Abstract
Functional analysis of mammalian genes in vivo is primarily achieved through analysing knockout mice. Now that the sequencing of several mammalian genomes has been completed, understanding functions of all the genes represents the next major challenge in the post-genome era. Generation of knockout mutant mice has currently been achieved by many research groups but only by making individual knockouts, one by one. New technological advances and the refinements of existing technologies are critical for genome-wide targeted mutagenesis in the mouse. We describe here new recombineering reagents and protocols that enable recombineering to be carried out in a 96-well format. Consequently, we are able to construct 96 conditional knockout targeting vectors simultaneously. Our new recombineering system makes it a reality to generate large numbers of precisely engineered DNA constructs for functional genomics studies.
INTRODUCTION
Sequencing and analysis of the mouse and human genomes have led to the identification of ∼25 000 genes as well as hundreds of conserved non-coding regions (1–3). Functional characterization of the genes and the potential regulatory elements genome-wide represents a major challenge in the post-genome sequencing era. While there are a range of experimental and informatics tools that address gene function, analysing knockout (KO) mice produced through gene targeting in mouse embryonic stem (ES) cells is still the most widely used approach to understand mammalian gene function (4). KO mutant lines have been generated for hundreds of mouse genes (5). Analyses of these mutant mice have provided fundamental insights into mammalian gene functions. In general, there are several phenotypic outcomes in the KO mice. The mutant mice may have the exact phenotypes anticipated based on a gene's expression pattern, in vitro assays, bioinformatics predictions or symptoms in human patients. Yet, in some cases, the mutant mice may not have any obvious phenotype, which might be due to genetic redundancy, nature of the KO alleles, genetic background effects, investigator's knowledge or incomplete phenotyping spectrums. For other genes, the mutants die in uteri owing to the critical roles of these genes in embryonic development, thereby precluding studying their functions in late development or in specific tissues of adult mice.
To circumvent the embryonic lethality problem and to investigate gene function temporally and spatially, conditional knockout (cko) approaches have been developed (6,7). The current cko strategy takes advantage of the Cre-loxP site-specific recombination system that functions well in mouse cells (8). In a typical cko allele, the critical exon(s) of a gene is flanked by two loxP sites so that it can be deleted by spatial and temporal Cre expression. With the advances of DNA manipulation technologies, it is now possible to generate a multi-purpose allele of a gene that can serve as a conventional KO, a conditional KO and a reporter allele (9).
The first step in generating a cko allele is to construct a targeting vector in E. coli that is subsequently transfected into mouse ES cells for homologous recombination. We and others have previously described methods for constructing targeting vectors using recombineering, which is based on highly efficient homologous recombination systems from bacteriophages (10–15). In the system using homologous recombination encoded by λ phage that we developed, a temperature-dependent repressor (cI857) tightly controls λ prophage expression, and, thus, the recombination functions of λ phage can be transiently supplied by shifting cultures to 42°C for 15 min (14,16). Three proteins encoded by λ are required for the homologous recombination function in recombineering (17–19): Exo, Beta and Gam. Exo is a 5′–3′ exonuclease that acts on linear double-strand DNA (dsDNA) to generate 3′ single-strand DNA (ssDNA) overhangs for recombination. Beta binds to the ssDNA overhangs that are longer than 35 nt and stimulates complementary strand annealing. The recombination is further enhanced by Gam, which inhibits RecBCD-dependent, linear DNA degradation activities of the host cell. The λ prophage-based recombineering systems have proven to be very efficient due to the fact that the three genes are expressed in single copy from their natural operon and from the strong promoter, pL, and that the three proteins are in appropriate molar ratio as they form a complex in vivo. The tightly regulated expression also ensures that the Beta and Exo proteins are present only after induction. This, together with the short induction time (15 min) for the expression of the recombination activities, help to minimize the potential genomic rearrangements caused by prolonged presence of the phage Red recombination proteins. To make a cko vector from a BAC, the first step is usually to transform the BAC DNA into recombineering-competent E. coli cells. Subsequently, selection cassettes with loxP sites are inserted via recombineering into genomic regions that flank the exon to be deleted (10).
We describe here a high-throughput recombineering system, which can generate conditional targeting vectors in a rapid efficient manner. A key reagent for this new system includes a complete λ phage that is replication defective in BAC-harbouring DH10B E. coli cells but still retains its full heat-inducible homologous recombination functions. A second reagent useful for high-throughput recombineering is a set of low-copy plasmids (pSim) that contain exo, bet and gam in their native phage operon, pL, under λ CI repressor control (20). By using cells carrying either of these two new reagents, thousands of BAC clones can be easily made recombineering competent by either simple λ infection or plasmid transformation. Further improvement of recombineering in these systems allowed all the steps for making targeting vectors to be performed in 96-well plates, making the whole process suitable for high-throughput operations. Our new recombineering system described here has solved a key technical bottleneck for the genome-wide targeted mutagenesis programmes in the mouse (5,21).
MATERIALS AND METHODS
Construction of the replication-defective phage
The replication-defective λ phage, λ cI857 ind1 CroTYR26amber PGLN59amber rex< >tetRA, was created by combining several different mutations to mimic the defective prophage strain that we have used for most of our recombineering studies. The cI857 mutation allows temperature induction by inactivating the CI repressor. The Cro mutation enhances pL operon and recombination gene expression analogous to the Cro deletion of the defective prophages. Wild-type λ lysogens die within 7 min after a normal prophage induction due to in situ λ DNA replication in the bacterial chromosome carried out by O and P functions of λ (22). The P mutation causes a defect in λ replication allowing induction of the prophage for at least 15 min. Replication-competent lysogens kill the host with less than a 10-min induction. The tetRA genes are added to select for the presence of the lysogenic prophage in the strain using tetracycline resistance.
The construction of λ cI857 CroTYR26amber was described previously (23). The amber mutation in Cro is at codon 26 changing it from a tyrosine TAT codon to the amber TAG codon. A high-titre lysate was prepared from this mutant phage on strain N720, which contains the supF amber suppressor for tyrosine tRNA allowing Cro expression by the phage. Note that this phage mutant does not form plaques on a non-suppressor strain like W3110 or DH10B or even on a supE glutamine suppressor tRNA strain; presumably because glutamine cannot be substituted for tyrosine at this position in Cro.
The mutation PGLN59amber was first created by Campbell (24) as an amber (sus) mutation in the λ replication gene P at glutamine codon 59. This phage mutant called λ P80 forms plaques on the C600 supE suppressor strain but not on DH10B or the N720 supF strain, i.e. just the reverse of the Cro mutation above. This mutation is a C > T change in position 39 759 of the λ DNA sequence. A high-titre lysate was prepared from this mutant phage on strain C600. From the pure phage preparation, we amplified the entire P gene carrying the amber mutation by PCR using Invitrogen Platinum HiF polymerase and cleaned the product with a Qiagen kit. We next did recombineering with the phage λ cI857 CroTYR26amber by crossing it with the linear DNA PCR product carrying the P80 mutation using the protocol outlined in the Oppenheim's article above (23).
We detected the recombinant phage containing both the Cro and P amber mutations on a tryptone agar Petri plate overlaid with two bacterial top agar layers of 2.5 ml each. In the first agar layer was the N720 bacterial strain on which the Cro amber mutant parent phage could form plaques but on which the double Cro P amber mutant would not form plaques because of the P80 mutation. In the top layer was the LE392 strain, which allows both amber mutations to be functionally suppressed and which is used for infection by phage from the recombineering cross described above. The desired double mutant, recombinant λ cI857 ind1 CroTYR26amberPGLN59amber is distinguished from the parental λ cI857 CroTYR26amber because the P mutant forms cloudy plaques due to its inability to grow on the bottom layer, and the P+ parental phage forms clear plaques by growing on both layers. These cloudy recombinant plagues were purified on LE392 and tested for plaquing on LE392, N720 and C600. As expected, they formed plaques efficiently only on the LE392 double suppressor strain. We amplified the DNA of the P gene from the recombinant and demonstrated that it contained the mutation P80 creating a BfaI site.
