Abstract
Genetic analysis of fission yeast suggests a role for the spHop2–Mnd1 proteins in the Rad51 and Dmc1-dependent meiotic recombination pathways. In order to gain biochemical insights into this process, we purified Schizosaccharomyces pombe Hop2-Mnd1 to homogeneity. spHop2 and spMnd1 interact by co-immunoprecipitation and two-hybrid analysis. Electron microscopy reveals that S. pombe Hop2–Mnd1 binds single-strand DNA ends of 3′-tailed DNA. Interestingly, spHop2-Mnd1 promotes the renaturation of complementary single-strand DNA and catalyses strand exchange reactions with short oligonucleotides. Importantly, we show that spHop2-Mnd1 stimulates spDmc1-dependent strand exchange and strand invasion. Ca2+ alleviate the requirement for the order of addition of the proteins on DNA. We also demonstrate that while spHop2-Mnd1 affects spDmc1 specifically, mHop2 or mHop2-Mnd1 stimulates both the hRad51 and hDmc1 recombinases in strand exchange assays. Thus, our results suggest a crucial role for S. pombe and mouse Hop2-Mnd1 in homologous pairing and strand exchange and reveal evolutionary divergence in their specificity for the Dmc1 and Rad51 recombinases.
INTRODUCTION
Diploid eukaryotes produce genetically recombined haploid gametes or spores through meiosis. This is achieved by one round of DNA replication followed by two successive rounds of nuclear division, meiosis I and meiosis II. These divisions are preceded by a distinctive meiotic prophase during which homologous chromosomes synapse and undergo genetic recombination between allelic and non-allelic positions (1,2). Reciprocal recombination events, named crossovers, establish chiasmata, which are physical connections between homologs that ensure proper chromosome segregation at meiosis I. Meiosis II involves the segregation of sister chromatids and is therefore analogous to a mitotic division. Meiotic interhomolog recombination leads to an exchange of genetic material, contributes to genetic diversity and is required for proper chromosome segregation (3).
Meiotic recombination in Saccharomyces cerevisiae is initiated by the creation of double-strand breaks by Spo11 (Rec12 in Schizosaccharomyces pombe), a type II topoisomerase (4). Spo11 knock-out mice display chromosome synapsis defects suggesting that the initiation of recombination precedes and is required for normal chromosome synapsis during meiosis I (5,6). In S. pombe, the Rec6, Rec7, Rec12, Rec14 and Rec15 proteins are essential for the creation of DNA double-strand breaks and meiotic recombination (7). The Mre11, Rad50 and Nbs1 proteins are involved in the processing of DSBs to form recombinogenic 3′-single-stranded tailed DNA (8). The resected DSBs are then used to invade homologous duplex DNA, leading to the exchange of genetic information, a process that requires the RAD51 and DMC1 genes. Rad51 and the meiosis-specific protein Dmc1 are homologs of the bacterial recombinase RecA. In vitro, these proteins are, however, less efficient than their bacterial counterpart. Hence, there exist accessory cofactors that assist Dmc1 and Rad51. Dmc1 is helped specifically by Mei5 and Sae3 (9) and Rad51 is aided by the members of the Rad52 group including Rad52, Rad54, Rad55 and Rad57 (10–12), whereas Tid1/Rdh54 assists both recombinases (13,14).
Several lines of evidence implicate Hop2 and Mnd1 in meiotic recombination. Saccharomyces cerevisiae Hop2 plays a crucial role in the proper alignment of homologs during meiotic prophase (15). Hop2 knock-out mice and S. cerevisiae mutants show resemblance in their phenotypes such as spermatocyte arrest at the pachytene-like chromosome condensation and the failure in meiotic DSBs to be repaired (16). Although the recombination defects of the Hop2 and Mnd1 mutants were thought to be an indirect consequence of the lack of chromosome pairing, studies have indicated that Hop2 and Mnd1 are directly involved in recombination. In budding yeast, hop2 and mnd1 mutants, meiotic cells arrest at the pachytene stage, double-strand breaks are not repaired properly and the Dmc1 and Rad51 recombinases accumulate on meiotic chromosomes at a high level but neither strand invasion, heteroduplex DNA or joint molecules are detected (15,17–19). A decrease in intragenic recombination and homologous pairing is observed in S. pombe meu13 mutants (a Hop2 homolog) (20). Mnd1 is suggested to be involved in strand invasion since mnd1-1 mutant cells initiate recombination but do not form heteroduplex DNA or Holliday Junctions (19). Genetic analysis of the fission yeast ortholog Mcp7, revealed functions necessary for meiotic recombination (21). Because of the very similar phenotypes of the hop2 and mnd1 mutants, the Hop2–Mnd1 proteins are thought to be involved in a common pathway leading to homology search during meiotic recombination. In fact, several lines of evidence suggest that Hop2 and Mnd1 both functions in a Dmc1-dependent pathway. First, the recombination phenotypes of the hop2 and mnd1 mutants are very similar to those of dmc1. Second, overexpression of RAD51 largely suppresses the meiotic defects of the dmc1 and hop2 mutants (22). Third, in every genome analyzed to date, the presence of the HOP2 and MND1 genes correlates with that of DMC1. In particular, the genome of C. elegans or Drosophila does not contain Dmc1, Hop2 or Mnd1 while Rad51 is present.
Biochemical studies of Hop2-Mnd1 have led to several paradoxes. Purified budding yeast Hop2-Mnd1 only stimulates the strand invasion activity of Dmc1 by a modest 3-fold (23). Purified mHop2-Mnd1 stimulated strand invasion of hRad51 by about 10-fold and hDmc1 by 35-fold (24) while in another study mouse Hop2 had no effect on human Rad51 in vitro (25). Finally, S. cerevisiae and human Hop2-Mnd1 was found to bind dsDNA preferentially (23,26), which is not consistent with its role in strand invasion.
