (gpD) and particles which displayed only the CD40-binding peptide (gpV-CD40) did not mediate levels of luciferase gene expression that were significantly above background, when compared to control cell lysates; a modest but non-statistically significant trend towards increased gene transduction was observed in the case of the gpV-CD40 phage at the high dose level. In contrast, surface display of gpD-UBHA produced a 10-fold increase in gene expression above baseline at the high dose level (P < 0.01, when compared to unmodified phage) and a modest (1.7-fold) but non-statistically significant increase at the intermediate dose level. Gene transfer efficiency was also tested in cells that were exposed to a simple mixture of phage particles that displayed either gpV-CD40 or gpD-UBHA on their surface (designated as ‘gpD-UBHA + gpV-CD40’). In this case, the full-indicated dose of each phage was added to the cells (i.e. at the high dose levels, cells received 1 × 106 p.f.u. of gpV-CD40 phage plus 1 × 106 p.f.u. of gpD-UBHA phage). Exposure of cells to this mixture of phage particles resulted in levels of gene transfer that were very similar to those elicited by the gpD-UBHA phage alone. There was a statistically significant 7-fold increase in gene transfer efficiency at the high dose group level (P < 0.01, compared to unmodified phage) and a modest (2-fold) but non-statistically significant increase at the intermediate dose level. Finally, cells were exposed to the bi-functional phage, which displayed both gpV-CD40 with gpD-UBHA on its surface (designated gpD-UBHA/gpV-CD40). This resulted in a further increase in phage-mediated gene expression (by ∼2-fold) when compared to the gpD-UBHA head modification alone, or to the simple mixture of gpD-UBHA and gpV-CD40 phage, at both the high and intermediate dose levels. At the intermediate dose level, this result achieved statistical significance (P < 0.01, compared to cells exposed either to gpD-UBHA phage alone or to the mixture of gpD-UBHA + gpV-CD40 phage). At the high dose level, the result achieved statistical significance when compared to the mixture of gpD-UBHA + gpV-CD40 phage (P < 0.05), but not when compared to the gpD-UBHA phage alone. Statistical analysis was performed using one-way ANOVA with Tukey's post-test; significance was taken as P < 0.05.
In contrast, phage that displayed the ubiquitinylation motif (UBHA) on their surface were able to successfully mediate gene transfer in RAW 264.7 cells, leading to luciferase levels that were roughly 10-fold greater than background, at the high dose level (P < 0.01); gene transfer was not significantly above background at the intermediate dose level. Very similar results were obtained following exposure of cells to a simple mixture of phage particles that displayed either gpV-CD40 or gpD-UBHA on their surface (groups designated as ‘gpD-UBHA + gpV-CD40’ in Figure 3).
Gene transfer was further enhanced (by ∼2-fold) when the UBHA motif was co-displayed on the phage surface together with the CD40-binding peptide (groups designated as ‘gpD-UBHA/gpV-CD40’ in Figure 3). At the intermediate dose level, this result achieved statistical significance (P < 0.01) when compared either to gpD-UBHA phage alone or to the mixture of gpD-UBHA + gpV-CD40 phage.