We next modified the phage by replacing the rexArexB genes with tetRA encoding tetracycline resistance. This was done following the same procedure as outlined previously (23) using recombineering with the λ cI857 ind1 CroTYR26amberPGLN59amber recombinant phage and a PCR product of the tetRA genes with flanking homologies designed to delete the rexArexB genes. The recombineering was done in the LE392 derivatives. Following the recombineering cross, the resultant phage lysate was used to infect LE392, and tetracycline-resistant lysogens were selected on LB-tet plates at 32°C. Lysogens were purified and tested for sensitivity to phages λ, λimm21, T4, and T4 rII. The strain was resistant (immune) to λ but not λimm21 or T4 as expected for a λ lysogen. This λ lysogen was sensitive to T4 rII which is expected if the T4 rII exclusion functions RexAB are absent. The lysogen was induced to produce a phage lysate by shifting an exponentially growing culture in LB to 42°C for 15 min, and then shifting to 39°C until cell lysis (60–90 min). For long term storage, the λ lysate was prepared by centrifuging the lysed culture with a couple drops of added chloroform. We routinely obtained λ lysates with titres greater than 109 p.f.u./ml.
Construction of pSim plasmids
Plasmid pSim6 was constructed by recombination the λ defective prophage into pSC101 plasmid backbone (20). The AmpR coding sequence was present as a substitute for the rexAB genes. We made pSim17 and pSim18 by replacing amp in pSim6 with either Blasticidin-or Hygromycin-resistant coding sequences using the recombineering functions expressed from the plasmid.
PCR products for recombineering
All DNA oligos in this study were purchased from Sigma. Linear templates to be used for PCR amplification were prepared as follows: The retrieval vector (PL611) was digested with EcoRI and BamHI, the loxP-F3-PGK-EM7-Neo-F3 selection cassette was cut with NotI and SalI, and the I-SceI-Bsd-I-CeuI cassette was digested with EcoRI and BamHI. The digestion mixtures were run on a gel and bands corresponding to Bsd (526 bp), Neo (2 kb) and PL611 (3 kb) were excised and purified using Qiagen purification kit. To avoid the background from uncut plasmids carrying these cassettes, we tested the gel-purified PL611 and the two selection cassettes by transforming 1.0 ng of them into DH10B cells. If there were any background drug-resistant colonies, the cassettes were re-purified and tested again until they were clean, thereby ensuring that all the BsdR or KanR cells selected out in liquid media after recombineering should be the recombinant ones. The restriction maps of the selection cassettes and PL611 are depicted in Supplementary Figures 2–6.
To generate products for targeting or retrieving, 1.0 ng of each of the three linear purified products above was used as a template for PCR reaction. PCR amplification was carried out using Extensor Hi-Fidelity PCR Master Mix 2 (2X, ABgene). Twenty-five microlitres of the master mix was added to 1 μl of template (1.0 ng), 2 μl of each primer (10 μM) and 20 μl of PCR-grade water. PCR was performed using PTC-225 PCR machine (Peltier Thermal Cycler) with the following settings: 94°C for 4 min, this was followed by 35 cycles of 94°C for 30 s, 60°C for 30 s and 68°C for 1 min (Bsd) or 2–3 min (Neo and retrieval backbone). This was then followed by 68°C for 5 min. After PCR reactions, 0.5 μl of exonuclease I (10 U, from either New England Biolabs or Epicentre) was added per 50 μl of PCR products and incubated at 37°C for 1 h followed by heat inactivation at 80°C for 20 min. The PCR products were then purified using Qiagen mini-preparation columns and eluted in 50 μl of PCR-grade water.
For 96-well operations, all primers were synthesised in 96-well plates so that they could be used directly in a 96-well PCR reaction plate. After Exo I treatment and heat inactivation, PCR products were precipitated with ethanol in 96-well PCR plates. Here, 100 μl ethanol with 2 μl 5 N NaCl were added to each well of 50 μl PCR reaction. DNA was precipitated by centrifuging the plate at 3700 r.p.m. (2250 g) for 30 min and washed once with 70% ethanol. Air-dried PCR products were dissolved in 100 mM CaCl2 for chemical transformation. Alternatively, for electroporation, DNA was dissolved in 50 μl water. Three micrograms of amplified DNA was usually obtained from one PCR reaction.
Making one or a few targeting vectors using the mobile recombineering reagents
Targeting selection cassettes to BACs
(1) Using replication-defective λ phage
Day 1. Inoculate BAC cells into 1 ml LB with Chloramphenical (12.5 μg/ml), 1% maltose, for overnight growth at 37°C with shaking.
Day 2. Cells were collected and washed once with 1 ml 10 mM MgSO4 and resuspended in 100 μl 10 mM MgSO4. One microlitre containing greater than 1 million lambda phage was added to 100 μl of the BAC cells and the mixture was incubated at 32°C for 20 min. One millilitre LB was added to the tube that was incubated for another hour before the cells are plated to a Tet-LB plate. The plate was incubated at 32°C for overnight. The lysogenization frequency was ∼1%, as expected, and infection yielded ∼10 000 TetR colonies.
(2) Using pSim plasmids
Day 1. Inoculate BAC cells into 1 ml LB with chloramphenical (12.5 μg/ml), for overnight growth at 37°C with shaking.
Day 2. Cells were collected and washed with cold water three times and were electroporated with pSim18 (1.0 ng) in 50 μl water. Electroporation condition: 200 Ohms (capacitance, 25 microFD, 0.1 cm gap, 1.8 kV). The transformation mixture was added with 1 ml LB and was incubated for 1 h at 32°C. The cells were plated onto a LB-Hygro (75 μg/ml) plate and incubated at 32°C for overnight.
If chemical transformation was used, cells were collected and were washed with 100 mM MgCl2, 100 mM CaCl2 and then with 100 mM CaCl2. Finally, cells were resuspended in 50 μl 100 mM CaCl2 with 1.0 ng pSim18. The cell–DNA mixture was heat shocked at 42°C for 2 min. The transformation mixture was added with 1 ml LB and was incubated for 1 h at 32°C before plated onto a LB-Hygro plate. The plate was incubated at 32°C for overnight.
Day 3. Pick up one TetR or HygroR colony and inoculate into 1 ml LB with chloramphenical and tetracycline (12.5 μg/ml each) or hygromycin (75 μg/ml), for overnight growth at 32°C with shaking.
Day 4. Inoculate 25, 35, 45 and 55 μl of the overnight culture into four 15-ml tubes (or into four wells of a 96-well deep plate) with 1 ml fresh LB in each tube. Shake at 32°C for 2 h. Without measuring OD, transfer the cells from the four tubes into wells of a 42°C heat block (Grant Instrument, Cambridge, UK). Incubate for 15 min without shaking. Transfer the heat block to an ice bucket. The temperature of heat block dropped to 0°C within 2 min. The metal heat block was chilled on ice for 5 min. Cells were transferred to four 1.5-ml eppendorf tubes and centrifuged at maximum speed for 25 s and combined at the washing steps. For using electroporation, cells were washed three times with cold water, electroporated with Bsd PCR product (3 μg) in 50 μl water. For using chemical transformation, cells were collected and were washed once with 100 mM MgCl2, 100 mM CaCl2 and then with 100 mM CaCl2. Finally, cells were resuspended in 50 μl 100 mM CaCl2 with 3 μg Bsd PCR product. Here, 1 ml LB was added to the transformation mixture, which was incubated for 1 h at 32°C before plated onto a LB-Bsd (75 μg/ml) plate. Alternatively, the transformation mixture was added to one well of a 96-well plate that had 1 ml Bsd-LB media (75 μg/ml).