In this manuscript, we present biochemical evidence that fission yeast Hop2-Mnd1 binds ssDNA of 3′-tailed molecules specifically by electron microscopy. spHop2-Mnd1 show properties reminiscent of recombinases, it can anneal long complementary single-strand DNAs and promote strand exchange. Moreover, D-loop and strand exchange assays revealed that spHop2-Mnd1 can activate spDmc1 efficiently. In addition, we show that mouse Hop2 and Hop2-Mnd1 can stimulate both human Rad51 and Dmc1 recombinases in strand exchange assays in accordance with the Yokohama group (26). These results show that Hop2-Mnd1 participates in both the homologous pairing and strand exchange steps of homologous recombination.
MATERIALS AND METHODS
Nucleic acids and S. pombe techniques
Single-strand and double-strand øX174 DNA was purchased from New England Biolabs (Beverly, MA, USA). pPB4.3 (27) was prepared using Qiafilter plasmid maxi kit (Qiagen, Valencia, CA, USA). Linear duplex DNA with 3′-single-stranded tails at both ends were prepared as described previously (28). For D-loop analysis, pUC18 plasmid DNA was purified by Triton X-100 lysis followed by CsCl banding (24). DNA concentrations are expressed in moles of nucleotides.
Schizosaccharomyces pombe methods were performed according to Moreno et al. (29). Schizosaccharomyces pombe mRNA was isolated using RNeasy (Qiagen). The diploid strain JN628 (kindly provided by K. Tanaka) was induced in meiosis and cells were collected at 2 h intervals. Protein extracts were subjected to western blot analysis with anti-mouse Hop2 that cross-recognize the S. pombe protein. The Meu13+ cDNA (S. pombe hop2+) was amplified by RT-PCR from S. pombe mRNA isolated from meiotic cells using primers JYM110 (5′-tcgcaattgtagcatatggctaaggcgaaag) and JYM122 (5′-agttggatccttagtttaagtcaattggtccctccgt) bearing NdeI and BamHI sites, respectively. The PCR product was subjected to digestion with NdeI and BamHI and cloned into pET16b and pET28b (Novagen, Madison, WI, USA) in which the Meu13 protein sequence was linked to a decahistidine and hexahistidine tag, respectively. The sequence of S. pombe Meu13+ was identical to accession number SPAC222.15. Mcp7+ (herein Mnd1+) was isolated from S. pombe genomic DNA using JYM123 (5′-gggaattccatatgcctcccaagggactatcgcttgc) and JYM124 (5′-cgcggatcctcacaaaatcggtagttgcagatcgtc) bearing NdeI and BamHI sites, respectively.
An intronless derivative was generated by removing a 94 bp intron essentially as published by Wang and Malcolm using primers JYM161 (5′ aggccattttccatgactctaaagatttttttcaactaaa) and JYM162 (5′-tttagttgaaaaaaatctttagagtcattgaaaatggcct) (30). The Mcp7+ gene was amplified by PCR with primers bearing NdeI and BamHI sites. The PCR product was cloned into pET16b (Novagen) to generate pET16b-Mcp7. To generate Myc-tagged Mcp7, Mcp7 was PCR amplified using primers JYM580 (5′-ccatggatggaggagcagaagctgatctcagaggaggacctgatgcctcccaagggactatcg) and JYM581 (5′-ggatcctcacaaaatcggtagttgcag) and the PCR product was cloned in pET28b using NcoI and BamHI sites. The sequence of S. pombe mcp7+ was identical to accession number SPAC13A11.03.
Purification of S. pombe Hop2–Mnd1 complex
Recombinant spHop2–Mnd1 proteins were purified from 800 ml of Escherichia coli BL21(DE3) RP (Stratagene, La Jolla, CA, USA) carrying plasmids pET28b-Meu13 and pET16b-Mcp7 grown at 37°C in tryptone phosphate media supplemented with 100 μg/ml ampicillin, 50 μg/ml kanamycin and 25 μg/ml chloramphenicol. At OD600 = 0.4, Hop2 and Mnd1 synthesis was induced by the addition of 0.1 mM IPTG, incubated at 30°C, after 4 h, the cells were harvested by centrifugation, frozen in dry ice and stored at −80°C. The cell paste was resuspended in 80 ml of P5 buffer (50 mM sodium phosphate pH 7.0, 500 mM NaCl, 10% glycerol, 0.02% Triton X-100) containing 5 mM imidazole and the protease inhibitors PMSF (1 mM), aprotinin (0.019 TIU/ml) and leupeptin (1 μg/ml). The suspension was divided into 40 ml aliquots, which were sonicated three times for 30 s. Insoluble material was removed by centrifugation at 35 000 r.p.m. for 1 h in a Sorvall Ultra Pro 80 T647.5 rotor. The supernatant was loaded on a 5 ml Talon column (BD Biosciences, Palo Alto, CA, USA).
The column was washed successively with 50 and 10 ml of P buffer containing 30 and 40 mM imidazole, respectively before Hop2-Mnd1 were eluted with 25 ml of P buffer containing 500 mM imidazole. Fractions of Hop2-Mnd1, were identified by SDS-PAGE, pooled and dialyzed against QS buffer (10 mM Tris–HCl pH 8.0, 10% glycerol, 1 mM DTT) containing 100 mM KCl (QS100) and loaded on a 1 ml MonoS column (HR 5/5) using a Pharmacia FPLC system. The column was washed with 20 ml of QS buffer containing 150 mM NaCl before Hop2-Mnd1 was eluted using a linear gradient of 10 ml of 0.1–0.8 M KCl in QS buffer. The protein eluted around 400 mM KCl and was dialyzed against 1 l of QS100 buffer and stored in aliquots at −80°C. The concentration of Hop2-Mnd1 was determined by SDS-PAGE using purified BSA as a standard and verified by Bradford assay. This fraction did not contain monomers of Hop2-Mnd1.
