Abstract
TREX2 is an autonomous nonprocessive 3′ → 5′ exonuclease, suggesting that it maintains genome integrity. To investigate TREX2's biochemical and cellular properties, we show that endogenous TREX2 is expressed widely in mouse tissues and human cell lines. Unexpectedly, endogenous human TREX2 is predominantly expressed as a 30-kDa protein (not 26 kDa, as previously believed), which is likely encoded by longer isoforms (TREX2L1 and/or TREX2L2) that possess similar capacity for self-association, DNA binding and catalytic activity. Site-directed mutagenesis analysis shows that the three functional activities of TREX2 are distinct, yet integrated. Mutation of amino acids putatively important for homodimerization significantly impairs both DNA binding and exonuclease activity, while mutation of amino acids (except R163) in the DNA binding and exonuclease domains affects their corresponding activities. Interestingly, however, DNA-binding domain mutations do not impact catalytic activity, while exonuclease domain mutations diminish DNA binding. To understand TREX2 cellular properties, we find endogenous TREX2 is down regulated during G2/M and nuclear TREX2 displays a punctate staining pattern. Furthermore, TREX2 knockdown reduces cell proliferation. Taken together, our results suggest that TREX2 plays an important function during DNA metabolism and cellular proliferation.
INTRODUCTION
To a cell, faithful replication and accurate repair of genomic DNA are daunting tasks necessary for maintaining genomic integrity. Problems with replication fidelity or DNA repair may cause genomic mutations that could result in hereditary and sporadic human diseases such as cancer and accelerating aging (1–4). To maintain genome integrity, cells have evolved a built-in DNA quality control network that consists of three highly coordinated components: DNA damage checkpoints, DNA repair and DNA replication. During the last decade, 3′ → 5′ exonuclease activity has been identified in DNA damage checkpoint proteins (hRad1 and hRad9) (5,6), DNA repair proteins (MRE11, WRN, APE1, APE2, XPF/ERCC1 and Dna2) (7–12), DNA replication polymerases (polδ, polγ and polε) (13–15) and the well-known tumor suppressor p53 (16). In yeast, homologs corresponding to these human genes (except p53) have also been identified, illustrating that this 3′ → 5′ exonuclease activity is evolutionarily conserved (17). Functional studies by gene inactivation in yeast and mouse models have demonstrated that mutation in any one of these genes directly leads to genomic instability (17). In fact, mutations in some of these genes such as MRE11, WRN, XPF, polγ and p53 cause a variety of pathologies including cancer and/or age-related diseases (18–23).
In 1999, two distinct mammalian nucleases, TREX1 (Three prime repair exonuclease, also called DNase III) and TREX2, were found to account for the majority of exonuclease activity in mammalian cell extracts (24,25). Highly purified endogenous and/or recombinant TREX1 and TREX2 showed a robust 3′ → 5′ exonuclease activity that favors DNA substrates with 3′ mismatches (25–27). In addition, TREX1 enhances ligation efficiency in a polβ-mediated base excision repair assay (24) and TREX2 may interact with polδ to increase replication accuracy (28). Protein sequence analysis shows that TREX1 and TREX2 share homology to the bacterial DNA polymerase III holoenzyme ε subunit, which exhibits 3′ → 5′ exonuclease (proofreading) activity. In addition, X-ray structure of the TREX2 homodimer shows strong structural similarity to this subunit, providing direct evidence for the structural relationship between TREX2 functional domains and its biochemical activities (29). Thus, both TREX1 and TREX2 appear to be important for ensuring genomic integrity. However, Trex1-mutant mice do not exhibit a phenotype predicted for impaired editing of 3′ termini, since spontaneous mutations and cancer incidence remained unchanged as compared to controls (30). Unexpectedly, Trex1-mutant mice succumb to inflammatory myocarditis. This phenotype could be explained by the recent observation that TREX1 processes anomalous DNA structures to prevent an abnormal innate immune response that causes Aicardi-Goutieres syndrome (31). Additionally, TREX1 exonuclease activity induces Granzyme A-mediated cell death in concert with the endonuclease NM23-H1 (32). At this time, the prediction about TREX2 function is based mainly on in vitro studies; therefore, the cellular and biological significance of TREX2 remains unclear.
For this study, we investigate the biochemical and cellular properties of TREX2. We find that TREX2 is widely expressed in a variety of tissues and cell lines, suggesting it has an important cellular function. In addition to the previously reported 26-kDa TREX2, we unexpectedly found that endogenous human TREX2 is predominantly expressed as a 30-kDa protein, thus leading to the isolation of two longer alternatively spliced isoforms that maintain the same basic biochemical function as the 26-kDa isoform. One of these isoforms, TREX2L1, is the predominant transcript for all samples tested (five cancer-derived human cell lines and kidney). Observation of a series of multiple and single amino acid mutations in the 26-kDa isoform shows that domains predicted to be important for homodimerization, DNA binding and exonuclease activity by X-ray structure have separate, yet integrated functions. We find that single amino acids predicted to be important for homodimerization do not measurably alter self-association, but greatly impair both DNA binding and catalytic activity. In contrast, single amino acid mutations predicted to be important for either DNA-binding or exonuclease activity abolish their corresponding activities. In addition, exonuclease domain mutations reduce DNA-binding activity while DNA-binding mutations do not affect catalytic activity. Our cellular studies show that endogenous TREX2 is distributed in both the nucleus and the cytoplasm, and that the nuclear fraction exhibits a punctate staining pattern. TREX2 is regulated during cell cycle, with lowest expression at the G2/M phase. Functional studies illustrate that depletion of endogenous TREX2 reduces cell proliferation. Taken together, our results suggest that TREX2 plays a key role in DNA metabolism.
MATERIALS AND METHODS
Plasmid construction, site-directed mutagenesis and cloning of TREX2 alternative splicing forms
GST-TREX2 fusion expression vectors were generated using the human TREX2 26-kDa isoform. We generated DNA binding defective (TREX2BD, with triple mutations of R163A/R165A/R167A), and exonuclease catalytic defective (TREX2CD, with double mutations of H188A/D193A) open-reading frames were amplified from the E. coli-based expression plasmids (generous gifts from Dr Fred W. Perrino, Wake Forest University) (29) by hTX25 forward primer and hTX23 reverse primer (Table 1). PCR products were digested by EcoRI and XhoI, and ligated with pGEX-6P-1 or pCMV-Tag2B to create in-frame GST- and Flag-tagged recombinant expression vectors. pGEX-TREX2 was used for site-directed mutagenesis to generate dimerization defective TREX2 (TREX2DD, E29A/K59A/D94A/R107A/E191A) and to generate single amino acid mutants to alanine with primers listed in Table 1 using the QuickChange® Multi Site-Directed Mutagenesis Kit according to the manufacturer's instructions (Stratagene). TREX2 alternative splicing forms were customarily amplified and cloned into pCVM6-XL based on an RNA pool of 33 human tissues (OriGene Technologies, Inc.) by RT-PCR using primers (Table 1) corresponding to exons 15 and 16 of TREX2 (26). Four resultant clones were sequenced; two clones encode DNA sequence for DQ650792 (called TREX2L2); one clone is identical to NP_542431 (called TREX2L1); the last one contains partial TREX2.
Table 1.