Day 5. Pick up 10 Bsd colonies to test whether they still retained the replication-defective λ phage or pSim18 by growing them either in LB-Tet or LB-Hygro. The positive cells were used for the next round of recombineering-targeting Neo cassette to the BACs. If BsdR cells were growing in a 96-well plate, 150 μl of the cells were transferred to another well in the same 96-well plate that had 1 ml of Bsd-Tet-LB or Bsd-Hygro-LB media to select for retaining the prophage or pSim plasmid.
Recombineering Neo cassette was performed exactly as described above except recombinants are selected on LB-Kan plates (20 μg/ml).
Retrieving
Day 1. BsdR-KanR-TetR (λ lysogen) or BsdR-KanR-HygroR (if using pSim18) BAC cells were inoculated into 1 ml LB with the antibiotics for overnight growth at 32°C.
Day 2. Recombineering was performed as described above using either electroporation or chemical transformation with PCR-amplified PL611 (∼3 μg). Transformants were plated on an Amp plate (50 μg/ml) and grew at 32°C for overnight. Alternatively, transformant mixture was inoculated into 1 ml LB with 50 μg/ml ampicillin and grew overnight at 32°C.
Day 3. Individual AmpR colonies were inoculated into LB-Amp media. Alternatively, if transformation mixture was inoculated into LB, plasmid was prepared from the AmpR liquid culture and 1.0 ng plasmid was transformed into DH10B or recombineering-competent E. coli cells (EL350 and DY380). The transformants were selected in 1 ml Kan-LB media.
Day 4. Plasmid mini-preparation.
Recombineering for making 96 targeting vectors
The procedure for manipulating 96 BACs is essentially the same as described above for making the Meox1 targeting vector. All BACs in this study except Bcl11a BAC (C3) were from the end-sequenced and indexed mouse 129 BAC library (25) and were ordered from the Sanger Institute Archive group (http://www.sanger.ac.uk/cgi-bin/software/archives/new_clone_login.cgi). The flow chart in Figure 4 illustrated the recombineering steps for vector construction in 96-well plates. We used a 12-channel pipette (Matrix) that has 850 μl capacity in the experiments. Therefore, all recombineering steps in 96-well plates used 850 μl LB for the 2 h growth of diluted overnight cultures.
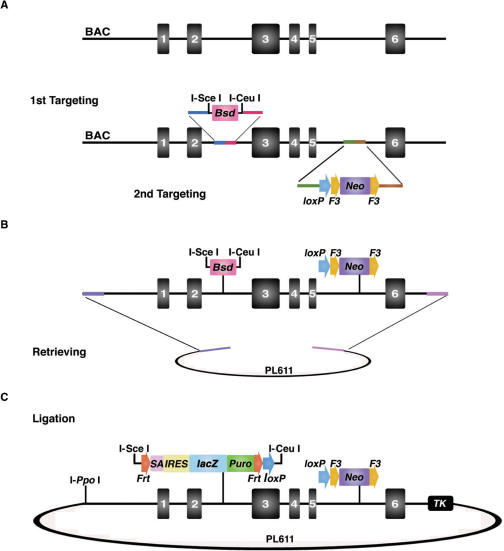
Figure 4.
Schematic diagram of recombineering steps in 96-well plates. Ninety-six BACs corresponding to the 96 genes on the mouse chromosome 11 were seeded into a 96-well plate. The BAC cells were infected by the replication-defective λ phage and lysogens were selected in Tet-LB media. The Bsd and Neo cassettes were consecutively targeted to the BACs in recombineering reactions performed in 96-well plates. Retrieved plasmid preparations were transformed back into DH10B cells to eliminate rearranged retrieval plasmid backbone. MC1TK, with a Chloramphenicol (Cm) cassette, was targeted to PL611 to provide a negative selection in ES cells.
Day 1. Individual BACs were inoculated into 850 μl LB medium with 12.5 μg/ml Chloramphenical, 1% maltose in a 96-well deep well plate and grew at 37°C overnight.
Day 2. Cells were collected by centrifugation at 3700 r.p.m. (2250 g) at 4°C for 5 min and were resuspended in 50 μl 10 mM MgSO4 solution. Here, 1–10 μl phage lysate was added to the cell suspension. The mixture was incubated at 32°C for 20 min. This was followed by the addition of 500 μl LB and 1 h incubation at 32°C. Lysogens were selected in Tet-LB media (by adding 500 μl Tet-LB at 25 μg/ml) at 32°C.
Day 3. 25, 35, 45 and 55 μl of the TetR overnight culture (OD600 ∼ 1.2–1.4) was transferred to 850 μl fresh LB in 96-well plates so that cultures from one original 96-deep-well plate were eventually transferred to four 96-well plates (Figure 5).
Figure 5.
Increasing the recombineering efficiency by inoculating different amounts of cells in 96-well plates. Escherichia coli cells harbouring BACs are growing differently due to the nature and the sizes of genomic DNA inserts. Efficient recombineering and transformation require cells growing in log phase. To increase the likelihood that cells of a particular BAC reach the optimal growth condition prior to heat induction and recombineering, four 96-well plates were used for the 2 h culture. Each plate had different amounts of the overnight culture. Cells from the four plates were combined after heat induction and were transformed with PCR products.
After 2-h growth at 32°C, cells were transferred to four 96-well metal heat-blocks at 42°C. Cells were kept at 42°C for 15 min for induction of recombination functions and then the heat block was immediately put into wet ice so that the temperature of the block was lowered to 0°C within 2 min. The cooled induced cell cultures were transferred back to regular 96-well plates that were centrifuged at 3700 r.p.m. for 5 min at 0°C.
In the chemical transformation protocol, the cells from four individual 96-well plates were combined and suspended in 850 μl 100 mM MgCl2, 100 mM CaCl2, pelleted again and washed with 100 mM CaCl2. These cells were finally suspended in 50 μl ice-cold 100 mM CaCl2 containing the PCR product. The DNA–cell mixture was heat shocked at 42°C for 2 min in a PCR machine and transferred to a 96-well plate with 500 μl fresh LB. The plate was incubated at 32°C for at least 2 h before 500 μl Bsd (150 μg/ml)-LB media was added to each well.
Day 4. 150 μl overnight culture was transferred into fresh Bsd (75–100 μg/ml)-Tet (12.5 μg/ml)-LB media and cultured for another 24–48 h for cell growth to reach saturation.
Day 5. cultures were ready for targeting the Neo cassette to the BACs. The protocol of targeting the Neo cassette to BACs in a 96-well plate was performed using the same procedure for targeting the Bsd cassette as described above, except Kan was used in the selection.
Once the BsdR/KanR/TetR BAC cells were obtained, the specific genomic DNA fragments were retrieved to PL611.
Day 1. Overnight cultures of BsdR/KanR/TetR BAC cells at 32°C were inoculated into fresh LB for 96-well recombineering using PCR-amplified PL611. Recombinant cells carrying the retrieved plasmids were selected in Ampicillin LB media. Because retrieving a genomic fragment from a BAC was generally less efficient than targeting a small selection cassette to a BAC, we obtained fewer colonies after retrieving. As a result, it may take up to 2 days to grow enough AmpR cells for plasmid preparation from 850 μl culture in each well.