Display of the CD40-binding peptide on the surface of lambda phage results in an enhancement of phage-mediated DNA uptake into cells
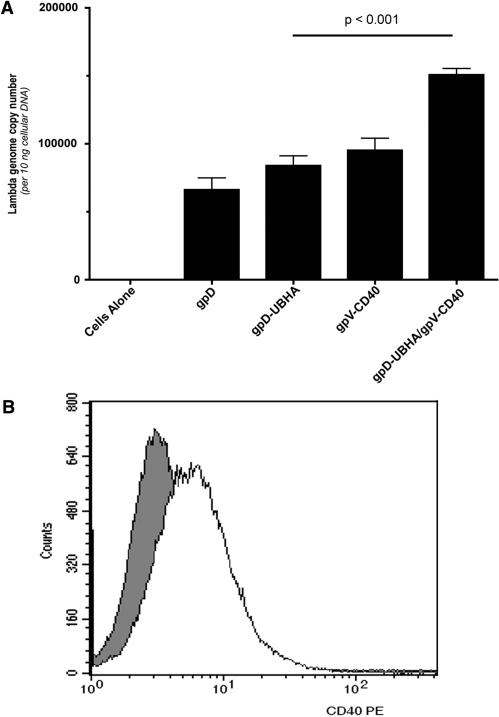
To further examine the effect of displaying the CD40-binding peptide on the phage surface, the efficiency of phage genome uptake into cultured RAW 264.7 cells was analyzed. The cells were confirmed to express CD40 by flow cytometric analysis (Figure 4B), and were then exposed to each of the phage constructs at a fixed MOI (1 × 106). Forty-eight hours later, cells were washed thoroughly, and lysates prepared. Analysis of lambda genome copy number in lysates from transduced cells revealed that DNA copy number was slightly, but significantly (P < 0.05), greater in cells which were transduced with phage particles that displayed gpV-CD40 versus unmodified phage (Figure 4A). Lambda genome copy number was further elevated in cells which were transduced with phage particles that displayed gpD-UBHA plus gpV-CD40 versus phage particles that displayed either gpD-UBHA or gpV-CD40 alone (Figure 4A; P < 0.001). This suggests that surface display of the CD40-binding peptide enhances phage-mediated gene transfer at the level of either phage binding or internalization (leading to an increase in the number of phage genome copies in the cell).
Figure 4.
Display of a CD40-targeting peptide on the phage capsid results in an enhancement of phage-mediated DNA uptake transfer into RAW 264.7 cells. (A) RAW 264.7 cells were incubated with phage particles at an MOI of 1 × 106 p.f.u. After an initial centrifugal enhancement step (1200 g, 1 h), cells were incubated with the phage for 48 h at 37°C, and then washed and lysed. Analysis of lambda genome copy number was conducted using 10 ng of total input cellular DNA prepared from the cell lysates. DNA copy number was slightly, but significantly (P < 0.05), greater in cells which were transduced with phage particles that displayed gpV-CD40 versus unmodified phage (gpD). The bi-functional phage, combining gpV-CD40 with gpD-UBHA, further increased lambda genome copy number (P < 0.001, when compared to phage displaying gpD-UBHA or gpV-CD40 alone). Surface display of the UBHA peptide did not lead to a statistically significant increase in the number of phage genome copies per cell, when compared to unmodified phage particles (gpD). Statistical analysis was performed using one-way ANOVA with Tukey's post-test; significance was taken as P < 0.05. (B) RAW 264.7 cells were stained with an anti-CD40-PE antibody or an isotype control antibody, and stained cells were analyzed by flow cytometry. The darkly shaded curve represents staining with the isotype control antibody, and the unshaded curve represents staining of cells with the CD40-specific antibody.
Analysis of phage genome copies in transduced cells also revealed that surface display of the UBHA peptide did not lead to an increase in the number of phage genome copies per cell, when compared to unmodified phage particles (Figure 4A; compare gpD and gpD-UBHA). Since surface display of the UBHA peptide resulted in a profound increase in phage-mediated gene expression (Figure 3), we conclude that surface display of the UBHA peptide enhances phage-mediated gene transfer at a post-internalization step. This may reflect an effect on intracellular trafficking of phage particles, on phage uncoating or on another ubiquitin/proteasome-dependent pathway (29).
DISCUSSION
Multiple phage capsid proteins suitable for peptide display have been identified in both lambda and M13. Despite this, simultaneous display from two or more protein platforms has rarely been described, even in the more widely studied filamentous phage display system (37–40). Recently, a bifunctional filamentous phage intended for delivering biological agents was described (40). The phage combined an integrin-targeting moiety at pIII with a streptavidin-binding sequence at pVIII. The authors tested their phage in vitro for receptor specific cell binding and internalization. In addition, they complexed their phage with quantum dots and then demonstrated tumor-specific accumulation after intravenous injection in mice. Despite these few successes, the design of bifunctional phage remains underexplored and underutilized. In 1996, Dunn proposed a bifunctional lambda phage combining gpD and gpV display, yet, until now, a bifunctional lambda phage has never been described (8).