Purification of Dmc1, Rad51, mouse Hop2, mouse Hop2-Mnd1 and E. coli SSB
Schizosaccharomyces pombe Rad51 and Dmc1 were purified as described (31). Mouse Hop2 or Hop2-Mnd1 was purified as described previously (24). Human RAD51 and Dmc1 were purified as described (32,33). Purified E. coli single-stranded binding protein (SSB) was purchased from USB. Purified BSA was obtained from Sigma (St. Louis, MO, USA).
Solubility tests, immunoprecipitations and affinity pull-downs
Escherichia coli total and soluble extracts expressing spMnd1, spHop2 or spMnd1 and spHop2 were prepared as follows. Escherichia coli BL21(DE3) RP (Stratagene) carrying either pET16b-spMnd1, pET28b-Hop2 or both plasmids were grown at 30°C in 50 ml LB with antibiotics. At OD600 = 0.4, spHop2 and spMnd1 synthesis was induced by the addition of 0.1 mM IPTG, incubated at 30°C, after 4 h, the cells were harvested by centrifugation, frozen in dry ice and stored at −80°C. The cell paste was resuspended in 5 ml of P5 buffer (50 mM sodium phosphate pH 7.0, 500 mM NaCl, 10% glycerol, 0.02% Triton X-100) containing 5 mM imidazole and the protease inhibitors PMSF (1 mM), aprotinin (0.019 TIU/ml) and leupeptin (1 μg/ml). The suspension was divided into 40 ml aliquots, which were sonicated three times for 30 s. An aliquot was collected and insoluble material was removed by centrifugation at 13 000 r.p.m. for 20 min. The supernatant was kept as the soluble fraction.
Immunoprecipitations were conducted with soluble extracts from E. coli BL21(DE3) RP (Stratagene) carrying pET16b-spHop2 and pET28b-spMnd1-Myc prepared as above. Protein complexes in the supernatant were pulled down for 1.5 h at 4°C using anti-Myc antibody (Santa Cruz, California, CA, USA) or beads also. Complexes were washed four times in P5 buffer, and visualized by western blotting using anti-mHop2 or anti-His antibody.
For affinity pull-downs, purified BSA, spDmc1 or spRad51 were covalently bound to NHS-activated Sepharose (Amersham, Piscataway, NJ, USA) to generate the affinity columns. spHop2-Mnd1 was incubated with affinity matrix in 20 μl buffer containing 20 mM Tris–HCl, pH 7.5, 150 mM KCl, 10% glycerol, 1 mM DTT and protease inhibitors. After 15 min of incubation at 37°C, the samples were washed four times with the same buffer. The proteins were eluted with SDS-PAGE loading buffer and visualized by western blotting using anti-His antibody.
Molecular mass estimation of spHop2-Mnd1
The molecular mass of purified spHop2-Mnd1 (100 μg) was determined by comparison with gel filtration standards [250 μg; bovine thyroglobulin (670 kDa), bovine gamma globulin (158 kDa), chicken ovalbumin (44 kDa), horse myoglobin (17 kDa) and vitamin B-12 (1.35 kDa)]. Proteins were analyzed on a FPLC Explorer 10 system fitted with a 24 ml Superdex 200 PC 3.2/30 column (Pharmacia, Peapack, NJ, USA) equilibrated in R150 buffer (20 mM Tris–Cl pH 8.0, 150 mM NaCl, 10% glycerol, 1 mM EDTA, 0.5 mM DTT). Fractions (250 μl) were collected and analyzed by SDS-PAGE followed by western blotting with a monoclonal anti-histidine antibody (BD Biosciences) or an antibody against mouse Hop2 that recognize S. pombe Hop2. Native (non-denaturing) electrophoresis was performed using the NativePAGE Novex Bis–Tris gel system (Invitrogen).
Yeast two-hybrid analysis
The full-length S. pombe Meu13+ and Mcp7+ genes were cloned into pGBK-T7 and pGAD-T7 (BD Biosciences), to produce fusions to the Gal4 DNA-binding and activation domains. Plasmids spHop2-NAD (amino acids 1–106), spHop2-CAD (107–211) and spMnd1-NAD (amino acids 1–110), spMnd1-CAD (amino acids 111–216) were constructed by cloning the appropriate PCR products in pGBK-T7. All fusions were confirmed by sequencing. AH109 was transformed with the indicated plasmids. Colony growth on media lacking tryptophan and leucine was observed with all constructions tested herein. Interactions between partners were assayed by growth on synthetic media lacking tryptophan, leucine, adenine and histidine supplemented with 2.5 mM 3-aminotriazole. Transformations were carried out according to the matchmaker kit manual (BD Biosciences).
DNA-binding assay
Reactions (10 μl) contained 100 nM DNA in binding buffer (20 mM triethanolamine–HCl, pH 7.5, 2 mM ATP, 1 mM Mg (CH3COO)2, 1 mM DTT and 100 μg/ml BSA). After 5 min at 37°C, the indicated amount of spHop2-Mnd1 was added (2 μl) and incubation was continued for a further 10 min. Complexes were fixed by addition of 0.2% glutaraldehyde followed by 15 min incubation at 37°C. Protein–DNA complexes were analyzed by 6% PAGE using TBE buffer. The gels were dried on DE81 filter paper followed by autoradiography. DNA substrates were prepared by annealing a 32P-labeled oligonucleotide (100 nt in length) with appropriate complementary sequences. The 100-mer double-stranded, and the corresponding 5′-tailed and 3′-tailed DNA (containing both 50 nt of ssDNA) were purified by 10% PAGE. The sequence of the 100-mer is 5′-GGGCGAATTGGGCCCGACGTCGCATGCTCCTCTAGACTCGAGGAATTCGGTAC CCCGGGTTCGAAATCGATAAGCTTACAGTCTCCATTTAAAGGACAAG-3′. DNA concentrations are expressed in terms of moles of nucleotides. Percentage of DNA binding were quantified on a Storm 860 phosphorimager (Molecular Dynamics, Piscataway, NJ, USA).