Day 3. 96-Well plasmid preparation. We prepared plasmids using a simple isopropanol/ethanol precipitation protocol for the crude DNA preparations that were sufficient for re-transformation to remove the background plasmids. Briefly, AmpR cells in each well of a 96-well plate were pelleted and suspended in 250 μl P1 solution (from Qiagen). This was followed by the addition of 250 μl P2 solution to each well. Finally, 350 μl N3 solution was added to each well and mixed. The plate was centrifuged at 3700 r.p.m. for 20 min at 4°C. The supernatant from each well was transferred to a new 96-well plate with 750 μl isopropanol in each well. DNA was pelleted by centrifuging the plate at 3700 r.p.m. for 30 min at 4°C. Plasmid DNA was washed with 70% ethanol, air dried and finally dissolved in 50 μl water. One microlitre of the plasmid DNA was transformed into either customarily or commercially made preparations of chemical competent DH10B cells. Transformants were selected in 850 μl Kan-LB media (20 μg/ml).
Day 4–5. Plasmid DNA was isolated using a commercial 96-well plasmid kit. To confirm the identities of the retrieved plasmids, they were usually either sequenced at the plasmid backbone-genome insert junctions or digested for restriction patterns.
Replacement of the Bsd cassette with the lacZ reporter
The lacZ reporter cassette was isolated from PL613 respectively with I-SceI and I-CeuI double digestion (purchased from New England Biolabs). The purified fragment was used for ligating to the retrieved plasmid digested with I-SceI and I-CeuI.
In a typical ligation, 10 μl of purified digested retrieved plasmids, 12 μl of digested PL613, 2.5 μl of T4 DNA ligase buffer and 1 μl of T4 DNA ligase (NEB). This reaction was incubated at room temperature for 2 h. Here, 5 μl of the ligation products was transformed into commercially purchased chemical competent cells, or to DH10B-λ competent cells for the next recombineering step to introduce the MC1TK cassette to the plasmid backbone. The transformed cells were selected on Puro/Kanamycin plates or in Hygromycin/Kanamycin LB medium, depending on which reporter was chosen. Puromycin media (powder) was purchased from InvivoGen.
Recombineering of the MC1TK cassette
To add MC1TK to the targeting vectors for negative selection in ES cells, we constructed a cassette where MC1TK-Cm cassette that is flanked by ∼600 bp sequences identical to pBR322. This enabled us to target MC1TK to the retrieval plasmid backbone by simple recombineering. MC1TK-Cm plasmid DNA was digested with NotI and SalI. The 4.2-kb fragment was purified as described above for recombineering.
The targeting vectors were transformed into DH10B-λ cells that were selected in Kan/Amp-LB media in 96-well plates. Overnight cultures were diluted into fresh LB and incubated for a further 2 h. These cells were then heat shocked at 42°C for 15 min to induce recombination activities and were processed for chemical transformation. We used 100 ng purified MC1TK-Cm cassette for each recombineering reaction. The recombinant cells were selected out in Kan/Cm-LB.
Cre or Flpe recombinase expression
To confirm the functionality of loxP, FRT and F3 sites in the targeting vectors, the targeting vector plasmid DNA was transformed into EL350 and EL250 E. coli cells that express Cre and Flpase respectively upon l-arabinose induction. The procedure was performed as described previously (10,14).
Genotyping ES cells by long-range PCR
AB2.2 ES cells transfected with the linearized targeting vectors were selected in 150 μg/ml G418 (active component) and 2 μM ganciclovir (Ganc). Genotyping targeted ES cell clones by long-range PCR was performed as following. ES cell colonies were picked into 96-well feeder plates. After 3 days, cells were trypsinized in 50 μl trypsin for 10–15 min with subsequent addition of 50 μl ES cell media. Here, 30 μl was disposed into two 96-well cell culture plates that were gelatinized for DNA preparation. For the remaining cells (∼30 μl), 30 μl 2× freezing media (DMEM, 20% FCS, 20% DMSO) was added and mixed. Cells in the plate were kept in a sealed styrofoam box at −80°C. Cells in the DNA plates were allowed to grow for further 2 days and DNA were prepared as previously published (26). DNA was dissolved in 30 μl water. Here, 2 μl of DNA was diluted in 18 μl water in a 96-well PCR plate and heated at 99°C for 10 min using a PCR machine to denature DNA. The plate was centrifuged at 3700 r.p.m. for 5 min. Here, 1 μl of the denatured DNA was used for the long-range PCR. The remaining DNA in the original DNA plate was used for restriction digestion and Southern blotting.
PCR amplification was carried out using Extensor Hi-Fidelity PCR Master Mix 2 (2×, ABgene). Here, 12.5 μl of the master mix was added to 1 μl of template, 1 μl of each primer (10 μM) and 9.5 μl of PCR-grade water. PCR was performed using PTC-225 PCR machine (Peltier Thermal Cycler) with the following settings: 94°C for 4 min, this was followed by 35 cycles of 94°C for 30 s, 60°C for 30 s and 68°C for 6 min (6–7 kb) or 3 min (2–3 kb). This was then followed by 68°C for 10 min.
Primers for genotyping Meox1 targeted ES cell clones: For 5′ side diagnosis:
Meox1-5′-F: 5′-CTGATGCTCCACCTCTGTTGCTAGCACACT
LacZ reverse: 5′-CAAGGAAACCCTGGACTACTGCGCCCTA
For 3′ side diagnosis:
BpA F: GAAAGAACCAGCTGGGGCTCGACTAGAG
Meox1-3′-R: 5′-CAGGTGCCTGTGTTCTTCTTGAAGAGATAC
RESULTS
The λ phage based recombineering is currently the most commonly used method for engineering DNA in E. coli. This is achieved, in many cases, by first transforming BAC DNA into special E. coli strains that harbour a defective λ prophage (14,16). For obscure reasons, some BACs are more difficult to transform, or the initial transformation leads to unwanted DNA rearrangements. Furthermore, re-transformation of BAC DNA from the original library isolates is not suitable for high-throughput operations. To address these problems with the current λ prophage recombineering systems, we made new mobile recombineering reagents and used them in a high-throughput recombineering protocol for making mouse KO targeting vectors.
Construction of a replication-defective λ phage and pSim plasmids
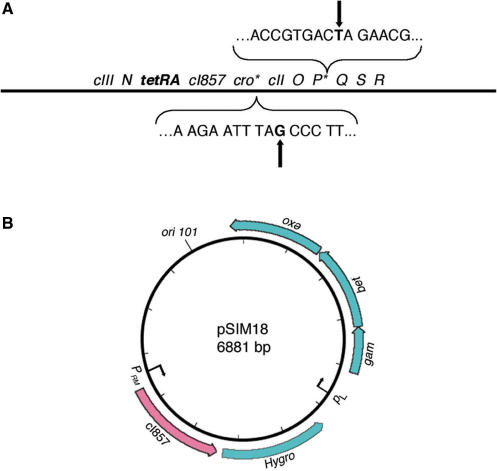
To create a mobile system that is suitable to efficiently deliver recombineering function to a large number of indexed BAC clones or to convert entire libraries, we constructed a replication-defective λ phage λ cI857 ind1 CroTYR26amber PGLN59amber rex< >tetRA, which has two amber mutations in genes Cro and P at codons 26 and 59, respectively, and a tetracycline-resistance cassette in rex (Figure 1A).
Figure 1.