Oligonucleotides used in this study
| Name | Sequence | Purpose |
|---|---|---|
| hTX25 | 5′-AAAAAAGAATTCTCCGAGGCACCCCGGGCCGAGACCTTT-3′ | TREX2 subcloning |
| hTX23 | 5′-AAAAAAAACTCGAGTCAGGCCTCCAGGCTGGGGTCATCA-3′ | TREX2 subcloning |
| hTX2-Kozak | 5′-CCGGAATTCGCCACCATGTCCGAGGCACCCCGGGCCGAG-3′ | TREX2 subcloning |
| hTX2L5 | 5′-TGGCCTACGCCAAAGCACAGGATGG-3′ | TREX2 splicing form cloning |
| hTX2L3 | 5′-TCAGGCCTCCAGGCTGGGGTCATC- 3′ | TREX2 splicing form cloning |
| hTX2-MIV5 | 5′-CCGGAATTCATGATTGTGGCAGCAGAAGCCGTTGC-3′ | TREX2L1 cloning |
| hTX2L2-5 | 5′-CGCGGATCCATGATTGTGGCAGCAGAAGCCGTTGC-3′ | TREX2L2 cloning |
| hTX2-945 | 5′-ATGCCGGAAGGTGGCTTTGCTGGCGCCGTGGTGCGGACGC-3′ | D94A mutalion |
| hTX2-1075 | 5′-GCAGGCCTTCCTGAGCGCCCAGGCAGGGCCCATCTGCC-3′ | R107A mutation |
| hTX2-191 | 5′-AAGCGCAGCCCACTCAGCCGCGGGCGACGTGCACACCCTGC-3′ | H191A mutation |
| hTX2-295 | 5′-GTGTGGAGCCCGAGATTGCCGCGCTGTCCCTCTTTGCTGTCCACC-3′ | E29A mutation |
| hTX2-595 | 5′-CCTAGTATTGCCCGCGGTCCTGGACGCGCTCACGCTGTGCATG-3′ | K59A mutation |
| hTX2-55595 | 5′-GTATTGCCCCGGGTCCTGGACGCGCTCACGCTGTGCATGTGC-3′ | R55A mutation |
| hTX2-R163A | 5′-CCCACAGCCACGGCACCGCGGCCCGGGGCCGCCAGGGTTAC-3′ | R163A mutation |
| hTX2-R165A | 5′-CAGCCACGGCACCCGGGCCGCGGGCCGCCAGGGTTACAGC-3′ | R165A mutation |
| hTX2-R167A | 5′-CGGCACCCGGGCCCGGGGCGCCCAGGGTTACAGCCTCGGC-3′ | R167A mutation |
| hTX2-H188A | 5′-CAGAGCCAAGCGCAGCCGCCTCAGCCGAGGGCGAC-3′ | H188A mutation |
| hTX2-D193A | 5′-CAGCCCACTCAGCCGAGGGCGCCGTGCACACCCTGCTCC-3′ | D193A mutation |
| hTX1-322 | 5′-GTATGAGCTGCAGTTCCTCAGCAT-3′ | Substrate for exonuclease assay |
| Biotin-mTXl-322 | Biotin-5′-GTATGAGCTGCAGTTCCTCAGCAT-3′ | Substrate for SPR analysis |
| TX2siRNAlF | 5′-CGACGAGUCUGGUGCCCUAUU-3′ | TREX2 knockdown |
| TX2siRNAlR | 5′-UAGGGCACCAGACUCGUCGUU-3′ | TREX2 knockdown |
| TX2siRNA2F | 5r-CCGGAAGGCUGGCUUUGAUUU-3′ | TRHX2 knockdown |
| TX2siRNA2R | 5′-AUCAAAGCCAGCCUUCCGGUU-3′ | TR1-X2 knockdown |
| TX2siRNA3F | 5′-ACAAUGGCUUUGAUUAUGAUU-3′ | TREX2 knockdown |
| TX2siRNA3R | 5′-UCAUAAUCAAAGCCAUUGUUU-3′ | TREX2 knockdown |
| hTX2LlF | 5′-AAGATCGAGTTGGCCGAGGATGG-3′ | RT-PCR |
| hTXL2F | 5′-GCTCCCAGAGCCAAAGGTCAC-3′ | RT-PCR |
| hTX2shortF | 5′-TTTGTCTTCCTGGACCTGGAA-3′ | RT-PCR |
| hTX2R | 5′-CCTGCAGCGTCCGCACCACG-3′ | RT-PCR |
| hTX2Ri | 5′-AGCGTGAGCTTGTCCAGGACC-3′ | RT-PCR |
Expression and purification of GST-fusion protein from Escherichia coli
BL21 cells were transformed with one of the pGEX-TREX2 expression vectors. GST-fusion proteins were produced and purified according to the manufacturer's instructions (Amersham Bioscience). To cleave TREX2 protein from GST-TREX2 fusion protein, beads-bound GST-TREX2 was digested by PreScission protease according to the manufacturer's instructions (Amersham Pharmacia). The purity and quantity of the eluted protein were determined by SDS-PAGE/Coomassie staining and Bradford assay.
Development of anti-TREX2 and anti-TREX1 antibodies
Beads-bound GST-fusion human 26-kDa TREX2 or TREX1 was prepared as described previously, and was used for immunization. Briefly, before immunization, a small amount of pre-immune serum was collected by retro-orbital bleeding. For the primary immunization, 100 μg (per mouse) of Sepharose 4B beads-bound GST-TREX2 or GST-TREX1 was injected into three 8-week-old C57BL/6 mice subcutaneously according to the standard procedure (33). After 2 weeks, the continued boosting of beads-bound GST-TREX2 or GST-TREX1 was performed weekly. Upon the third boost, the anti-TREX2 or anti-TREX1 serum was collected every 2 weeks. The titer and specificity of the anti-TREX2 or anti-TREX1 polyclonal antibodies were evaluated by direct- and immunoprecipitation -western blot. Anti-β-actin antibody (clone AC-15) and anti-Flag antibody (M2 monoclonal) were purchased from Sigma. Anti-Myc antibody was purchased from BD Bioscience Clontech.
Cell transfection, immunoblotting and immunoprecipitation
Transfection was performed using FuGENE6 Transfection Reagent (Roche Applied Science) according to the manufacturer's instructions. After 48 h, cells were collected and lysed in ice-cold lysis buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40) supplemented with appropriate amount of protease inhibitor cocktails (Sigma). Cell lysates containing 20–50 µg of protein were subjected to 10 or 12% SDS-polyacrylamide gel electrophoresis (PAGE) followed by electro-transfer of resolved proteins to nylon membrane. After blocking with non-fat milk (5%) in TBST buffer (Tris-HCl, pH 7.5, 100 mM NaCl and 0.1% Tween 20), the membrane was incubated with the corresponding primary and secondary antibodies. The immune complex on the membrane was detected using an enhanced chemiluminescence kit (Amersham Pharmacia Biotech). For immunoprecipitation, 500–1000 µg of total protein (µg/µl) was incubated with 2 µl of mouse polyclonal antibodies at 4°C for 2 hours, followed by 1-hour incubation with protein G-Sepharose. After five washes with ice-cold lysis buffer containing protease inhibitors, the immunoprecipitation complexes were subjected to western blot analysis as described above.
TREX2 small interference RNA (siRNA) assay
Prior to transfection (∼24 h), HeLa cells were seeded at 3 × 105/well (6-well plate) to obtain ∼60% confluence the following day. Three different TREX2–siRNA duplexes (Table 1, from DHARMACON) corresponding to human TREX2 open-reading frames or control siRNA (Cat #: D-001210-01, DHARMACON, CO) was transfected into cells by TransIT-TKO® transfection reagent (Cat No: MIR 2154, Mirus, Inc.) according to the manufacturer's instructions. Briefly, 6 μl of transfection reagent was added to 250 μl of serum-free media. After 5–20 min incubation, 6.25 nM of each pair of siRNA was added and mixed well for an additional 5–20 min incubation. The TransIT-TKO® transfection reagent–siRNA complex mixture was subsequently added to the 6-well plate containing 1 ml of complete growth medium. After 72 h of transfection, cell lysates were prepared and subjected to western blot using anti-TREX2 antibodies.
Cell culture and immunostaining
All cell lines used in this study were from the American Type Culture Collection (ATCC, Camden, NJ). HCT116 (human colon cancer epithelial cells); LoVo (human colonrectal adenocarcinoma epithelial cells); Caco-2 (human colon cancer epithelial cells); CV-1/EBNA1 (human kidney normal fibroblast transformed with Epstein–Barr nuclear antigen 1); PC-3 (human prostate epithelial cells); LNCap (human prostate epithelial cell); HepG2 (human hepatocellular carcinoma cells); BG1 (ovarian cancer). The HeLa cells were derived from a human cervical epitheloid carcinoma; HEK293 cells were derived from an E1A-transformed human embryonic kidney cell line. All the cell lines were cultured according to ATCC instructions. For immunostaining, cells were seeded at ∼20–30% confluence on the chamber slides (Nalge Nuc International Corp. Nalerville, IL). After 48 h, cells were rinsed with PBS once and then fixed with 4% of paraformaldehyde (dissolved in PBS) by incubating at room temperature for 10 min. After three washes with PBS, cells were permeabilized with 0.3% Triton in TBST for 10 min at room temperature. Following permeabilization, the cells were blocked with the blocking buffer (5% non-fat milk in TBST) for 1–2 h at room temperature, and primary antibody (mouse anti-serum, 1:1000 dilution) was subsequently incorporated to continue incubation for 1 h at room temperature. Upon the completion of primary antibody incubation, the cells were washed three times with TBST and were then incubated with blocking buffer containing a fluorescent-labeled secondary antibody (Alexa Fluor®488 F(AB′)2 fragment of goat anti-mouse IgG (H + L), Molecular Probe, OR, working dilution—1:5000) for 1 h at room temperature. After rinsing with TBST for three to four times, one drop of DAPI-containing mounting medium (VectorShield® mounting medium, Vector Laboratory, Inc.) was added to the culture slide, and a coverslip was placed on top of the mounting medium. Cells were immediately observed under a Zeiss fluorescent microscope.