Single-strand annealing reactions
The substrate was prepared as follows. pPB4.3 was digested with NdeI and HindIII, dephosphorylated with calf alkaline phosphatase (NEB), followed by gel purification of the corresponding 400 bp fragment using Quiaquick gel purification kit (Qiagen). The 400 bp fragment was end-labeled with g-ATP and T4 polynucleotide kinase (NEB). Reactions (10 μl) contained denatured 5′-end labeled 400 bp fragment (450 nM) and spH2M1 (1.2 μM) in HEPES buffer (20 mM HEPES pH 7.5, 6.5 mM Mg (CH3COO)2, 2 mM ATP, 5 mM DTT, 100 mM NaCl and 100 μg/ml BSA). Incubation was at 20°C for 5 min or the indicated time. The reaction products were deproteinized by addition of one-tenth volume of stop buffer (10% SDS and 10 mg/ml proteinase K) followed by 15 min incubation at 20°C. Labeled DNA products were analyzed by electrophoresis through a 4% TBE1X/PAGE gel run at 150 V for 2 h 15 min, dried onto DE81 filter paper and visualized by autoradiography.
Strand exchange reactions
Reactions were performed according to Lio et al. (34). Reactions (10 μl) contained a 63-mer single-stranded oligonucleotide (0.7 μM) with the indicated concentrations of spHop2-Mnd1 and spDmc1 in PTUK buffer (20 mM Tris–HCl, pH 7.5, 2.5 mM MgCl2, 2 mM ATP, 1 mM DTT, 7.5 mM creatine-phosphate and 30 U/ml creatine kinase). After 5 min at 37°C, the corresponding 32P-end labeled 63 bp dsDNA (0.7 μM) was added and incubation was continued for 90 min. Reaction products were deproteinized by addition of one-fifth volume of stop buffer (0.1 M Tris–HCl, pH 7.5, 0.1 M MgCl2, 3% SDS, 5 μg/ml ethidium bromide and 10 mg/ml proteinase K) followed by 30 min incubation at 37°C. Labeled DNA products were analyzed by electrophoresis through 8% TBE1X/PAGE. For DNA-melting assays, 13.1 μM of a 79-mer ssDNA was included in the reaction stop buffer.
Strand exchange with longer substrates contained purified single-stranded pPB4.3 DNA (15 μM) with the indicated concentrations of either human Dmc1 or Rad51 and/or mouse Hop2 or Hop2-Mnd1 in standard buffer (50 mM triethanolamine–HCl, pH 7.5, 1 mM Mg (OAc)2, 2 mM ATP, 1 mM DTT and 100 μg/ml BSA). Otherwise indicated, hRad51 or hDmc1 was added before mHop2 or mHop2-Mnd1 on the ssDNA. After 5 min at 37°C, 32P-end labeled pPB4.3 DNA (400 bp fragment) was added and incubation was continued for 90 min. Reaction products were deproteinized as above. Labeled DNA products were analyzed by electrophoresis through 0.8% TAE agarose gels containing 1 μg/ml ethidium bromide, run at 4.3 V/cm, dried onto filter paper and visualized by autoradiography.
Strand invasion assays
Strand invasion assays were performed as published previously (24).
Electron microscopy
Schizosaccharomyces pombe Hop2-Mnd1 or human Rad51 reactions (10 μl) contained 5 μM ϕX174 single-stranded DNA, or pPB4.3 tailed DNA in 20 mM triethanolamine–HCl, pH 7.5, 1 mM DTT, 2 mM ATP, 2.5 mM Mg (CH3COO)2. After 5 min at 37°C, the indicated amount of protein was added and incubation was continued for a further 10 min. When fixation was required, protein–DNA complexes were fixed by addition of glutaraldehyde to 0.2% followed by 15 min incubation at 37°C. Samples were diluted and washed in 5 mM Mg (CH3COO)2 prior to uranyl acetate staining (35). Complexes were visualized at a magnification of 22000× using a Philips CM12 electron microscope.
RESULTS
Expression and purification of S. pombe Hop2 and Mdn1
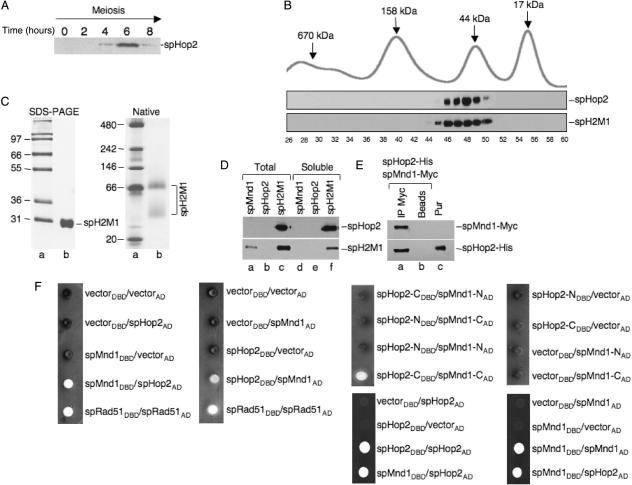
The expression of Meu13 (herein, S. pombe Hop2) during meiosis was monitored in order to amplify the gene by RT-PCR since it contains four introns. Meiosis was induced and the expression of spHop2 was observed by western blot analysis. Maximal expression of S. pombe Hop2 protein was observed after 6 h (Figure 1A), which is consistent with published northern blotting experiments (20). Hence, the Meu13+ gene was amplified by RT-PCR from mRNA extracted 6 h after the induction of meiosis. The mcp7+ gene (Mnd1+) was amplified from genomic DNA and the intron was removed by a modified Quickchange site-directed mutagenesis protocol (30). S. pombe hop2+ was cloned into pET28b and S. pombe mnd1+ was cloned in pET16b (Novagen) to add, respectively, a 10- and 6-histidine tag at the N-terminus of the proteins. Escherichia coli BL21 (DE3) RP carrying both plasmids was induced by IPTG, the solubilized proteins were then subjected to Talon affinity chromatography. Individual fractions were subjected to SDS-PAGE and Coomassie blue staining. By carefully selecting the fractions, the Hop2–Mnd1 proteins were generally pure after this step. When necessary, a Mono S chromatography was performed and co-elution of spHop2-Mnd1 was observed. The purified proteins were free of endo- and exonuclease activities (data not shown). The predicted molecular weight of his-tagged Hop2 is 28 652 Da and his-tagged Mnd1 is 26 761 Da. Hence, the two proteins co-migrate on a SDS-PAGE (Figure 1C, SDS-PAGE). Mass spectrometry was performed to confirm the purity of our preparation and the presence of both Hop2 and Mnd1 (data not shown).