The new mobile recombineering reagents. (A) A schematic diagram of the replication-defective λ phage with the two amber mutations in Cro and P genes. Arrows point to the nucleotide changes in the amber mutations. (B) pSim plasmids provide an alternative convenient route for recombineering. The Amp coding sequence in pSim6 plasmid is engineered to either Blasticidin- (pSim17) or Hygromycin (pSim18)-resistant gene-coding sequences, which are driven by PRM promoter. All the pSim plasmids have the pSC101 temperature sensitive, low-copy replication origin.
High-titre lysates of this multiply mutant phage can be made on strain LE392 carrying tRNA suppressor alleles for the amber mutants. The resultant mutant λ phage lysate was infectious but did not lyse non-suppressive strains such as DH10B where most BAC libraries are constructed and maintained. The lysogens can be easily selected with tetracycline at 32°C.
Besides the new λ phage, we have developed and tested an alternative plasmid-based recombineering system, pSim, which is a temperature-sensitive, low-copy plasmid (pSC101) that carries the exo, bet and gam genes under the control of their native pL operon in a mini-prophage (20) (Figure 1B). Three versions of this pSim plasmid, pSim-6 (Amp), pSim-17 (Blasticidin) and pSim-18 (Hygromycin), were constructed and were used for high-throughput studies. The only difference among these three plasmids is that they carry different antibiotic-resistant gene-coding sequences driven by the pRM promoter that also directs the CI repressor gene transcription. pSim plasmids are easier to use than the mini-lambda we made previously (27) since integration of mini-lambda into the E. coli genome is relatively inefficient. BAC cells harbouring pSim plasmids showed efficient recombineering in each type of experiment described in this study (data not shown). Details of using pSim plasmids in recombineering can be found in the Materials and methods section. Since it is easier to introduce recombineering functions into 96 BACs in a 96-well plate, or into an entire BAC library, by simple λ phage infection rather than plasmid transformation, we describe here the new high-throughput recombineering procedures, focussing on using the replication-defective λ phage.
Recombineering with short homology using chemical transformation in 96-well plates
To test recombineering using the replication-defective λ, we infected BAC-C3 (containing mouse Bcl11a gene) with the phage and used the resultant TetR lysogen in the experiments. The goal was to insert a Neo/Kan cassette into this BAC by using a 2.8-kb DNA fragment (from plasmid PL440) in which the Neo/Kan cassette is flanked by two 400-bp genomic DNA homology arms. The heat-induced lysogenic cells were electroporated with the DNA fragment using a conventional E. coli transformation condition (16). We obtained thousands of Kanamycin-resistant (KanR) recombinant colonies, demonstrating that the new λ was efficient in recombineering. We also noticed more cells lysed during electroporation compared to using either regular BAC cells or heat-induced recombineering competent cells with a defective λ prophage (EL350), suggesting that upon heat induction the slight leakiness of lysis genes S and R might contribute to the sensitivities of these cells to high voltages (28). Plating post-electroporation cells on LB plates demonstrated that 30% of the DH10B-λ lysogens survived electroporation compared to about half of the cells that survive when regular DH10B cells are used. Importantly, no noticeable DNA rearrangements were detected in the BACs after heat induction (data not shown).
To find out whether chemical transformation was sufficient for recombineering in the heat-induced λ lysogens, we compared the recombineering efficiencies in BAC-C3 BAC lysogen cells using either chemical transformation (see Materials and methods section) or conventional electroporation. With 50 ng of the gel-purified 2.8-kb fragment from PL440, similar numbers of Kanamycin-resistant (KanR) colonies were obtained from either chemical transformation (2004 KanR/3 × 109 surviving cells) or electroporation (3400 KanR/6 × 108 surviving cells), although the absolute recombineering efficiency of recombinants (KanR) versus total survival cells was eight times lower in chemical transformation. Nevertheless, chemical transformation is simpler, does not require any special equipment, and can easily be adapted into a 96-well format.
In general, 50-bp homology is routinely used for targeting a selection cassette to the E. coli genome by recombineering. This is usually achieved by using two 70-mer PCR primers that have 50 nt for homology and 20 nt for PCR amplification. To examine whether the new λ prophage system is sufficient to recombine with short homologies, we replaced the Neo coding sequence in PL451 (PGK-EM7-Neo-bpA) plasmid (10) with the coding sequence of a Puromycin resistance gene (Puro), indicating that the new λ phage recombineering system had provided high enough recombineering efficiencies with relatively short homology arms.
Next, we investigated whether it is possible to obtain enough recombinants using cells cultured in a small volume. We repeated the above experiment of replacing the Neo coding sequence with Puro. We inoculated 50 μl overnight culture into 1 ml LB in a 96-well plate for the 2-h incubation. After chemical transformation, we obtained ∼200 PuroR colonies, demonstrating that it was indeed feasible to scale down the culture volume to perform recombineering in a 96-well plate.
Construction of a conditional targeting vector for the Meox1 gene in 96-well plates
We next made a cko targeting vector using the new recombineering reagents and the 96-well recombineering protocol. The vector design was a cko strategy, where a reporter is integrated into the targeted allele to allow the detection of endogenous gene expression (9), and combined with our past experience in gene targeting (Figure 2).
Figure 2.
Construction of conditional knockout targeting vectors using the new recombineering reagents. (A) The genomic structure of a locus with exons 3–5 to be deleted in the cko allele. The Bsd cassette flanked by two rare cutter sites, I-SceI and I-CeuI, is targeted to the 5′ side of the intended deletion region. Subsequently, the loxP-F3-PGK-EM7-Neo-F3 (Neo) cassette is targeted to the 3′ side of the deletion region. The point mutation present in the Neo coding sequence of PL452 and PL451 plasmids (10,36) was corrected in this Neo cassette which resulted in higher resistance to Kanamycin in E. coli and a 2-fold increase in the number of G418-resistant ES colonies. Coloured lines represent the short homology arms in recombineering. (B) The genomic DNA fragment is retrieved from the BAC to PL611, which has the AmpR gene. In a typical cko vector, we choose 4–5-kb genomic DNA as the left homology arm (5′), and 2–3 kb as the right homology arm (3′). The genomic DNA region to be deleted is generally between 1 and 7 kb. (C) The Bsd cassette can conveniently be replaced by a reporter, i.e. lacZ, in a simple ligation reaction. The final targeting vector has the reporter flanked by two FRT sites followed by a loxP site at the 5′ side of the intended deletion region, and a F3 flanked Neo cassette providing positive selection in ES cells. The negative selection marker TK is added to the vector backbone by recombineering. The vector is linearized with the rare-cutter I-PpoI.
To construct a targeting vector, we first incorporated a 526-bp Blasticidin selection marker (Bsd) flanked by two rare cutter sites (I-SceI/I-CeuI) to the 5′ of the region to be deleted (Figure 2A). Empirically, we find that smaller selection cassettes generally give rise to better recombineering efficiencies. After the Bsd is integrated to the BAC, the loxP-F3-Neo-F3 cassette is subsequently targeted to the 3′ side of the region to be deleted (Figure 2A). F3 is a mutant variant of the wild-type FRT site. It can recombine with another F3 site to excise Neo but not with wild-type FRT sites (29). The Neo cassette has a PGK promoter and an EM7 promoter so it is functional in both mammalian cells and E. coli. Next, the modified genomic DNA fragment with the two selection markers (Bsd and Neo) was retrieved from the BAC to a modified pBR322 plasmid (PL611) (Figure 2B). We used pBR322 rather than a pUC19 backbone to reduce the instability problems associated with cloning mouse genomic DNA into the very high-copy pUC-type plasmid. Finally, the Bsd cassette was removed in vitro with I-SceI/I-CeuI double digestion and replaced with a reporter (lacZ) as illustrated in Figure 2C. The lacZ reporter, with its splicing acceptor site (SA) and polyadenylation site, serves to detect transcription expression of the targeted gene and at the same time may disrupt the gene and possibly create a loss of function allele. To produce a cko allele, the lacZ and Neo cassettes can be excised by expressing Flp either in ES cells, or preferably in the mouse germline (Figure 3A). After Cre-loxP-mediated deletion of the genomic region, FRT and F3 sites remain at the targeted locus, providing an anchor point for FRT-F3-mediated cassette exchange in cultured cells (29).