Semi-quantitative RT-PCR
Total RNA was prepared from human cells with Trizol Reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions and reverse transcribed using oligo dT (SuperscriptII, Invitrogen, Carlsbad, CA, USA). Human kidney mRNA was obtained from Ambion. The conditions for all reactions are the same: 1 cycle of 98°C for 5 min followed by 35 cycles of 98°C for 1 min, 65°C for 1 min, 72°C for 10 s followed by 1 cycle of 72°C for 10 min.
To amplify a 410-bp mRNA fragment specific to TREX2L1, we used hTX2L1F (5′-AAGATCGAGTTGGCCGAGGATGG-3′) and hTX2R (5′-CTGCAGCGTCCGCACCACG-3′). The TREX2L1 mRNA product was re-amplified using hTX2L1F with an internal reverse primer, hTX2Ri (5′-AGCGTGAGCTTGTCCAGGACC-3′) to amplify a 288-bp fragment.
To amplify a 364-bp mRNA fragment specific to TREX2L2, we used hTX2L2F (5′-GCTCCCAGAGCCAAAGGTCAC-3′) and hTX2R (5′-CCTGCAGCGTCCGCACCACG-3′). Since these primers failed to amplify a spliced sequence from mRNA, genomic DNA was successfully amplified to verify the quality of these primers and conditions (not shown).
To amplify a 280-bp non-specific mRNA fragment common to all isoforms, we used hTX2shortF (5′-TTTGTCTTCCTGGACCTGGAA-3′) and hTX2R (5′-CTGCAGCGTCCGCACCACG-3′). The TREX2 mRNA product was re-amplified using hTX2shortF with an internal reverse primer, hTX2Ri (5′-AGCGTGAGCTTGTCCAGGACC-3′) to amplify a 158-bp fragment.
GST pull-down assay
The cell lysate (∼500 µg of total protein) extracted from HEK293 cells 48 h post-transfection with Flag-tagged TREX2 was incubated with purified beads-bound GST-fusion proteins (∼10 µg) at 4°C for 2 h. Pull-down products were washed three times by lysis buffer containing protease inhibitors. Bound proteins were separated by SDS-PAGE, GST- and Flag-tagged protein, interaction was detected by Coomassie staining and western blotting with anti-Flag antibody.
In vitro exonuclease assay
A synthetic 24-mer oligonucleotide substrate (mTX1-322, Table 1) was synthesized and subsequently radiolabeled by γ-32P-ATP (PerkinElmer, Boston, MA, 250 μCi, 5.0 mCi/ml) by the 5′-end labeling protocol; first, 50 μl of labeling reaction mixture containing 5 μl of 10× T4 kinase buffer, 2 μl (10 units/μl) of T4 kinase, 3 μl of γ-32P-ATP, 1 μl of 5 μM oligo and 29 μl of distilled water was incubated at 37°C for 30 min. The labeling reaction was stopped by the addition of stop buffer (a mixture of 400 μl of 100% ethanol alcohol, 150 μl of ddH2O, 40 μl of 3M NaAcetate and 2 μl of 10 μg/μl yeast-tRNA), and incubated at −80°C for 30 min. After centrifugation at 14 000 r.p.m. for 10 min, the supernatant was removed and the pellet was further washed by 100% ethanol alcohol once to remove any free γ-32P-ATP. After air drying for a brief period, the oligo was dissolved in 200 μl of distilled water. The relative radiation activity (∼2500 c.p.m./pmol) of the probe was determined by using liquid scintillation counter (LS6000IS, Beckman). For a ‘standard’ in vitro exonuclease assay using purified TREX2 from E. coli, a reaction mixture (20 µl) consisting of 1 µl (2.5 pmol) of γ-32P-ATP-labeled probe, and 4 µl of 5× exonuclease buffer (100 mM Tris-HCl, pH 7.5, 10 mM dithiothreitol, 25 mM MgCl2, and 500 µg/ml bovine serum albumin) and 1 µl of the appropriate TREX2 dilution was used. TREX2 dilutions were prepared in 1 mg/ml bovine serum albumin at 4°C. Reactions were performed at 37°C for 15 min and were subsequently quenched by the addition of 200 µl of cold 95% ethanol. Samples were dried in vacuo and re-suspended in 10 µl of DNA sequencing loading dye. Samples were heated at 72°C for 5 min, and 3 µl of the sample was subjected to electrophoresis on a 23% polyacrylamide denaturing gel. After fixing and drying, the radiolabeled bands within the gel were visualized and quantified by phosphorimager (Molecular Dynamics).
Measurement of TREX2–ssDNA interaction by surface plasmon resonance
TREX2–ssDNA-binding analyses were performed using Biacore2000 instrument (Biacore International AB, Sweden). The biotinylated oligo (Biotin-5′-GTA-TGA-GCT-GCA-GTT-CCT-CAG-CAT-3′) was synthesized and diluted to a final concentration of 10 nM by the running buffer (10 mM Tris-HCl, pH 7.5, 2.5 mM EDTA, 2.5 mM DTT, 250 mM NaCl, 10% glycerol and 0.02% Triton X-100). The biotinylated ssDNA was immobilized onto the sensor chip SA (streptavidin chip, Biacore International AB, Sweden) to a final density of 100–120 response units (RUs). One response unit corresponds to a surface density of 0.73 pg/mm2 DNA (34). The chip surface was regenerated with an injection of 20 µl of 2 M NaCl. Purified wild-type or mutant TREX2 proteins were diluted to nanomolar concentration by the running buffer and injected over ssDNA-immobilized and control flow-cells for 360 s at a flow rate of 30 μl/min. TREX2–ssDNA-binding ability and binding kinetic analysis were performed using BIAevaluation4.1 global analysis software. To estimate the relative DNA-binding capacity of each mutant TREX2 compared to wild-type TREX2, the maximum DNA-binding value (at 60 second after injection) of each mutant protein is divided by the value of wild-type TREX2 and then multiplied by 100%.
Cell synchronization and cell cycle analysis by FACS
HeLa cells were grown to 60% confluency in a DMEM medium containing 10% calf serum and 1% GPS (2 mM glutamine, 30 µg penicillin/ml and 50 µg streptomycin/ml). For double thymidine block (35), cells were blocked for 18 h with 2 mM thymidine and released for 9 h by washing out the thymidine. Cells were blocked again with 2 mM thymidine for 17 h to arrest all the cells at the beginning of S phase. The cells were released by washing out the thymidine. Upon release from the thymidine block, >95% of the cells progress into S phase (0–4 h), entered G2 (5–6 h) and underwent a synchronous mitosis (M phase) for 7–8 h. Cells were collected at the following time point: G1/S cells—0 h; S-phase cells—2 h and late S/G2 cells—5.5 h. Mitotic cells were collected every 10 min with mild shaking and flushing with PBS. The cells enriched from each cell cycle phase were confirmed by flow cytometry and the corresponding lysates were prepared for western blot analysis.
Cell proliferation assay by TREX2–siRNA knockdown
Twenty-four hours before siRNA, HeLa cells (1 × 105 cells/well, 6-well plate) were seeded in a DMEM medium containing 10% calf serum and 1% GPS. By the next day, cells are at ∼60% confluency, and the siRNA mix (including three siRNA duplexes, Table 1) was transfected by the same protocol as described in the previous sections. Forty-eight hours post-transfection, complete DMEM medium and 10% fetal bovine serum was replaced, and cells were counted at 96 hours post-transfection.
RESULTS
Expression of endogenous TREX2 and identification of two alternatively spliced isoforms
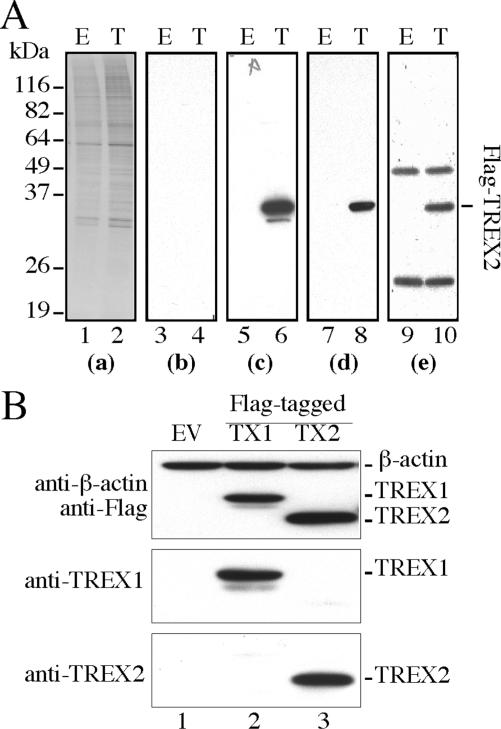
Mouse anti-TREX2 antibodies were generated to better understand the cellular characteristics of TREX2. Prior to the examination of endogenous TREX2, the specificity of these polyclonal antibodies was evaluated. Using direct- and anti-Flag immunoprecipitation-western blot analysis with mouse anti-TREX2 antibodies, a single protein band with a molecular mass of ∼28 kDa was clearly detected in extracts prepared from HEK293 cells transiently transfected with the pCMV-Flag-TREX2 expression vector (Figure 1A, lane 6, 8 and 10). The same band was absent in either extracts prepared from empty vector-transfected HEK293 cells or the immunoprecipitation complex by pre-immune serum (Figure 1A, lines 5, 7, and 9), indicating that anti-TREX2 antibodies specifically recognize recombinant Flag-TREX2 with nearly no background (Figure 1A-c). We observed that the level of endogenous TREX2 in HEK293 cells, but not in other human cell lines (see below), is too low to be detected (Figure 1A, lanes 5 and 7). To verify the potential cross-reactivity of anti-TREX2 antibodies to TREX1 because of their peptide sequence homology, the HEK293-expressed Flag-TREX1 or Flag-TREX2 was subjected to western blot with anti-Flag, anti-TREX1 or anti-TREX2 antibodies. Anti-TREX2 antibodies specifically detect recombinant Flag-TREX2 (Figure 1B), but not Flag-TREX1 or other cellular proteins, showing that anti-TREX2 antibodies are highly specific to TREX2.