Figure 1.
Purification and interaction of fission yeast Hop2-Mnd1. (A) Expression of Hop2 during S. pombe meiosis. Meiosis was induced by nitrogen starvation and the expression of spHop2 was observed by western blot analysis using anti-Hop2 at the indicated times. (B) Native molecular mass of purified spH2M1 as determined by gel filtration through Superdex 200. Top part: size standards, bottom part: western blotting of spHop2 and spH2M1 using anti-mHop2 and anti-His, respectively. (C) Left: SDS-PAGE of purified spHop2-Mnd1 (spH2M1). Lane a, molecular weight markers; lane b, purified spHop2-Mnd1 (5 μg). Proteins were loaded on a 10% SDS-PAGE and stained with Coomassie blue. Right: native electrophoresis of purified spHop2-Mnd1. Lane a, NativeMark protein standard; lane b purified spHop2-Mnd1 (2 μg). Proteins were loaded on a 4–16% NativePAGE Bis–Tris gel and stained with Coomassie blue. (D) Solubility of spMnd1, spHop2 and co-expressed spHop2 and spMnd1 in E. coli. Thirty microgram of total (lanes a–c) or soluble extracts (lanes d–f) of E. coli BL21 (DE3) RP (Stratagene) expressing the indicated proteins were prepared and subjected to western blotting of spHop2 and spH2M1 using anti-mHop2 and anti-His, respectively. (E) Co-immunoprecipitation of spHop2 and spMnd1. SpMnd1-Myc and spHop2-His were co-expressed in E. coli and soluble extracts were prepared followed by immunoprecipitation with anti-Myc (lane a) or beads alone (lane b). The proteins were detected by western blotting using anti-Myc and anti-His, respectively. Lane c, purified H2M1 (100 ng). (F) Fission yeast Hop2 interacts with Mnd1 by two-hybrid analysis. AH109 strain was co-transformed with the indicated plasmids and positive interactions were monitored by growth and coloration on media lacking tryptophan, leucine, histidine, adenine and supplemented with 2.5 mM aminotriazole. The annotations DBD and AD design fusions to the Gal4 DNA-binding domain and Gal4 activation domain, respectively.
Gel filtration and interactions between S. pombe Hop2 and Mnd1
The native molecular weight of the spHop2-Mnd1 protein preparation was determined by gel filtration through Superdex 200. The bulk of spHop2 and spMnd1 eluted in fractions 46–50 suggesting that these proteins could form complexes containing a variable number of molecules (Figure 1B). The corresponding molecular mass of these fractions range from 47 to 80 kDa with a peak observed at 60 kDa (Figure 1B, lanes 54–56). Native electrophoresis experiments revealed that our preparation contained two complexes of 40 and ∼66 kDa (Figure 1C, native). Given the predicted molecular weight of his-tagged Hop2 (28.6 kDa) and his-tagged Mnd1 (26.7 kDa), this is indicative of multimeric forms composed of homo- or hetero-complexes. Monomeric Hop2 or Mnd1 were not detected (Figure 1B, lane 56 and Figure 1D).
In order to characterize these complexes, three experiments were conducted. First, we expressed spMnd1, spHop2 or both proteins in E. coli and total and soluble extracts were prepared (Figure 1D). When expressed on their own, spMnd1 and spHop2 were not soluble (lanes d–e). Solubility was only achieved if both proteins were co-expressed (lane f). Similar results were obtained in insect cells (data not shown). This result suggests that our protein preparation is composed of an hetero-complex of spHop2-Mnd1. Second, immunoprecipitations were conducted using cells extracts expressing Myc-tagged spMnd1 and His-tagged spHop2. Anti-Myc antibodies pulled down a complex between spMnd1 and spHop2 (Figure 1E, lane a). Third, the interactions of Hop2 and Mnd1 were further characterized by two-hybrid analysis. The AH109 strain (BD Bioscience) was co-transfected with plasmids encoding fusions to the Gal4 DNA binding and activation domains, respectively, and spotted on media lacking tryptophan, leucine, histidine and adenine with 3-aminotriazole. Strong interactors are white in color because of the expression of the ADE gene. Co-expression of full-length spMnd1 fused to the Gal4 DNA binding and full-length spHop2 fused to the activation domains in yeast lead to growth of white colonies on omission media (Figure 1F). Interactions were also observed when spHop2 fused to the Gal4 DNA binding and Mnd1 fused to the activation domain were co-transformed. The strength of interactions between spHop2 and Mnd1 was similar to spRad51 self-interactions, which indicates a strong interaction between the proteins. Truncations of spHop2 and spMnd1 in N-terminal and C-terminal portions revealed that the C-terminus of spHop2 (amino acids 111–216) interacts with the C-terminal portion of spMnd1 (amino acids 107–211). The N-terminus of spHop2 (amino acids 1–110) and spMnd1 (amino acids 1–106) did not interact with each other or the C-terminus of the proteins. Interestingly, spHop2 and spMnd1 self-interactions were also observed suggesting that these complexes can form in vivo although we were unable to get these proteins soluble when expressed on their own.