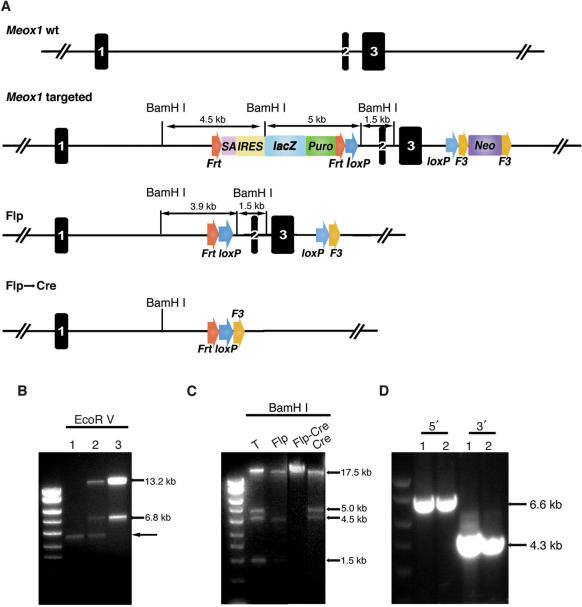
Figure 3.
Targeting at the mouse Meox1 locus. (A) Construction of the Meox1 conditional knockout (cko) allele using the design depicted in Figure 2. The lacZ reporter is targeted to the intron 1 and the Neo cassette is in intron 3. Flp excises both lacZ reporter and the Neo cassette, leaving behind a cko allele. Cre recombinase deletes the region between the two loxP sites. One FRT, one loxP and one F3 site still remain after Cre-loxP-mediated deletion. (B) Elimination of retrieving background by re-transformation. After the Bsd and Neo cassettes were targeted to the BAC, the genomic DNA was retrieved to PL611 (AmpR). Many of the AmpR colonies were the rearranged PL611 backbone (lane 1). When the plasmid preparation from pooled AmpR colonies was examined, there were some true retrieved fragments besides the PL611 rearranged band (lane 2). Once the plasmid mixture was re-transformed into DH10B cells that were selected in Kan-LB, only the correctly recombined plasmid survived and background plasmids were eliminated (lane 3). The correctly retrieved plasmid had the Bsd cassette and produced the 13.2-kb and 6.8-kb fragments after EcoRV digestion. Arrow points to the rearranged retrieval vector backbone (PL611). (C) Test of the functionality of FRT, F3 and loxP sites. The final targeting vector was digested BamHI which generated four fragments: 17.5, 5.0, 4.5 and 1.5 kb. The digestion pattern changed after Flp or Cre excision of the selection cassettes as anticipated. T: targeting vector. (D) Long-range PCR identification of targeted ES clones. The internal primers were from the lacZ or the Neo cassette, and the external ones from genomic DNA immediately outside the homology arms. The gel image shows PCR amplification of two targeted clones. For the 5′ diagnosis, a 6.6-kb fragment was amplified (lanes 1 and 2) and for the 3′ side, the 4.3-kb junction fragment was detected in the targeted clones.
Based on this design and using the 96-well recombineering reagents and protocol, we made a cko vector of the mouse Meox1 gene. The KO strategy for the Meox1 locus is to flank the last two exons with loxP sites to create a deletion of ∼3.5 kb (Figure 3A). The six primers for introducing the two selection markers and for retrieving the genomic DNA fragment were designed by a software for genome-wide targeting strategies (D.M. and P.L., unpublished data). These primers have 70–80 nt of mouse genomic sequences for recombineering homology and 20 nt complementary to the selection cassettes (Supplementary Table 2).
Cells of BAC bMQ-434G24 (containing Meox1), ordered from an end-sequenced and indexed 129S5 BAC library (25), were infected by the replication-defective phage and λ lysogenic cells were selected either on Tet-LB plates or in liquid Tet-LB in a 96-well plate. In the latter case, because of the high titre of the λ lysate, thousands of cells became λ lysogens and the TetR lysogenic liquid culture in the 96-well plate was used for recombineering the next day. After Meox1 BAC lysogens were heat induced to express red genes and chemically transformed with the PCR-amplified Bsd cassette, recombineering mixture was selected with 1 ml of Bsd-LB media at 32°C in a 96-well plate.
To introduce the 3′loxP site, the BsdR/TetR cells in a 96-well plate were heat induced and transformed with PCR-amplified loxP-F3-PGK-EM7-Neo-F3 cassette. The transformed cells were selected directly in LB-Kan media in a 96-well plate.
To retrieve the genomic DNA fragment, PCR-amplified retrieval vector (PL611) with 70-bp homology on each side was transformed into heat-induced BsdR/KanR/TetR cells. Up to 100 colonies were usually obtained for each retrieval experiment in chemical transformation. Analysis of these colonies showed that many of them were simple rearrangement or end-joining of the original retrieval vector and did not contain the retrieved fragment, an observation also made by other laboratories using recombineering to retrieve or to perform gap repair (Figure 3B, lane 1). To eliminate this problematic background, we selected the retrieval transformants directly in Amp-LB media in a 96-well plate and made a mini-preparation from AmpR cells that represented a mixture of true recombinants as well as the background (Figure 3B, lane 2). Because the correctly recombined plasmid should retain the Bsd and Neo selection markers targeted to the genomic DNA region, the background plasmids could easily be eliminated by transforming the plasmid DNA mixture into regular DH10B or its recombineering-ready sub-strains (DH10B-λ, EL350 or DY380) and selecting transformants in Kan-LB (Figure 3B, lane 3).
Once the Bsd cassette was replaced by the lacZ reporter, and the functionality of loxP, and the FRT/F3 sites was confirmed (Figure 3C), the Meox1 targeting vector was linearized with I-PpoI prior to being transfected into AB2.2 mouse ES cells.
Correct recombinant ES clones were identified by PCR using external and internal (lacZ or Neo) primers (Figure 3D), and were confirmed on Southern blots using external probes (data not shown). Twenty-five percent of the G418rGancr clones were correctly targeted at both the 5′ and the 3′ side, and had the desired lacZ reporter cko of the Meox1 locus.
Construction of multiple cko targeting vectors in 96-well plates
We selected 96 genes on the mouse chromosome 11 to make KO vectors simultaneously because it is one of the chromosomes sequenced and annotated at the Sanger Institute. The primers for the 96 KO vectors were generated by the vector design software (Supplementary Table 2), and were ordered through the Sanger's high-throughput oligo ordering pipeline. The intended deletion regions in these genes are generally in the range of 3–5 kb. BACs from the 129S5 BAC library corresponding to these genes were infected with λ phage and selected with tetracycline directly in a 96-well plate in liquid media. The procedure for recombineering in 96-well format is depicted in Figure 4.
Escherichia coli DH10B carrying BACs harbouring different genomic inserts grow at different rates. To ensure that at least some cells for any given BAC in the 96-well plate would achieve optimal growth density for recombineering and transformation, we inoculated various amounts of the overnight culture of TetR lysogens into 0.85 ml fresh LB for the 2 h incubation at 32°C prior to heat induction. For example, we usually inoculated 25, 35, 45, 55 μl of the overnight cell culture into 850 μl fresh LB in four wells of four 96-well plates respectively (Figure 5). This added step should increase the likelihood that some cells in at least one well for any BAC would reach optimal growth density (log phase) for heat induction and recombineering after 2 h incubation at 32°C.