The experiments reported here resulted in the simultaneous co-display of two different peptide modifications on the head and tail of lambda phage particles, as translational fusions to gpD and gpV, respectively. In both cases, a high copy number of the displayed peptide was achieved (full replacement, or roughly 400 copies/phage particle in the case of the gpD fusion protein and partial replacement, or roughly 100 copies/phage particle in the case of the gpV fusion protein). Moreover, because the gpD and gpV expression plasmids described here are compatible with the previously described, CDF-origin-based plasmid, pTrcCDF:gpD-Fusion (17), it should be possible to introduce up to three modifications to a single phage particle in the future (two to gpD, and one to gpV).
The long-term goal of our experiments is to develop a bacteriophage lambda vector system that is capable of mediating efficient gene transfer into mammalian cells. As a first step towards this goal, we evaluated phage-mediated gene transfer in a murine macrophage cell line using recombinant lambda phage particles that encoded a luciferase reporter gene. These experiments showed that the surface display of an ubiquitinylation motif resulted in a profound enhancement of phage-mediated gene transfer.
The ubiquitinylation motif may enhance phage-mediated gene transfer as a result of proteasome-mediated uncoating of the phage particle, or because of effects on the intracellular trafficking of internalized phage particles (30), and possible localization of internalized phage particles to multi-vesicular bodies (31), or due to effects on other intracellular pathways (29). It is noteworthy that certain mammalian viruses such as Vesicular Stomatitis Virus (VSV) rely on multi-vesicular bodies for endosomal escape and efficient infection of host cells (41,42). Attempts to resolve the mechanism by which the displayed ubiquitinylation motif enhances phage-mediated gene transfer were inconclusive, since the well-characterized proteasome inhibitors MG-132 and lactacystin proved toxic to the RAW 264.7 cells, even at the minimum effective dose. Thus, future studies will be needed to investigate the mechanism by which the ubiquitinylation motif enhances phage-mediated gene transfer.
Our proof-of-principle experiments also evaluated whether the simultaneous display of two different modifications on the phage surface may further enhance phage-mediated gene transfer. To do this, we generated luciferase-encoding phage particles that displayed a receptor (CD40)-binding peptide on their major tail protein (gpV) in addition to the ubiquitinylation motif on their major head protein (gpD). These bifunctional phage particles were able to mediate an enhanced efficiency of gene transfer into a cultured murine macrophage cell line, when compared to phage particles that displayed only a single peptide moiety on their surface. This effect was dose-dependent, with higher levels of gene transfer being detected when larger amounts of phage were added to cells. Furthermore, co-display of both motifs on the same phage particle was required for the observed enhancement of phage-mediated gene transfer; gene transfer efficiency was significantly improved when compared to a simple mixture of gpD-UBHA phage plus gpV-CD40 phage. Thus, the enhanced gene transfer effect by the dual-display construct cannot be attributed to a trans-effect.
Overall, the results reported provide strong support for the notion that it may be possible to rationally improve the efficiency of phage-mediated gene transfer by displaying several different peptides on the phage surface. In the future it should be possible to introduce and test other modifications with the intention of eventually designing a highly efficient phage-based gene delivery vector.
ACKNOWLEDGEMENTS
The authors would like to thank Ketna Volcy for her assistance in the production and purification of the gpV protein used for antibody production, Dr Stan Hattman for providing laboratory space, Drs Margaret Lieb and Ron Hoess for helpful discussions, Dr Andreas Plückthun and Dr Patrick Forrer (University of Zurich) for providing gpD-encoding plasmids, and Dr Mahito Nakanishi and DNAVEC Corporation for providing λ phage vectors [λD1180 (Luc)]. This work was supported by NIH grants T32 AI007362 and T32 GM007356 (to C.N.Z.), T32 DE007165 (to B.B.T.), R01 DE14914 (to R.S.) and R21 AI058791 (to S.D.). Funding to pay the Open Access publication charges for this article was provided by intramural funds from the University of Rochester School of Medicine and Dentistry.
Conflict of interest statement. None declared.