Figure 1.
Anti-TREX2 polyclonal antibody development. (A) Detection of Flag-TREX2 expression in HEK293 cells by anti-TREX2 polyclonal anti-serum. E: pCMV-Flag; T: pCMV-Flag-TREX2. (a) Coommassie stain for total loading of cell lysates; (b) immunoblot by pre-immune serum; (c) immunoblot by anti-TREX2 anti-serum; (d) immunoblot by anti-Flag; (e) anti-Flag immunoprecipitation followed by immunoblot with anti-TREX2 antibody. (B) Test cross-reactivity of anti-TREX2 antibody to TREX1. Empty vector: EV; TREX1: TX1; TREX2: TX2. β-Actin serves as an internal loading control.
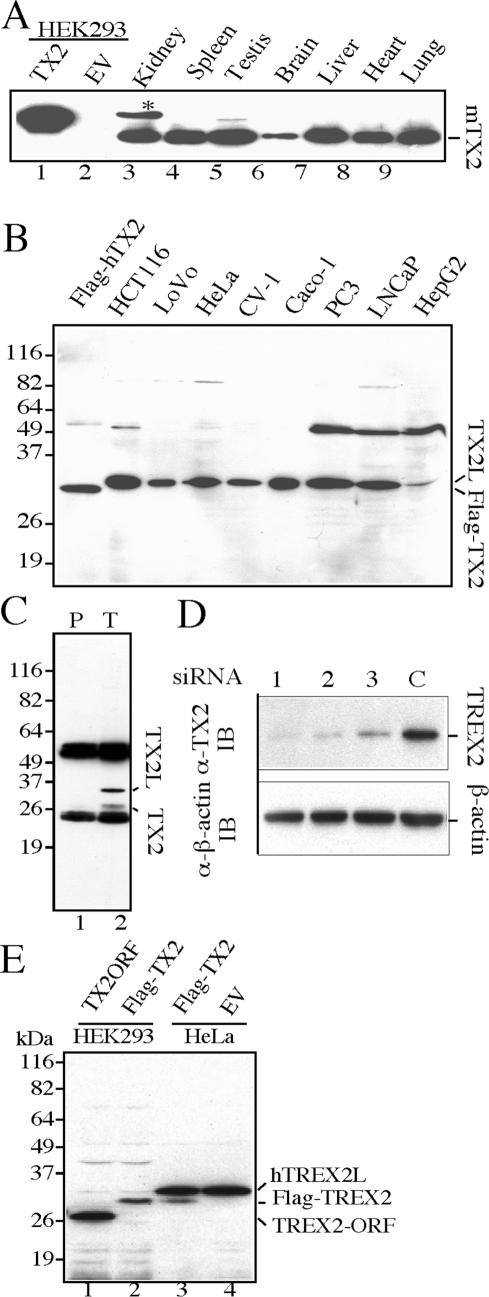
To detect endogenous TREX2, lysates prepared from mouse lung, liver, kidney, spleen, brain, testis and heart were separated by SDS-PAGE gel followed by western blot with anti-TREX2 antibodies. A protein band with the predicted molecular weight (∼26 kDa) was detected in all tissues (Figure 2A), displaying an expression pattern similar to the transcription pattern using human tissues (26). To extend this finding, extracts from human cell lines were examined by the same approach. To our surprise, a single major protein band with a molecular mass of 30 kDa, rather than 26 kDa, was detected in all human cell lines (Figure 2B) except HEK293 cells, in which no signal, neither 30- nor 26-kDa band, was detected (Figure 2A lanes 1 and 2). To further verify this result, we did immunoprecipitation (IP)-western blot using mouse anti-TREX2 antibodies or pre-immune serum. Our results show that the 30-kDa protein band is immunoprecipitated specifically by anti-TREX2 antibodies, but not by pre-immune serum, indicating that the 30-kDa protein band is likely the endogenous human TREX2 (Figure 2C). Nevertheless, we observed a weak protein band corresponding to 26 kDa in an anti-TREX2 immunoprecipitation-western blot while it is under detectable limits in a direct western blot (Figure 2C, lane 2 and Figure 2B), suggesting that it is likely 26-kDa TREX2. To confirm the identity of the 30-kDa protein band, we introduced human TREX2 small interference RNA (siRNA) into HeLa cells. All three independent TREX2–siRNA duplexes (Table 1), but not the control siRNA, significantly reduced the 30-kDa protein band but not endogenous β-actin (Figure 2D), thus providing strong evidence that the 30-kDa protein band is endogenous human TREX2.
Figure 2.
Expression of endogenous TREX2 in mouse tissues and human cell lines. (A) Mouse tissues. Lanes 1 and 2: lysates from human HEK293 cells transiently transfected with TX2 (pCMV-Flag-TREX2) or EV (pCMV empty vector). Flag-TREX2 is the fusion product of 19 amino acids (including Flag-peptide) fused with the 26-kDa human TREX2 ORF. The predicated molecular weight is 28 kDa. (B) Human cell lines. Flag-TREX2: lysate from pCMV-Flag-TREX2-transfected HEK293 cells. (C) Immunoprecipitation-western blot using HeLa cells. Immunoprecipitation complex by either pre-immune serum (lane 1) or anti-TREX2 antibodies (lane 2) was resolved by SDS-PAGE, followed by anti-TREX2 western blot. (D) Endogenous TREX2–siRNA knockdown. HeLa cells transfected with each of TREX2–siRNA duplexes (lanes 1, 2 and 3, Table 1) or control siRNA (lane C) were collected 72 h post-transfection. Cell lysates were prepared and subjected to western blot with anti-TREX2 antibody. β-Actin serves as a loading control. (E) Examination for post-translational modification of TREX2. Lane 1: 26-kDa TREX2 open-reading frame (ORF) (25) expressed in HEK293 cells. Lane 2: Flag-tagged 26-kDa TREX2 expressed in HEK293 cells. Lane 3: Flag-tagged 26 kDa TREX2 expressed in HeLa cells. Lane 4: empty vector-transfected HeLa cells.
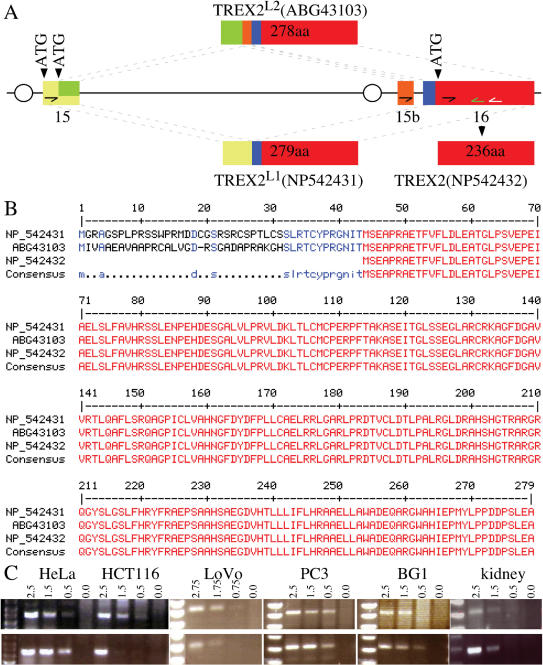
There are two possibilities to account for endogenous human TREX2 migrating at 30 kDa rather than 26 kDa: (1) post-translational modification or (2) expression of bigger isoforms. To determine the nature of this 30-kDa protein, Flag-tagged and non-tagged TREX2 expressed in either HEK293 or HeLa cells were compared to endogenous human TREX2 by SDS-PAGE gel electrophoresis followed by western blot using mouse anti-TREX2 antibodies (Figure 2E). Non-tagged and Flag-tagged recombinant TREX2 migrate at their predicted molecular weights, 26 and 28 kDa, respectively (Figure 2E, lanes 1, 2 and lane 3 lower band), smaller than endogenous human TREX2 (30-kDa, Figure 2E, lanes 3 and 4); thus, the slow migration of endogenous human TREX2 is not due to post-translational modification of the previously reported 26-kDa protein. To test whether the 30-kDa human TREX2 is a bigger isoform, we isolated longer TREX2 mRNA from a pool of 33 human tissues using primers corresponding to exons 15 and 16 of the reported TREX2 gene, AF319571 (26). As a result, two longer spliced isoforms were identified, called TREX2L1 and TREX2L2. TREX2L1 is spliced by two exons, encoding a 279-amino-acid peptide identical to NP_542431 (Figure 3A), while TREX2L2 is novel and results from three spliced exons encoding a 278-amino-acid peptide, ABG43103 (Figure 3A). Both of these alternatively spliced isoforms contain an extra peptide sequence (either 43 or 42 amino acids) fused to the N-terminus of the previously reported 26-kDa sequence (Figure 3B). The predicted molecular weight of these two long isoforms is 30 kDa, consistent with the molecular weight of endogenous human TREX2. Collectively, these long isoforms are predominantly expressed in human cells compared to the 26-kDa isoform (Figure 2B) while the short isoform is predominantly expressed in mouse tissues (Figure 2A).