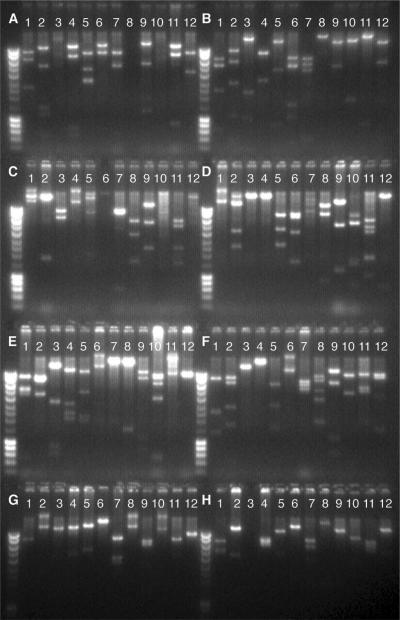
In our first attempt to manipulate 96 BACs simultaneously, we obtained 94 retrieved targeting vectors that were subsequently confirmed by sequencing the junctions between the plasmid backbone and the genomic DNA inserts, and by restriction digestion. Figure 6 shows the restriction digestion patterns of these KO vectors.
Figure 6.
Targeting vectors constructed in 96-well plates. We started with 96 BACs in a 96-well plate and, after three rounds of recombineering, obtained 94 vectors. Recombineering did not work for the two failed BACs (lanes A8 and H3) at the Bsd targeting step because these two wells were empty or clear after Bsd selection. The gel image shows the BamHI restriction digestion patterns of these plasmids. The vectors still carried the Bsd cassette, which could be replaced by various reporters.
DISCUSSION
To develop a robust high-throughput recombineering protocol, we developed two mobile recombineering-enabling systems. The λ phage provides a very efficient and convenient way to deliver recombineering functions to many BAC clones or an entire BAC library by simple phage infection. On the other hand, pSim plasmids serve as an effective alternative vehicle for the same purpose. Both of these recombination systems are robust. In this article, we primarily described the replication-defective λ system, but pSim plasmids delivered similar results and might be more convenient to use if one needs to manipulate only one or a few BACs using the standard recombineering protocols because most molecular biology laboratories are more familiar with using plasmid transformation.
We also improved the previous recombineering protocols at several steps. For example, we used a relatively large amount of PCR products (3 μg) in transformation in order to compensate for using fewer E. coli cells growing in 96-well plates and for the less efficient chemical transformation. To obtain clean PCR-amplified selection cassettes, we used E. coli Exo nuclease I to digest the excessive PCR primers and ssDNA so that any potential competition between the single-strand primers and the double-strand PCR products for the same target DNA in recombineering would be eliminated (see Materials and methods section). When a selection marker is targeted to single-copy BACs, the genomic region can have significant effects on expression of this marker in E. coli and thus on detection of recombineering events. Therefore, we used strong, clean selection markers Bsd and a codon-corrected PGK-EM7-Neo-bpA (Figure 2).
To increase the chance that for any given BAC at least some cells would be in the optimal growth phase for recombineering or transformation, we inoculated different amounts of overnight cultures from one 96-well plate to four 96-well plates for the 32°C–2 h incubation prior to heat induction (see Figure 5). In practice, this did not substantially increase the workload because the recombineering procedure itself requires a minimal amount of physical work. We found that many of the AmpR colonies in retrieval experiments were end-joined products of the linear PCR-amplified retrieval plasmid backbone. This background was completely eliminated by transforming mini-prepared DNA of the retrieved plasmid mixture to E. coli and selecting cells in Kan-LB, thus avoiding picking and characterizing individual colonies.
It is known that increased homology length improves recombineering efficiencies (12,16,30,31) and using PCR-amplified long homology arms (200–500 bp) further substantially improved recombineering reliability and efficiency (10,31). However, using PCR-amplified long homology arms is time consuming and is not suitable for a 96-well-plate-based recombineering protocol. Therefore, we chose 70–80 bp homology in this study even though 50 bp might be sufficient for a majority of the recombineering reactions. The relatively long homology (70–80 bp) should reduce the impact of SNPs among mouse strains and short stretches of identical sequences in the homology arms on recombineering efficiencies (32).
Using the new λ phage system, we demonstrated that chemical transformation yielded sufficient recombinants. This has enabled us to easily perform recombineering in 96-well plates. Not surprisingly, BAC cells containing the λ lysogen or harbouring the pSim plasmids were very efficient in recombineering using conventional electroporation protocols (16) (data not shown). A recent publication has also demonstrated that recombineering using λ red genes, together with a transiently supplied RecA function, worked well in manipulating Caenorhabditis elegans BACs using E. coli cells cultured in eppendorf tubes (33). The system we described here takes advantage of growing cells in 96-well plates, uses chemical transformation and is based on the very efficient λ recombination system where all three red genes are in their native operon and are controlled by the strong pL promoter. Importantly, our system does not require RecA for efficient recombineering, the function of which can lead to rearrangements of mammalian DNA in BACs. During the process of developing the new recombineering system, we made 103 targeting vectors out of the 105 mouse genes on chromosome 11 that were attempted (Supplementary Table 1). These vectors include the first vector (Meox1) made completely in 96-well plates, then the 8 vectors made simultaneously (data not shown) and finally 96 vectors constructed at once in a scaled-up operation.
The basic targeting vector design presented in this study comes from many years of gene targeting experience of the authors, even though the KO strategy, the selection markers and the use of loxP, FRT and F3 sites described in this article can all vary depending on individual research purposes. In general, we choose a long 5′ homology arm so as to survive cellular nuclease attacks, and a shorter 3′ homology arm to minimize the size of the vector and to facilitate PCR-mediated genotyping. In the plasmid vector, the negative selection marker (MC1TK) is adjacent to the 3′ arm and is protected by the rest of the plasmid backbone. Additionally, the use of PGK-EM7-Neo-bpA as the positive selection marker for homologous recombination in ES cells makes it possible to target genes that are both expressed and non-expressed in ES cells. For targeting in ES cells, the vectors are linearized at the 5′ side to ensure that the basal transcription of a locus and the initiation of homologous recombination are coordinated (34,35), although we have not tested whether this coordination have an effect on the targeting efficiency. Nevertheless, the targeting vectors configured in this way, for inserting a selection cassette, have generally given rise to correct targeting frequencies that are in the range of 30–95% of the stable mouse ES cell transfectants, regardless of the expression status of a locus in ES cells. The genomic DNA regions between the two loxP sites in our conditional targeting vectors can be as large as 7.0 kb without reducing the targeting frequencies in ES cells. Our vectors therefore can be used to delete multiple exons after Cre-loxP recombination and generate null alleles. Using the recombineering system described in this study, we have recently generated the lacZ-tagged cko mouse lines for Bcl11a and Bcl11b, and obtained the true cko alleles after Flpe excised lacZ and Neo in the germline (S.C.L. and P.L., unpublished data). X-gal staining of the heterozygotes with the lacZ has demonstrated distinct expression patterns of the genes in embryonic development (Supplementary Figure 1).