REFERENCES
- 1.Jepson CD, March JB. Bacteriophage lambda is a highly stable DNA vaccine delivery vehicle. Vaccine. 2004;22:2413–2419. doi: 10.1016/j.vaccine.2003.11.065. [DOI] [PubMed] [Google Scholar]
- 2.Chauthaiwale VM, Therwath A, Deshpande VV. Bacteriophage lambda as a cloning vector. Microbiol. Rev. 1992;56:577–591. doi: 10.1128/mr.56.4.577-591.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maniatis T, Hardison RC, Lacy E, Lauer J, O'Connell C, Quon D, Sim GK, Efstratiadis A. The isolation of structural genes from libraries of eucaryotic DNA. Cell. 1978;15:687–701. doi: 10.1016/0092-8674(78)90036-3. [DOI] [PubMed] [Google Scholar]
- 4.Baker ML, Jiang W, Rixon FJ, Chiu W. Common ancestry of herpesviruses and tailed DNA bacteriophages. J. Virol. 2005;79:14967–14970. doi: 10.1128/JVI.79.23.14967-14970.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoess RH. Bacteriophage lambda as a vehicle for peptide and protein display. Curr. Pharm. Biotechnol. 2002;3:23–28. doi: 10.2174/1389201023378481. [DOI] [PubMed] [Google Scholar]
- 6.Eguchi A, Akuta T, Okuyama H, Senda T, Yokoi H, Inokuchi H, Fujita S, Hayakawa T, Takeda K, et al. Protein transduction domain of HIV-1 Tat protein promotes efficient delivery of DNA into mammalian cells. J. Biol. Chem. 2001;276:26204–26210. doi: 10.1074/jbc.M010625200. [DOI] [PubMed] [Google Scholar]
- 7.Piersanti S, Cherubini G, Martina Y, Salone B, Avitabile D, Grosso F, Cundari E, Di Zenzo G, Saggio I. Mammalian cell transduction and internalization properties of lambda phages displaying the full-length adenoviral penton base or its central domain. J. Mol. Med. 2004;82:467–476. doi: 10.1007/s00109-004-0543-2. [DOI] [PubMed] [Google Scholar]
- 8.Dunn IS. Mammalian cell binding and transfection mediated by surface-modified bacteriophage lambda. Biochimie. 1996;78:856–861. doi: 10.1016/s0300-9084(97)84338-6. [DOI] [PubMed] [Google Scholar]
- 9.Yang F, Forrer P, Dauter Z, Conway JF, Cheng N, Cerritelli ME, Steven AC, Pluckthun A, Wlodawer A. Novel fold and capsid-binding properties of the lambda-phage display platform protein gpD. Nat. Struct. Biol. 2000;7:230–237. doi: 10.1038/73347. [DOI] [PubMed] [Google Scholar]
- 10.Buchwald M, Murialdo H, Siminovitch L. The morphogenesis of bacteriophage lambda. II. Identification of the principal structural proteins. Virology. 1970;42:390–400. doi: 10.1016/0042-6822(70)90282-5. [DOI] [PubMed] [Google Scholar]
- 11.Buchwald M, Steed-Glaister P, Siminovitch L. The morphogenesis of bacteriophage lambda. I. Purification and characterization of lambda heads and lambda tails. Virology. 1970;42:375–389. doi: 10.1016/0042-6822(70)90281-3. [DOI] [PubMed] [Google Scholar]
- 12.Casjens SR, Hendrix RW. Locations and amounts of major structural proteins in bacteriophage lambda. J. Mol. Biol. 1974;88:535–545. doi: 10.1016/0022-2836(74)90500-2. [DOI] [PubMed] [Google Scholar]
- 13.Larocca D, Kassner PD, Witte A, Ladner RC, Pierce GF, Baird A. Gene transfer to mammalian cells using genetically targeted filamentous bacteriophage. FASEB J. 1999;13:727–734. doi: 10.1096/fasebj.13.6.727. [DOI] [PubMed] [Google Scholar]
- 14.Larocca D, Witte A, Johnson W, Pierce GF, Baird A. Targeting bacteriophage to mammalian cell surface receptors for gene delivery. Hum. Gene Ther. 1998;9:2393–2399. doi: 10.1089/hum.1998.9.16-2393. [DOI] [PubMed] [Google Scholar]