Figure 3.
Human TREX2 isoforms. (A) Genomic structure of the human TREX2 gene. Human TREX2, located on chromosome X, contains 16 exons (26), of which only exons 15 and 16 are shown. TREX2L1 (NP_542431) is a spliced product of exons 15 and 16. TREX2L2 is a novel spliced product of exons 15, 15b and 16. The nucleotide sequence for TREX2L2 has been deposited in the GenBank database under accession number DQ650792. The amino acid sequence of TREX2L2 can be accessed through NCBI Protein Database under NCBI access number ABG43103. Oval indicates the predicated promoter region. (B) Protein sequence alignment of three TREX2 isoforms. TREX2L1 encodes a 279-amino-acid peptide and TREX2L2 is a 278-amino-acid peptide. Both TREX2L1 and TREX2L2 have a predicated molecular weight of ∼30 kDa. Red depicts identical homology among the three TREX2 isoforms; blue depicts identical homology between the two longer isoforms; black dots show the difference between the two longer isoforms. (C) Semi-quantitative RT-PCR to detect the TREX2 isoforms in cancer-derived human cells (HeLa, HCT116, LoVo, PC3 and BG1) and human kidney. The TREX2L1 isoform (upper panel) is amplified with primers hTX2L1F (half arrow in the yellow exon) and hTX2R (white half arrow in the red exon). As a control, hTX2short (black half arrow in the red exon) and hTX2R were used to amplify the product common to all isoforms (lower panel). These products were re-amplified with the same forward primer and a new internal primer, hTX2Ri (green half arrow in the red exon). This TREX2L1 product was sequenced to verify the results. We used hTX2L2F (half arrow in the orange exon) and hTX2R in our attempt to amplify the TREX2L2 isoform; however, no product was seen. PCR was performed with different concentrations of cDNA (μg).
To further evaluate these long isoforms, we performed a semi-quantitative RT-PCR on mRNA isolated from five human cell lines (HeLa, LoVo, HCT116, PC3 and BG1) and one tissue (kidney) using primers specific to TREX2L1 and TREX2L2. We also used primers located to the short isoform (thus, a control since all described isoforms contain this region). We find that TREX2L1 is the predominant and possibly only transcript for all samples since TREX2L1 levels are equal to that of the short isoform (Figure 3C). These transcripts were verified as being TREX2L1 by re-amplification using an internal reverse primer along with the original forward primer and then by sequencing this re-amplification product. Since TREX2L2 was not detected in these samples but present in a mixture of 33 human tissues, we anticipate that expression of TREX2L2 is likely restricted to a subset of specific tissues or cell types.
Structure–function analysis of TREX2 functional domains
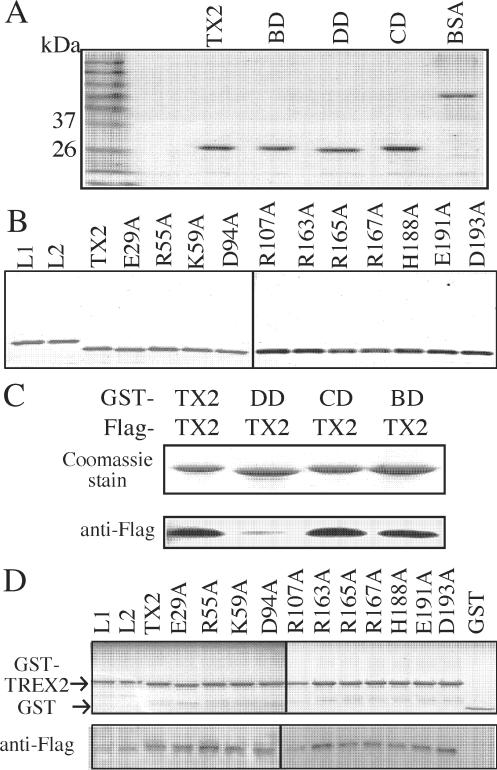
Based on the peptide sequence alignment and the X-ray structural similarity between TREX2 and bacterial DNA polymerase III ε subunit, TREX2 is proposed to possess three functional domains important for three distinct activities: homodimerization, DNA binding and exonuclease activity (29). In the same study, the amino acids critical for each functional activity have been proposed: arginine 163, 165 and 167 for DNA-binding ability; histidine 188 and aspartic acid 193 for exonuclease activity; and glutamic acid 29, arginine 55, lysine 59, aspartic acid 94, arginine 107 and glutamic acid 191 for dimerization (29). To further map the functional contribution and the coordination of these three functional domains, we first investigate TREX2 (the 26-kDa isoform) with multiple mutations in each domain: homo-dimerization defective (TREX2DD, E29A/K59A/D94A/R107A/E191A), DNA-binding defective (TREX2BD: R163A/R165A/R167A) and exonuclease catalytic defective (TREX2CD: H188A/D193A). We then investigate TREX2 with single amino acid changes for each domain using highly purified proteins (Figure 4A and B). In addition, we investigate the three in vitro activities of the long TREX2 isoforms, TREX2L1 and TREX2L2.
Figure 4.
TREX2 self-association using purified proteins. (A) Purification of TREX2 (TX2), TREX2BD (BD), TREX2DD (DD) or TREX2CD (CD). Here, 1 μg of bovine serum albumin (BSA) is loaded to estimate the concentration of purified TREX2 proteins. (B) Purification of TREX2, TREX2L1 (L1), TREX2L2 (L2) and TREX2 with single amino acid changes. (C and D) GST pull-down assay. The same amount (∼10 μg) of GST-TREX2 fusion protein is incubated with an equal amount (∼5 mg) of cell lysates prepared from HEK293 cells transiently transfected with Flag-tagged TREX2. The GST pull-down complexes were separated by SDS-PAGE followed by either Coomassie stain or anti-Flag western blot.
Homodimerization is measured by a GST pull-down assay (Figure 4C, D). We find TREX2, TREX2L1 and TREX2L2 are able to self-associate. In fact, self-association is not measurably diminished for any of the altered TREX2 proteins with the exception of TREX2DD (Figure 4C and D). These results indicate that the combination of amino acids in the homodimerization domain (E29, K59, D94A, R107A and E191) is essential for TREX2 dimerization.