DNA binding by S. pombe Hop2-Mnd1
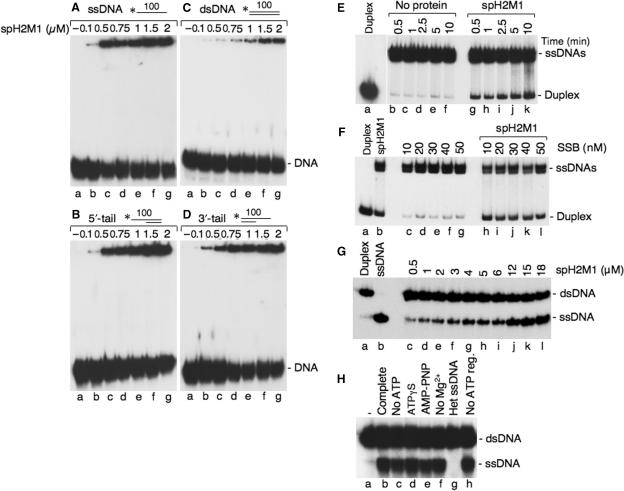
In the presence of ATP, RecA binds single-strand DNA preferentially over double-strand DNA because of a kinetic barrier to associate with dsDNA (36). This difference in affinity is critical for strand exchange reactions since pairing occurs between single-strand DNA and a naked duplex DNA. Electrophoretic Mobility Shift Assays were used to investigate the ssDNA- and dsDNA-binding properties of spHop2-Mnd1. spHop2-Mnd1 bound preferentially to ssDNA, over 3′- or 5′-tailed DNA or dsDNA (Figure 2A–D). We observed the formation of protein–DNA networks that tended to smear up the gel, which is characteristic of the formation of networks containing a variable number of protein and DNA molecules. Quantification of the DNA–protein complexes at 0.5 μM Hop2-Mnd1 indicated that 31% of the input ssDNA was stably bound compared with 10% for 3′-tailed and 5′-tailed DNA or 3% for dsDNA. The formation of protein–DNA complexes on ssDNA was optimal at 50 mM NaCl, supported in a wide range of Mg (CH3COO)2 concentrations (from 1 to 10 mM; with an optimal concentration at 2 mM) and was gradually reduced by inclusion of increasing concentrations of potassium chloride over 150 mM (data not shown). Binding to ssDNA was observed both in the absence of ATP or ATP analogs suggesting that DNA binding can occur without ATP or ATP hydrolysis.
Figure 2.
DNA binding, single-strand annealing and strand exchange properties of S. pombe Hop2-Mnd1. (A–D) DNA binding of spH2M1. DNA-binding reactions contained ssDNA (A), 5′-tailed duplex DNA (B), dsDNA (C) and 3′-tailed duplex DNA (D) and the indicated concentrations of spH2M1. (E) Time course of single-strand annealing by spH2M1. Lane a, purified 400 bp duplex DNA. Reactions contained denatured 400 bp duplex DNA and no protein (lanes b–f) or spH2M1 (1.2 μM, lanes g–k). The reactions were stopped at the indicated times. (F) spH2M1 promote single-strand annealing in the presence of a single-strand binding protein. Lane a, purified 400 bp duplex DNA; lane b, spH2M1 alone (1.2 μM); lanes c–g, SSB alone; lanes h–l, spH2M1 (1.2 μM) with the indicated concentrations of SSB. (G) spH2M1 promotes strand exchange. spH2M1 was mixed with 1.5 μM 63-mer ssDNA. DNA strand exchange was initiated by addition of homologous 3 μM of 63-bp dsDNA, in which the strand that would be displaced during DNA strand exchange was 32P-labeled.Lane a, duplex DNA; lane b, denatured duplex DNA; lanes c–l, DNA strand exchange as a function of spH2M1 concentration. (H) Effect of cofactors on spH2M1 strand exchange. Reactions were carried out in buffer containing an ATP regeneration system using 3 μM spH2M1 (lane b); in buffer without ATP (lane c); in standard buffer in which ATP was replaced with ATPγS (lane d); AMP-PNP (lane e); standard buffer lacking Mg2+ (lane f); reactions with heterologous ssDNA (lane g); in buffer without ATP regeneration (lane h); lane a, without protein.
Strand annealing and strand exchange catalyzed by Hop2-Mnd1
We next investigated their ability to promote single-strand annealing (Figure 2E). Long single-strand DNA of 400 bases was used instead of oligonucleotides, which can be prone to artifacts. Interestingly, spHop2-Mnd1 promotes annealing of long complementary single stranded DNAs. Single-strand annealing was observed rapidly, within 0.5 min (6%) and increased to a level of 16% after 10 min (Figure 2E, lanes g–k). Low levels of spontaneous strand annealing were observed in the control alone (lanes b–f). The renatured products were in the form of linear duplex as opposed to high molecular weight networks. Annealing by spHop2-Mnd1 could persist in the presence of a single-strand binding protein (Figure 2F). The functional analogy between RPA and SSB is supported by results indicating that SSB can substitute for budding yeast and human RPA in strand exchange mediated by S. cerevisiae and human Rad51, respectively (37,38). spHop2-Mnd1 annealing did not require ATP consistent with the fact that spHop2 or spMnd1 do not contain any known ATP-binding motif (data not shown).