- 15.Akuta T, Eguchi A, Okuyama H, Senda T, Inokuchi H, Suzuki Y, Nagoshi E, Mizuguchi H, Hayakawa T, et al. Enhancement of phage-mediated gene transfer by nuclear localization signal. Biochem. Biophys. Res. Commun. 2002;297:779–786. doi: 10.1016/s0006-291x(02)02282-9. [DOI] [PubMed] [Google Scholar]
- 16.Eguchi A, Furusawa H, Yamamoto A, Akuta T, Hasegawa M, Okahata Y, Nakanishi M. Optimization of nuclear localization signal for nuclear transport of DNA-encapsulating particles. J. Control Release. 2005;104:507–519. doi: 10.1016/j.jconrel.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 17.Zanghi CN, Lankes HA, Bradel-Tretheway B, Wegman J, Dewhurst S. A simple method for displaying recalcitrant proteins on the surface of bacteriophage lambda. Nucleic Acids Res. 2005;33:e160. doi: 10.1093/nar/gni158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Losick VP, Schlax PE, Emmons RA, Lawson TG. Signals in hepatitis A virus P3 region proteins recognized by the ubiquitin-mediated proteolytic system. Virology. 2003;309:306–319. doi: 10.1016/s0042-6822(03)00071-0. [DOI] [PubMed] [Google Scholar]
- 19.Richards JL, Abend JR, Miller ML, Chakraborty-Sett S, Dewhurst S, Whetter LE. A peptide containing a novel FPGN CD40-binding sequence enhances adenoviral infection of murine and human dendritic cells. Eur. J. Biochem. 2003;270:2287–2294. doi: 10.1046/j.1432-1033.2003.03596.x. [DOI] [PubMed] [Google Scholar]
- 20.Forestell SP, Dando JS, Bohnlein E, Rigg RJ. Improved detection of replication-competent retrovirus. J. Virol. Methods. 1996;60:171–178. doi: 10.1016/0166-0934(96)02052-6. [DOI] [PubMed] [Google Scholar]
- 21.Hudson JB, Misra V, Mosmann TR. Cytomegalovirus infectivity: analysis of the phenomenon of centrifugal enhancement of infectivity. Virology. 1976;72:235–243. doi: 10.1016/0042-6822(76)90326-3. [DOI] [PubMed] [Google Scholar]
- 22.O'Doherty U, Swiggard WJ, Malim MH. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J. Virol. 2000;74:10074–10080. doi: 10.1128/jvi.74.21.10074-10080.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katsura I. Structure and function of the major tail protein of bacteriophage lambda. Mutants having small major tail protein molecules in their virion. J. Mol. Biol. 1981;146:493–512. doi: 10.1016/0022-2836(81)90044-9. [DOI] [PubMed] [Google Scholar]
- 24.Maruyama IN, Maruyama HI, Brenner S. Lambda foo: a lambda phage vector for the expression of foreign proteins. Proc. Natl Acad. Sci. USA. 1994;91:8273–8277. doi: 10.1073/pnas.91.17.8273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dunn IS. Total modification of the bacteriophage lambda tail tube major subunit protein with foreign peptides. Gene. 1996;183:15–21. doi: 10.1016/s0378-1119(96)00400-3. [DOI] [PubMed] [Google Scholar]
- 26.Dunn IS. Assembly of functional bacteriophage lambda virions incorporating C-terminal peptide or protein fusions with the major tail protein. J. Mol. Biol. 1995;248:497–506. doi: 10.1006/jmbi.1995.0237. [DOI] [PubMed] [Google Scholar]
- 27.Yan Z, Zak R, Luxton GW, Ritchie TC, Bantel-Schaal U, Engelhardt JF. Ubiquitination of both adeno-associated virus type 2 and 5 capsid proteins affects the transduction efficiency of recombinant vectors. J. Virol. 2002;76:2043–2053. doi: 10.1128/jvi.76.5.2043-2053.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ros C, Kempf C. The ubiquitin-proteasome machinery is essential for nuclear translocation of incoming minute virus of mice. Virology. 2004;324:350–360. doi: 10.1016/j.virol.2004.04.016. [DOI] [PubMed] [Google Scholar]