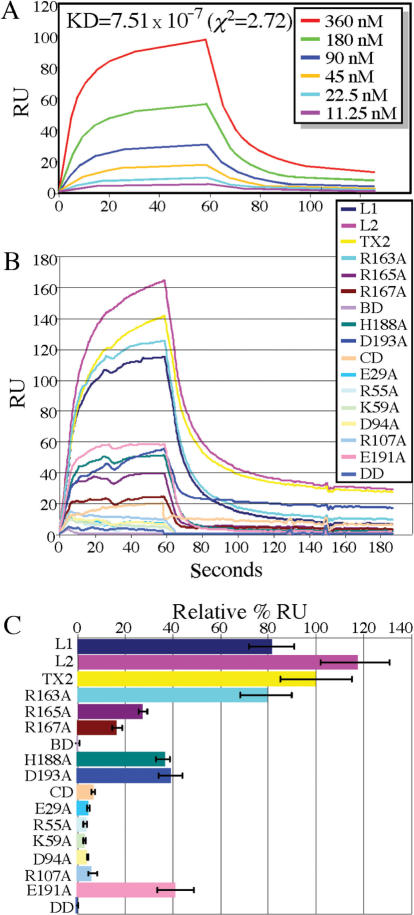
DNA binding is evaluated by a real-time DNA-binding assay based on surface plasmon resonance (SPR) that shows TREX2 directly binds to a single-strand DNA (ssDNA) substrate (Table 1) with a moderate binding affinity (Figure 5A; KD = 7.51 × 10−7, χ2 = 2.72). TREX2L1 and TREX2L2 bind to this ssDNA substrate with ∼80 and ∼120% affinity relative to TREX2, respectively. In contrast, TREX2BD (R163A, R165A and R167A) is completely unable to bind to the ssDNA substrate (Figure 5B and C) and differs from the previously published result, which showed that the same mutations reduced TREX2–DNA-binding activity by ∼90-fold (29). This discrepancy likely reflects the different methodologies used in these studies, the former study is based on the steady-kinetic analysis by calculating the kcat/Km while our study evaluates the real-time TREX2–DNA-binding kinetics by SPR technology. Using an electrophoretic mobility shift assay (EMSA), we find that TREX2BD, like TREX2, binds DNA substrate (data not shown), indicating that EMSA or kcat/Km calculation is less stringent than SPR analysis. To extend this finding, we further find single amino acid changes for two of these amino acids (R165A and R167A) impair DNA binding by ∼80%; however, changing the third amino acid (R163A) had only a marginal impact (∼20%). Like TREX2BD, TREX2DD is completely unable to bind to the ssDNA substrate, and the single amino acid changes in this domain also severely impair substrate binding (>90% defective) suggesting that homodimerization is essential for the TREX2–DNA substrate interaction. Unexpectedly, TREX2CD is also severely impaired (∼90%) while single amino acid changes are moderately impaired (∼60%) for substrate binding indicating that H188 and D193 are not only essential for catalytic activity, but also important for DNA binding.
Figure 5.
TREX2–ssDNA real-time binding kinetics by surface plasmon resonance. (A) Binding kinetics. A series of diluted 26-kDa TREX2 were injected into the streptavidin-coated sensor chip immobilized with 100 RU of ssDNA. Kinetic analyses of sensorgrams were performed using BIAeval 4.1 global analysis software, and manually using spreadsheet and graphing software. (B) Real-time ssDNA-binding capacity of 26-kDa TREX2 and mutant TREX2 at 480 nM. Calculation of the relative binding is described in Methods and Materials. (C) Graph demonstrating the average of three experiments. Maximum ssDNA-binding efficiency shown relative to TREX2 (TX2).
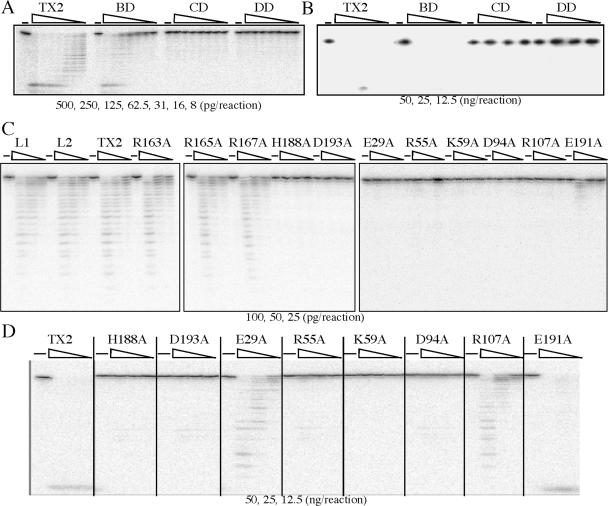
Exonuclease activity is evaluated by an in vitro exonuclease assay as previously described (25). TREX2, TREX2L1 and TREX2L2 exhibit the same level of catalytic activity while TREX2CD and TREX2 with single amino acid changes in this domain (H188A or D193A) are completely defective for exonuclease catalytic activity even at high protein concentrations (Figure 6A, B), consistent with previous results (29). Like TREX2CD, TREX2DD possesses no catalytic activity even at high protein concentrations and changing most of the amino acids in this domain greatly reduces catalytic activity (R55A, K59A and D94A—completely ablate activity; E29A and R107A—severely diminish activity; and E191A—moderately diminishes activity) (Figure 6C, D). Interestingly, TREX2BD displays moderately reduced catalytic activity compared to TREX2, but the single amino acid changes do not diminish activity.
Figure 6.
In vitro exonuclease assay. No TREX2 was used to serve as negative control (−). (A) Picograms (500, 250, 125, 62.5, 31, 16 and 8) and (B) nanograms (50, 25 and 12.5) of protein are shown for TREX2 (TX2), TREX2BD (BD), TREX2DD (DD) or TREX2CD (CD). (C) Picograms (100, 50 and 25) and (D) nanograms (50, 25 and 12.5) of protein are shown for TREX2, TREX2L1 (L1), TREX2L2 (L2) and TREX2 with single amino acid changes.
Cellular characteristics of endogenous TREX2
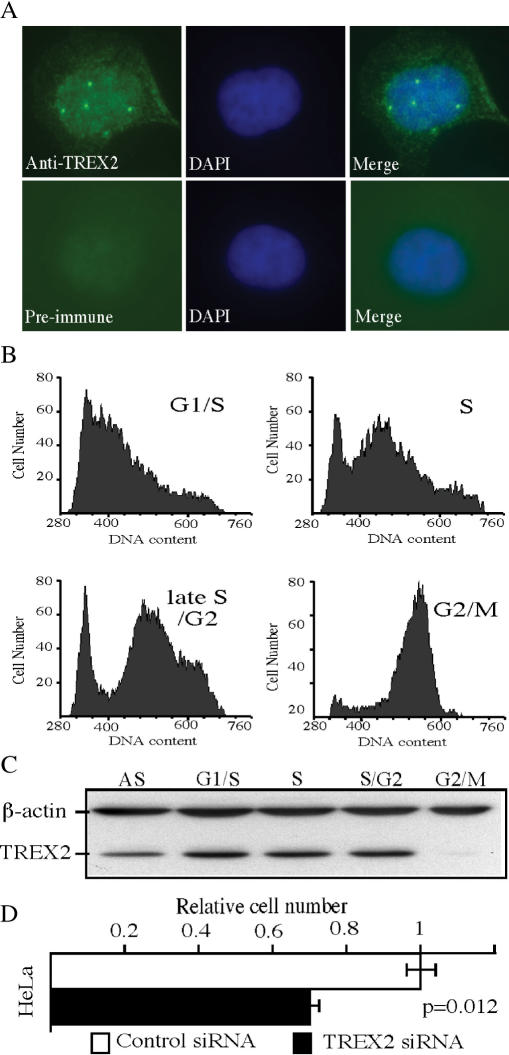
Endogenous TREX2 was investigated for subcellular localization, cell cycle distribution and RNAi knockdown. To determine TREX2 subcellular localization, HeLa cells were immunostained using mouse anti-TREX2 antibodies. We find endogenous human TREX2 is localized in both the nucleus and cytoplasm. Interestingly, nuclear TREX2 displays a punctuate staining pattern with ∼3–5 foci/cell (Figure 7A), a common characteristic shared by proteins participating in DNA damage checkpoints, DNA repair and replication. To investigate whether endogenous TREX2 expression is regulated during the cell cycle, HeLa cells were synchronized at G1/S by double-thymidine block (35), as confirmed by flow cytometry (Figure 7B). Cells form each phase of the cell cycle were collected and cell lysates were analyzed by western blot using mouse anti-hTREX2 antibodies. Our results show that endogenous TREX2 levels are regulated through the cell cycle with the lowest expression level in G2/M (Figure 7C), indicating that TREX2 plays a unique function in specific cell phases such as G1/S and S. RNAi knockdown was used to determine the impact loss-of-TREX2 function has on cell proliferation. The depletion of endogenous TREX2 in HeLa cells significantly reduces cell proliferation (P < 0.012) (Figure 7D). Taken together, these results suggest that TREX2 plays an important role in regulating cell proliferation.
Figure 7.
Cellular characteristics for TREX2. (A) Subcellular localization of TREX2 in HeLa cells that were fixed, permeablized and incubated with either anti-TREX2 antibodies (upper panel) or pre-immune serum (lower panel). (B) HeLa cells in different cell cycle phases (G1/S, S, late S/G2, G2/M) were collected and verified by flow cytometry analysis of propidium-iodide-stained cells. (C) Cell lysates prepared from each cell phase were analyzed by SDS-PAGE followed by anti-TREX2 western blot. AS: asynchronized cells; S/G2: late S/G2. (D) HeLa cells (1 × 105) transfected with either TREX2–siRNA or control siRNA; 96-h post-transfection, cells were counted. Cell proliferation rate represents the ratio of the number of cells in experimental group divided by the number of cells in control group. This graph is calculated based on three independent transfected wells for each treatment.