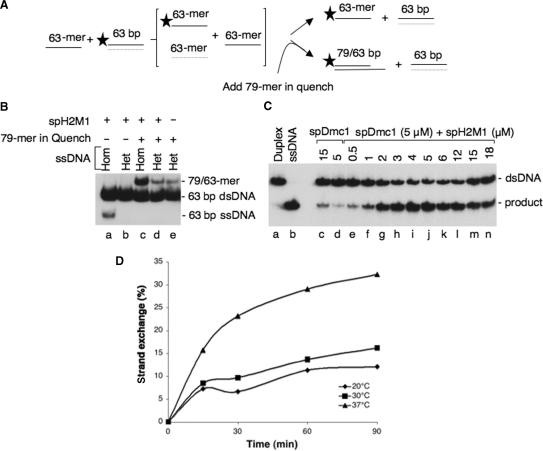
Since spHop2-Mnd1 promoted strand annealing, a characteristic shared by the S. pombe Rad51 and Dmc1 recombinases (31), we investigated whether spHop2-Mnd1 possessed other recombination activities. We then examined the ability of purified Hop2-Mnd1 to promote strand exchange in vitro using a linear single-stranded DNA and homologous linear duplex DNA. spHop2-Mnd1 could perform strand exchange with oligonucleotides (Figure 2G, lane c–l) but not with longer substrates (data not shown) suggesting that it has limited ability to do this reaction. The reaction did not require ATP hydrolysis but homologous single-strand DNA (Figure 2H, lanes b–f, g). DNA strand exchange activity with oligonucleotides can be due to thermal opening of the duplex followed by subsequent annealing, as demonstrated for the RAD51C protein (34). In order to eliminate this possibility, we performed experiments as described by Lio et al. (Figure 3A–B). First, no released ssDNA was produced when the DNA substrates were heterologous (Figure 3B, compare lanes a–b). This result demonstrates that spHop2-Mnd1 DNA strand exchange is homology-dependent. Next, the standard homology-dependent experiment was carried out, with an excess of homologous ssDNA added to the stop buffer. The oligonucleotide used is of a different length (79-mer). DNA pairing was observed with homologous DNA but not heterologous DNA (compare lane c–d). The level of pairing observed with heterologous DNA was similar to spontaneous reannealing (lane e). This result shows that spHop2-Mnd1 promotes strand exchange independently of a DNA melting activity. The temperature dependence of spHop2-Mnd1 strand exchange was also analyzed. Strand exchange increased over a period of 90 min and was more efficient at 37°C than 30°C or and 20°C (Figure 3D). This result is inconsistent with DNA melting which occurs within 5 min (34). Since spHop2-Mnd1 are expected to function in concert with the spRad51 and spDmc1 recombinases during meiotic recombination we investigated whether spHop2-Mnd1 was able to stimulate spDmc1 and spRad51 strand exchange. We have previously shown that spRad51 and spDmc1 have strand exchange abilities (31). When limiting concentrations of spDmc1 were used, only 2% of displaced ssDNA were produced (Figure 3C, lane d, 2% products). However, when spDmc1 was supplemented with spHop2-Mnd1 at concentrations of 2–5 μM, strand exchange occurred at a higher level than with either protein alone (compare Figure 3C, lanes g–j to Figure 2G, lanes e–h). Quantification of the products at 5 μM spDmc1 and 4 μM spHop2-Mnd1-Dmc1 revealed a 5-fold increase in activity compared to spHop2-Mnd1 alone. These results reveal that stimulation of spHop2-Mnd1 was specific for spDmc1, as the effect was not observed on Rad51 (data not shown).
Figure 3.
spHop2-Mnd1-promoted DNA strand exchange is not due to a DNA-melting activity. (A) Strategy to detect DNA melting. If spH2M1 melts the dsDNA, the free ssDNA strands would quickly re-anneal after deproteinization. The standard strand exchange with 63-mer oligonucleotides were carried out, but in addition, an excess amount of 79-mer homologous ssDNA was added to the stop buffer. Thus, pairing that might occur during the deproteinization step would be distinguished from DNA strand exchange with the intended partner as diagramed. (B) Strand exchange reactions were carried using homologous 63-mer ssDNA (lane a); heterologous 48-mer ssDNA (lane b); homologous 63-mer ssDNA and 79-mer ssDNA in the reaction stop buffer (lane c); heterologous ssDNA and 79-mer ssDNA in the reaction stop buffer (lane d); heterologous ssDNA and 79-mer ssDNA in the reaction stop buffer without protein (lane e). (C) Stimulation of S. pombe Dmc1 strand exchange by spH2M1. Lane a, duplex DNA; lane b, denatured duplex DNA; lane c, spDmc1 (15 μM); lane d, spDmc1 (5 μM); lanes e–n, effect of the indicated concentrations of spH2M1 (0.5–18 μM) on spDmc1-mediated strand exchange (5 μM). (D) Time-course and temperature dependence of spH2M1 strand exchange. spH2M1 (μM) strand exchange reactions were carried out at 20°C (diamonds), 30°C (squares) or 37°C (triangles) over a period of 90 min.