- 29.Agarwal S, Harada J, Schreifels J, Lech P, Nikolai B, Yamaguchi T, Chanda SK, Somia NV. Isolation, characterization, and genetic complementation of a cellular mutant resistant to retroviral infection. Proc. Natl Acad. Sci. USA. 2006;103:15933–15938. doi: 10.1073/pnas.0602674103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hicke L. A new ticket for entry into budding vesicles-ubiquitin. Cell. 2001;106:527–530. doi: 10.1016/s0092-8674(01)00485-8. [DOI] [PubMed] [Google Scholar]
- 31.Katzmann DJ, Babst M, Emr SD. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 2001;106:145–155. doi: 10.1016/s0092-8674(01)00434-2. [DOI] [PubMed] [Google Scholar]
- 32.Warskulat U, Zhang F, Haussinger D. Modulation of phagocytosis by anisoosmolarity and betaine in rat liver macrophages (Kupffer cells) and RAW 264.7 mouse macrophages. FEBS Lett. 1996;391:287–292. doi: 10.1016/0014-5793(96)00753-3. [DOI] [PubMed] [Google Scholar]
- 33.Saxena RK, Vallyathan V, Lewis DM. Evidence for lipopolysaccharide-induced differentiation of RAW264.7 murine macrophage cell line into dendritic like cells. J. Biosci. 2003;28:129–134. doi: 10.1007/BF02970143. [DOI] [PubMed] [Google Scholar]
- 34.Tone M, Tone Y, Babik JM, Lin CY, Waldmann H. The role of Sp1 and NF-kappa B in regulating CD40 gene expression. J. Biol. Chem. 2002;277:8890–8897. doi: 10.1074/jbc.M109889200. [DOI] [PubMed] [Google Scholar]
- 35.Aki D, Mashima R, Saeki K, Minoda Y, Yamauchi M, Yoshimura A. Modulation of TLR signalling by the C-terminal Src kinase (Csk) in macrophages. Genes Cells. 2005;10:357–368. doi: 10.1111/j.1365-2443.2005.00839.x. [DOI] [PubMed] [Google Scholar]
- 36.March JB, Clark JR, Jepson CD. Genetic immunisation against hepatitis B using whole bacteriophage lambda particles. Vaccine. 2004;22:1666–1671. doi: 10.1016/j.vaccine.2003.10.047. [DOI] [PubMed] [Google Scholar]
- 37.Light J, Lerner RA. PhoPhabs: antibody-phage-alkaline phosphatase conjugates for one step ELISA's without immunization. Bioorg. Med. Chem. Lett. 1992;2:1073–1078. [Google Scholar]
- 38.Bonnycastle LL, Brown KL, Tang J, Scott JK. Assaying phage-borne peptides by phage capture on fibrinogen or streptavidin. Biol. Chem. 1997;378:509–515. doi: 10.1515/bchm.1997.378.6.509. [DOI] [PubMed] [Google Scholar]
- 39.Gao C, Mao S, Lo CH, Wirsching P, Lerner RA, Janda KD. Making artificial antibodies: a format for phage display of combinatorial heterodimeric arrays. Proc. Natl Acad. Sci. USA. 1999;96:6025–6030. doi: 10.1073/pnas.96.11.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen L, Zurita AJ, Ardelt PU, Giordano RJ, Arap W, Pasqualini R. Design and validation of a bifunctional ligand display system for receptor targeting. Chem. Biol. 2004;11:1081–1091. doi: 10.1016/j.chembiol.2004.05.019. [DOI] [PubMed] [Google Scholar]
- 41.Uchil P, Mothes W. Viral entry: a detour through multivesicular bodies. Nat. Cell. Biol. 2005;7:641–642. doi: 10.1038/ncb0705-641. [DOI] [PubMed] [Google Scholar]
- 42.Clarke SR. The critical role of CD40/CD40L in the CD4-dependent generation of CD8+ T cell immunity. J. Leukoc. Biol. 2000;67:607–614. doi: 10.1002/jlb.67.5.607. [DOI] [PubMed] [Google Scholar]