SUPPLEMENTRY DATA
Supplementary data are available at NAR Online.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by The Wellcome Trust (P.L.), and in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research and by a Trans NIH/FDA Intramural Biodefense Program Grant from NIAID (D.C.). P.L. thanks Drs Allan Bradley, Neal Copeland and Nancy Jenkins for their encouragement for these experiments. Funding to pay the Open Access publication charge was provided by The Wellcome Trust.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 3.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 4.van der Weyden L, Adams DJ, Bradley A. Tools for targeted manipulation of the mouse genome. Physiol. Genomics. 2002;11:133–164. doi: 10.1152/physiolgenomics.00074.2002. [DOI] [PubMed] [Google Scholar]
- 5.Austin CP, Battey JF, Bradley A, Bucan M, Capecchi M, Collins FS, Dove WF, Duyk G, Dymecki S, et al. The knockout mouse project. Nat. Genet. 2004;36:921–924. doi: 10.1038/ng0904-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rajewsky K, Gu H, Kuhn R, Betz UA, Muller W, Roes J, Schwenk F. Conditional gene targeting. J. Clin. Invest. 1996;98:600–603. doi: 10.1172/JCI118828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glaser S, Anastassiadis K, Stewart AF. Current issues in mouse genome engineering. Nat. Genet. 2005;37:1187–1193. doi: 10.1038/ng1668. [DOI] [PubMed] [Google Scholar]
- 8.Sauer B, Henderson N. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc. Natl Acad. Sci. USA. 1988;85:5166–5170. doi: 10.1073/pnas.85.14.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Testa G, Schaft J, van der Hoeven F, Glaser S, Anastassiadis K, Zhang Y, Hermann T, Stremmel W, Stewart AF. A reliable lacZ expression reporter cassette for multipurpose, knockout-first alleles. Genesis. 2004;38:151–158. doi: 10.1002/gene.20012. [DOI] [PubMed] [Google Scholar]
- 10.Liu P, Jenkins NA, Copeland NG. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13:476–484. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Angrand PO, Daigle N, van der Hoeven F, Scholer HR, Stewart AF. Simplified generation of targeting constructs using ET recombination. Nucleic Acids Res. 1999;27:e16. doi: 10.1093/nar/27.17.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang P, Li MZ, Elledge SJ. Towards genetic genome projects: genomic library screening and gene-targeting vector construction in a single step. Nat. Genet. 2002;30:31–39. doi: 10.1038/ng797. [DOI] [PubMed] [Google Scholar]
- 13.Valenzuela DM, Murphy AJ, Frendewey D, Gale NW, Economides AN, Auerbach W, Poueymirou WT, Adams NC, Rojas J, et al. High-throughput engineering of the mouse genome coupled with high-resolution expression analysis. Nat. Biotechnol. 2003;21:652–659. doi: 10.1038/nbt822. [DOI] [PubMed] [Google Scholar]
- 14.Lee EC, Yu D, Martinez de Velasco J, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73:56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- 15.Cotta-de-Almeida V, Schonhoff S, Shibata T, Leiter A, Snapper SB. A new method for rapidly generating gene-targeting vectors by engineering BACs through homologous recombination in bacteria. Genome Res. 2003;13:2190–2194. doi: 10.1101/gr.1356503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl Acad. Sci. USA. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Court DL, Sawitzke JA, Thomason LC. Genetic engineering using homologous recombination. Annu. Rev. Genet. 2002;36:361–388. doi: 10.1146/annurev.genet.36.061102.093104. [DOI] [PubMed] [Google Scholar]
- 18.Murphy KC. Use of bacteriophage lambda recombination functions to promote gene replacement in Escherichia coli. J. Bacteriol. 1998;180:2063–2071. doi: 10.1128/jb.180.8.2063-2071.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Datta S, Costantino N, Court DL. A set of recombineering plasmids for Gram-negative bacteria. Gene. 2006;379:109–115. doi: 10.1016/j.gene.2006.04.018. [DOI] [PubMed] [Google Scholar]
- 21.Auwerx J, Avner P, Baldock R, Ballabio A, Balling R, Barbacid M, Berns A, Bradley A, Brown S, et al. The European dimension for the mouse genome mutagenesis program. Nat. Genet. 2004;36:925–927. doi: 10.1038/ng0904-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weisberg RA, Gallant JA. Dual function of the lambda prophage repressor. J. Mol. Biol. 1967;25:537–544. doi: 10.1016/0022-2836(67)90204-5. [DOI] [PubMed] [Google Scholar]
- 23.Oppenheim AB, Rattray AJ, Bubunenko M, Thomason LC, Court DL. In vivo recombineering of bacteriophage lambda by PCR fragments and single-strand oligonucleotides. Virology. 2004;319:185–189. doi: 10.1016/j.virol.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 24.Campbell A. Sensitive mutants of bacteriophage lambda. Virology. 1961;14:22–32. doi: 10.1016/0042-6822(61)90128-3. [DOI] [PubMed] [Google Scholar]
- 25.Adams DJ, Quail MA, Cox T, van der Weyden L, Gorick BD, Su Q, Chan WI, Davies R, Bonfield JK, et al. A genome-wide, end-sequenced 129Sv BAC library resource for targeting vector construction. Genomics. 2005;86:753–758. doi: 10.1016/j.ygeno.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 26.Ramirez-Solis R, Rivera-Perez J, Wallace JD, Wims M, Zheng H, Bradley A. Genomic DNA microextraction: a method to screen numerous samples. Anal. Biochem. 1992;201:331–335. doi: 10.1016/0003-2697(92)90347-a. [DOI] [PubMed] [Google Scholar]
- 27.Court DL, Swaminathan S, Yu D, Wilson H, Baker T, Bubunenko M, Sawitzke J, Sharan SK. Mini-lambda: a tractable system for chromosome and BAC engineering. Gene. 2003;315:63–69. doi: 10.1016/s0378-1119(03)00728-5. [DOI] [PubMed] [Google Scholar]
- 28.Campbell A. Comparative molecular biology of lambdoid phages. Annu. Rev. Microbiol. 1994;48:193–222. doi: 10.1146/annurev.mi.48.100194.001205. [DOI] [PubMed] [Google Scholar]
- 29.Schlake T, Bode J. Use of mutated FLP recognition target (FRT) sites for the exchange of expression cassettes at defined chromosomal loci. Biochemistry. 1994;33:12746–12751. doi: 10.1021/bi00209a003. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y, Buchholz F, Muyrers JP, Stewart AF. A new logic for DNA engineering using recombination in Escherichia coli. Nat. Genet. 1998;20:123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]
- 31.Murphy KC, Campellone KG. Lambda Red-mediated recombinogenic engineering of enterohemorrhagic and enteropathogenic E. coli. BMC Mol. Biol. 2003;4:11. doi: 10.1186/1471-2199-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muyrers JP, Zhang Y, Buchholz F, Stewart AF. RecE/RecT and Redalpha/Redbeta initiate double-stranded break repair by specifically interacting with their respective partners. Genes Dev. 2000;14:1971–1982. [PMC free article] [PubMed] [Google Scholar]
- 33.Sarov M, Schneider S, Pozniakovski A, Roguev A, Ernst S, Zhang Y, Hyman AA, Stewart AF. A recombineering pipeline for functional genomics applied to Caenorhabditis elegans. Nat. Methods. 2006;3:839–844. doi: 10.1038/nmeth933. [DOI] [PubMed] [Google Scholar]
- 34.Nickoloff JA. Transcription enhances intrachromosomal homologous recombination in mammalian cells. Mol. Cell. Biol. 1992;12:5311–5318. doi: 10.1128/mcb.12.12.5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aguilera A. The connection between transcription and genomic instability. EMBO J. 2002;21:195–201. doi: 10.1093/emboj/21.3.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yenofsky RL, Fine M, Pellow JW. A mutant neomycin phosphotransferase II gene reduces the resistance of transformants to antibiotic selection pressure. Proc. Natl Acad. Sci. USA. 1990;87:3435–3439. doi: 10.1073/pnas.87.9.3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.