DISCUSSION
Here, we investigate the cellular and biochemical properties of the 3′ → 5′ exonuclease, TREX2. We find that TREX2 is commonly expressed in a wide range of mouse tissues and human cell lines and have identified at least three TREX2 isoforms generated by alternative splicing that include a 26-kDa isoform and two 30-kDa isoforms. These isoforms function equally well for homodimerization, DNA binding and exonuclease activity. For the 26-kDa isoform, we find that both DNA binding and catalytic activity depend on efficient self-association while DNA binding and exonuclease activities may be partly separated from each other. These activities are likely important for genome metabolism as supported by our observations that TREX2 forms nuclear foci, is cell cycle regulated and important for cellular proliferation.
We did not expect to find TREX2 to be predominately expressed as a 30-kDa protein rather than the 26-kDa protein in human cells. We provide five lines of evidence that support this 30-kDa protein is TREX2. First, the 30-kDa protein band is not endogenous TREX1 (∼33 kDa) non-specifically recognized by anti-TREX2 antibodies because no cross-reactivity of anti-TREX2 antibodies against TREX1 is detected (Figure 1B). Second, the 30-kDa protein band is clearly detected as a single major band in both direct- and immunoprecipitation-western blot by using anti-TREX2 antibodies, but not pre-immune serum (Figure 2B and C). Third, the 30-kDa protein band is specifically knocked down by three independent human TREX2–siRNA duplexes, but not by a control siRNA duplex (Figure 2D). Fourth, 26-kDa TREX2 is not modified to result in a 30-kDa protein band since neither Flag-tagged nor non-tagged 26-kDa TREX2 migrates at 30-kDa when transfected into human cells (Figure 2E). Fifth, two alternatively spliced isoforms, TREX2L1 and TREX2L2, were isolated, and these isoforms code for proteins predicted to be ∼30 kDa (Figure 3). In addition, these long isoforms are completely functional for self-association, DNA binding and exonuclease activity (Figure 4–6). Taken together, our results support that the predominantly expressed endogenous 30-kDa protein is endogenous human TREX2. Because TREX2L1 and TREX2L2 were isolated based on an mRNA pool of 33 human tissues, we cannot conclude whether these isoforms display different tissue expression patterns at this time since this will require further investigation by using antibodies that specifically recognizing the 30-amino-acid peptide at the N-terminus. Unlike human TREX2, endogenous mouse TREX2 is predominantly detected as the 26-kDa isoform. Interestingly, a ∼30-kDa protein band was also observed in mouse kidney and testis, but not in other tissues, co-existing with 26-kDa TREX2 (Figure 2A), suggesting multiple TREX2 isoforms exist in mouse tissues as well.
The X-ray structure and amino acid conservation predict three domains for TREX2 that are important for homodimerization, DNA binding and exonuclease activity. We have developed three assays to evaluate each activity for a variety of mutants designed to disrupt only one domain. For the homodimerization domain, we find that the self-association is defective only when TREX2 contains five mutations. This highly altered protein is also severely defective for DNA binding and catalytic activities. Therefore, TREX2 was altered for only single amino acids within this domain and these TREX2 mutants are not obviously defective for self-association, but are severely defective for both DNA binding and catalytic activities demonstrating the importance for optimal self-association. Consistent with this finding, mutation of TREX1 R114 (equivalent to TREX2 R107) identified from Aicardi-Goutieres syndrome patients impairs catalytic activity (31). For the DNA-binding domain, TREX2 with three mutations has no observable impact on self-association but severely disrupts both DNA binding and catalytic activity. TREX2 with single amino acid mutations in this domain shows that two of these amino acids (R165 and R167) are critical for fully efficient DNA binding each displaying an ∼80% decline; however, both mutants exhibit strong exonuclease activity. Thus, efficient exonuclease activity does not require efficient DNA binding, at least in vitro. Contrary to the X-ray structural prediction, mutation of R163 barely affects DNA binding even though this amino acid may be required for fully optimal binding. For the exonuclease domain, TREX2 with two mutations has no impact on self-association but severely disrupts its predicted activity as shown previously (29) and also impairs DNA binding by ∼90%. TREX2 with single amino acid mutations in this domain show that each amino acid is critical for catalytic activity and important for fully efficient DNA binding since each mutant exhibits no catalytic activity and ∼60% reduction in DNA binding. Thus, amino acids essential for catalytic function are also important for efficient DNA binding. The genetic screen for TREX2 SNPs has led to the identification of S39F, R137 and R156L in prostate cancer clinical samples, but these mutations do not alter TREX2 in vitro exonuclease activity (36). It would be interesting to know whether DNA-binding activity or homodimer formation was affected by the mutation of S39F, R137 and R156L, thus altering the cellular function of TREX2.
TREX2 exhibits cellular characteristics that suggest its function supports genome integrity and cellular proliferation. We find that TREX2 forms nuclear foci and TREX2 levels are regulated during the cell cycle with the lowest expression during G2/M phase. Similarly, the other closely related 3′→5′ exonuclease, TREX1, is also cell cycle regulated, with the highest expression level in S-phase and the lowest in M phase (35). In accordance with this observation, TREX2 has been reported to associate with replication proteins that include polδ (28) and the virus replication machinery (37) suggesting a role during DNA replication. However, the exact role of TREX2 in DNA replication remains to be investigated.
In summary, we have characterized the biochemical and cellular characteristics of TREX2. We find that TREX2 is expressed in most tissues and cells, and endogenous human TREX2 is dominantly expressed as a 30-kDa protein, likely encoded by either TREX2L1 and/or TREX2L2. TREX2 forms nuclear foci and its levels are cell cycle regulated. TREX2 possesses at least three basic biochemical functions: homodimerization, DNA binding and enzymatic activity, and these functions are co-dependent, in particular, fully efficient homodimerization seems to be required for both DNA binding and exonuclease activity, and the exonuclease domain seems to be required for fully efficient DNA binding. Even though these domains are co-dependent, the DNA catalytic activity does not require fully efficient DNA binding by our in vitro assays. Finally, cellular studies show that TREX2 is important for cell proliferation. Collectively, our results suggest that TREX2 plays an important role in DNA metabolism and cell proliferation.
ACKNOWLEDGEMENTS
We thank Drs Hai Rao, Dave Sharp, Tom Boyer, Yousin Suh, Jan Vijg and Arlan Richardson for critical comments and Dr Eileen Lafer for helping with the Biacore results. We thank Dr Fred Perrino for providing TREX2 expression vectors and for critical review of the manuscript and Drs Tom Boyer and Yousin Suh for providing human cells and kidney mRNA. We thank Dr Teresa Marple for helping with the FACS analysis. We are grateful to Charnae Williams for her general lab support and all the members in the Hasty lab for their help and encouragement. This work was supported by UO1 ES11044, DAMD17-02-1-0587, NIH P01 AG17242 and R01 CA76317-05A1 to P.H. T32 CA86800-03 to M.J.C. Funding to pay the Open Access publication charge was provided by NIH R01 CA123203-01A1.
The nucleotide sequence for TREX2L1 peptide sequence is identical to exons 15 and 16 of GenBank Access Number AF319571. The amino acid sequence of TREX2L1 can be accessed through NCBI Protein Database under NCBI Access Number NP_542431; the nucleotide sequence for TREX2L2 has been deposited in the GenBank database under GenBank Accession Number DQ650792. The amino acid sequence of TREX2L2 can be accessed through NCBI Protein Database under NCBI Access Number ABG43103.
Conflict of interest statement. None declared.
REFERENCES
- 1.NCBI. On-line Mendelian Inheritance in Man. NCBI; 2005. OMIM Statistics. [Google Scholar]
- 2.Matsuyama A, Croce CM, Huebner K. Common fragile genes. Eur. J. Histochem. 2004;48:29–36. [PubMed] [Google Scholar]
- 3.Heinen CD, Schmutte C, Fishel R. DNA repair and tumorigenesis: lessons from hereditary cancer syndromes. Cancer Biol. Ther. 2002;1:477–485. doi: 10.4161/cbt.1.5.160. [DOI] [PubMed] [Google Scholar]
- 4.Hasty P, Vijg J. Accelerating aging by mouse reverse genetics: a rational approach to understanding longevity. Aging Cell. 2004;3:55–65. doi: 10.1111/j.1474-9728.2004.00082.x. [DOI] [PubMed] [Google Scholar]
- 5.Parker AE, Van de Weyer I, Laus MC, Oostveen I, Yon J, Verhasselt P, Luyten WH. A human homologue of the Schizosaccharomyces pombe rad1+ checkpoint gene encodes an exonuclease. J. Biol. Chem. 1998;273:18332–18339. doi: 10.1074/jbc.273.29.18332. [DOI] [PubMed] [Google Scholar]
- 6.Bessho T, Sancar A. Human DNA damage checkpoint protein hRAD9 is a 3′ to 5′ exonuclease. J. Biol. Chem. 2000;275:7451–7454. doi: 10.1074/jbc.275.11.7451. [DOI] [PubMed] [Google Scholar]
- 7.Paull TT, Gellert M. The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol. Cell. 1998;1:969–979. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- 8.Huang S, Li B, Gray MD, Oshima J, Mian IS, Campisi J. The premature ageing syndrome protein, WRN, is a 3′ → 5′ exonuclease [letter] Nat. Genet. 1998;20:114–116. doi: 10.1038/2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chou KM, Kukhanova M, Cheng YC. A novel action of human apurinic/apyrimidinic endonuclease: excision of l-configuration deoxyribonucleoside analogs from the 3′ termini of DNA. J. Biol. Chem. 2000;275:31009–31015. doi: 10.1074/jbc.M004082200. [DOI] [PubMed] [Google Scholar]
- 10.Mu D, Bessho T, Nechev LV, Chen DJ, Harris TM, Hearst JE, Sancar A. DNA interstrand cross-links induce futile repair synthesis in mammalian cell extracts. Mol. Cell. Biol. 2000;20:2446–2454. doi: 10.1128/mcb.20.7.2446-2454.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burkovics P, Szukacsov V, Unk I, Haracska L. Human Ape2 protein has a 3′–5′ exonuclease activity that acts preferentially on mismatched base pairs. Nucleic Acids Res. 2006;34:2508–2515. doi: 10.1093/nar/gkl259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Masuda-Sasa T, Imamura O, Campbell JL. Biochemical analysis of human Dna2. Nucleic Acids Res. 2006;34:1865–1875. doi: 10.1093/nar/gkl070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chung DW, Zhang JA, Tan CK, Davie EW, So AG, Downey KM. Primary structure of the catalytic subunit of human DNA polymerase delta and chromosomal location of the gene. Proc. Natl Acad. Sci. USA. 1991;88:11197–11201. doi: 10.1073/pnas.88.24.11197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ropp PA, Copeland WC. Cloning and characterization of the human mitochondrial DNA polymerase, DNA polymerase gamma. Genomics. 1996;36:449–458. doi: 10.1006/geno.1996.0490. [DOI] [PubMed] [Google Scholar]
- 15.Kesti T, Frantti H, Syvaoja JE. Molecular cloning of the cDNA for the catalytic subunit of human DNA polymerase epsilon. J. Biol. Chem. 1993;268:10238–10245. [PubMed] [Google Scholar]
- 16.Mummenbrauer T, Janus F, Muller B, Wiesmuller L, Deppert W, Grosse F. p53 Protein exhibits 3′-to-5′ exonuclease activity. Cell. 1996;85:1089–1099. doi: 10.1016/s0092-8674(00)81309-4. [DOI] [PubMed] [Google Scholar]
- 17.Shevelev IV, Hubscher U. The 3′ 5′ exonucleases. Nat. Rev. Mol. Cell Biol. 2002;3:364–376. doi: 10.1038/nrm804. [DOI] [PubMed] [Google Scholar]
- 18.Carney JP, Maser RS, Olivares H, Davis EM, Le Beau M, Yates JR, III, Hays L, Morgan WF, Petrini JH. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93:477–486. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- 19.Stewart GS, Maser RS, Stankovic T, Bressan DA, Kaplan MI, Jaspers NG, Raams A, Byrd PJ, Petrini JH, et al. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell. 1999;99:577–587. doi: 10.1016/s0092-8674(00)81547-0. [DOI] [PubMed] [Google Scholar]
- 20.Yu CE, Oshima J, Fu YH, Wijsman EM, Hisama F, Alisch R, Matthews S, Nakura J, Miki T, et al. Positional cloning of the Werner's syndrome gene [see comments] Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 21.Thompson LH, Brookman KW, Weber CA, Salazar EP, Reardon JT, Sancar A, Deng Z, Siciliano MJ. Molecular cloning of the human nucleotide-excision-repair gene ERCC4. Proc. Natl Acad. Sci. USA. 1994;91:6855–6859. doi: 10.1073/pnas.91.15.6855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Goethem G, Dermaut B, Lofgren A, Martin JJ, Van Broeckhoven C. Mutation of POLG is associated with progressive external ophthalmoplegia characterized by mtDNA deletions. Nat. Genet. 2001;28:211–212. doi: 10.1038/90034. [DOI] [PubMed] [Google Scholar]
- 23.Soussi T, Kato S, Levy PP, Ishioka C. Reassessment of the TP53 mutation database in human disease by data mining with a library of TP53 missense mutations. Hum. Mutat. 2005;25:6–17. doi: 10.1002/humu.20114. [DOI] [PubMed] [Google Scholar]
- 24.Hoss M, Robins P, Naven TJ, Pappin DJ, Sgouros J, Lindahl T. A human DNA editing enzyme homologous to the Escherichia coli DnaQ/MutD protein. EMBO J. 1999;18:3868–3875. doi: 10.1093/emboj/18.13.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mazur DJ, Perrino FW. Identification and expression of the TREX1 and TREX2 cDNA sequences encoding mammalian 3′ → 5′ exonucleases. J. Biol. Chem. 1999;274:19655–19660. doi: 10.1074/jbc.274.28.19655. [DOI] [PubMed] [Google Scholar]
- 26.Mazur DJ, Perrino FW. Structure and expression of the TREX1 and TREX2 3′ → 5′ exonuclease genes. J. Biol. Chem. 2001;276:14718–14727. doi: 10.1074/jbc.M010051200. [DOI] [PubMed] [Google Scholar]
- 27.Mazur DJ, Perrino FW. Excision of 3′ termini by the Trex1 and TREX2 3′ → 5′ exonucleases. Characterization of the recombinant proteins. J. Biol. Chem. 2001;276:17022–17029. doi: 10.1074/jbc.M100623200. [DOI] [PubMed] [Google Scholar]
- 28.Shevelev IV, Ramadan K, Hubscher U. The TREX2 3′ → 5′ exonuclease physically interacts with DNA polymerase delta and increases its accuracy. Scientific World Journal. 2002;2:275–281. doi: 10.1100/tsw.2002.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perrino FW, Harvey S, McMillin S, Hollis T. The human TREX2 3′ → 5′-exonuclease structure suggests a mechanism for efficient nonprocessive DNA catalysis. J. Biol. Chem. 2005;280:15212–15218. doi: 10.1074/jbc.M500108200. [DOI] [PubMed] [Google Scholar]
- 30.Morita M, Stamp G, Robins P, Dulic A, Rosewell I, Hrivnak G, Daly G, Lindahl T, Barnes DE. Gene-targeted mice lacking the Trex1 (DNase III) 3′ → 5′ DNA exonuclease develop inflammatory myocarditis. Mol. Cell. Biol. 2004;24:6719–6727. doi: 10.1128/MCB.24.15.6719-6727.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crow YJ, Hayward BE, Parmar R, Robins P, Leitch A, Ali M, Black DN, van Bokhoven H, Brunner HG, et al. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 cause Aicardi-Goutieres syndrome at the AGS1 locus. Nat. Genet. 2006;38:917–920. doi: 10.1038/ng1845. [DOI] [PubMed] [Google Scholar]
- 32.Chowdhury D, Beresford PJ, Zhu P, Zhang D, Sung JS, Demple B, Perrino FW, Lieberman J. The exonuclease TREX1 is in the SET complex and acts in concert with NM23-H1 to degrade DNA during granzyme a-mediated cell death. Mol. Cell. 2006;23:133–142. doi: 10.1016/j.molcel.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 33.Harlow E, Lane D. Using Antibodies: A Laboratory Manual. Package edition. Cold Spring Harbor Laboratory Press; [Google Scholar]
- 34.Speck C, Weigel C, Messer W. ATP- and ADP-dnaA protein, a molecular switch in gene regulation. EMBO J. 1999;18:6169–6176. doi: 10.1093/emboj/18.21.6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whitfield ML, Sherlock G, Saldanha AJ, Murray JI, Ball CA, Alexander KE, Matese JC, Perou CM, Hurt MM, et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol. Biol. Cell. 2002;13:1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perrino FW, Krol A, Harvey S, Zheng SL, Horita DA, Hollis T, Meyers DA, Isaacs WB, Xu J. Sequence variants in the 3′ → 5′ deoxyribonuclease TREX2: identification in a genetic screen and effects on catalysis by the recombinant proteins. Adv. Enzyme. Regul. 2004;44:37–49. doi: 10.1016/j.advenzreg.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 37.Liao G, Huang J, Fixman ED, Hayward SD. The Epstein-Barr virus replication protein BBLF2/3 provides an origin-tethering function through interaction with the zinc finger DNA binding protein ZBRK1 and the KAP-1 corepressor. J. Virol. 2005;79:245–256. doi: 10.1128/JVI.79.1.245-256.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]