Dmc1 strand invasion is stimulated by Hop2-Mnd1
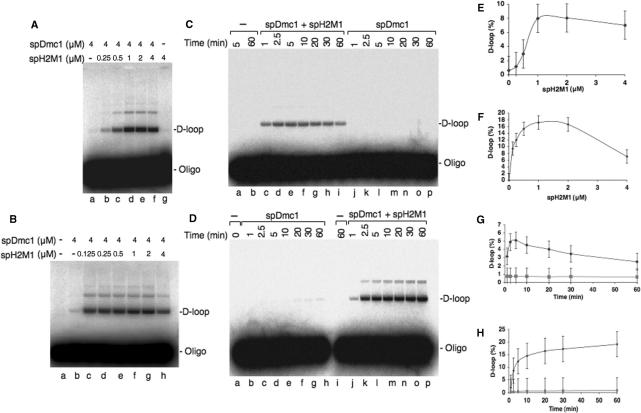
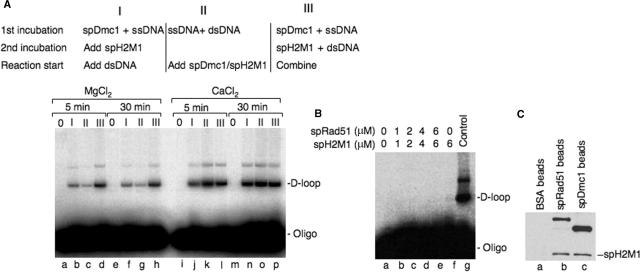
spHop2-Mnd1 are also expected to help the spRad51 and spDmc1 recombinases during strand invasion. First, purified spHop2-Mnd1 was unable to perform strand invasion on its own (Figure 4A, lane g). Strikingly, spHop2-Mnd1 stimulated spDmc1 D-loop formation in a buffer containing magnesium (Figure 4A, lanes b–f). At 1 μM Hop2-Mnd1 a 8-fold increase was observed (Figure 4E). This spDmc1-dependent spHop2-Mnd1 stimulatory effect was even greater in calcium buffer (Figure 4B, lanes c–h) and quantification of the products revealed a 17-fold increase in D-loop formation at 1 μM spHop2-Mnd1 (Figure 4F). D-loop formation is a reversible reaction, known as the D-loop cycle (39). Therefore, we studied the reversibility of the D-loop assay using 1 μM spHop2-Mnd1, which was optimal for the magnesium and calcium reactions (Figure 4E and F). In magnesium buffer, we observed a constant diminution of the D-loop reaction from 5 to 60 min (Figure 4C and G). Apparent reversibility of the D-loops may be due to nicking of the supercoiled plasmid DNA by contaminating nucleases. This possibility was ruled out since 4 μM concentrations of spDmc1 or spH2M1 did not reveal any increase in the nicked DNA (data not shown). D-loop cycles were not observed in calcium buffer as D-loop products increased over a period of 60 min (Figure 4D and H). The order of addition of the proteins to the DNA in D-loop was assayed in a magnesium buffer. Stimulation of spHop2-Mnd1 on spDmc1 was maximal when Dmc1 was added to the ssDNA and spHop2-Mnd1 to the dsDNA and the two mixtures were combined (Figure 5A, lanes d and h). Intermediate levels of stimulation were observed when spDmc1 was loaded on the ssDNA, followed by spHop2-Mnd1 and the dsDNA (Figure 5A, lanes b and f). When the ssDNA and dsDNA were premixed, followed by the addition of spDmc1 and spHop2-Mnd1, stimulation occurred at lower efficiency (Figure 5A, lanes c and g). Notably, spHop2-Mnd1 stimulated spDmc1 regardless of the order of addition in calcium buffer (Figure 5A, lanes j–p). These results suggest that calcium is an important regulator of spHop2-Mnd1 functions. As for strand exchange, SpHop2-Mnd1 enhanced the activity of spDmc1 specifically, as the effect was not observed on spRad51 (Figure 5B). No stimulation of the D-loop formation was observed at concentrations of Rad51 and Hop2-Mnd1 up to 6 and 8 mM, respectively. Negative results were also observed when various order of addition was used as well as different salt conditions and the use of non-hydrolysable ATP analogs ATPgS and AMP-PNP (data not shown). A number of different Rad51: Hop2-Mnd1 ratios were tested and at least two time points (8 and 30 min) were taken for all reactions. Specific stimulation of Dmc1 but not Rad51 by Hop2–Mnd1 complex was also observed in budding yeast S. cerevisiae [(23) and Ting-Fang Wang, personal communication]. Therefore, we investigated whether spHop2-Mnd1 interacted with spDmc1 and spRad51 (Figure 5C). Pull-down assays revealed an interaction with both spRad51 and spDmc1. Hence, the specificity of spHop2-Mnd1 for spDmc1 is not due to protein–protein interactions in solution.
Figure 4.
Fission yeast Hop2-Mnd1 stimulates D-loop formation by spDmc1. (A) Effect of spH2M1 in a buffer containing Mg2+ (2.5 mM). Lane a, spDmc1 alone (4 μM); lanes b–f, effect of spH2M1 (0.25–4 μM) on spDmc1-mediated D-loop formation (4 μM); lane g, reaction with spH2M1 (4 μM) alone. (B) Effect of spH2M1 in a buffer containing Ca2+ (2.5 mM). Lane a, DNA substrates (ssDNA and dsDNA) without proteins; lane b, spDmc1 alone (4 μM); lanes c–h, effect of spH2M1 (0.125–4 μM) on spDmc1-mediated D-loop formation (4 μM). (C) Analysis of the reversibility of the D-loop reaction by S. pombe Dmc1 (4 μM) and spH2M1 (1 μM) in buffer containing Mg2+ (2.5 mM). (A) Lane a, no protein (5 min reaction); lane b, no protein (60-min reaction); lanes c–i, reactions with spDmc1 and spH2M1 stopped at 1, 2.5, 5, 10, 20, 30 and 60 min, respectively; lanes j–p, spDmc1-mediated D-loop formation stopped at 1, 2.5, 5, 10, 20, 30 and 60 min, respectively. (D) Absence of reversibility of the D-loop reaction by spDmc1 (4 μM) and spH2M1 (1 μM) in buffer containing Ca2+ (2.5 mM). Lane a, no protein (reaction was stopped immediately); lanes b–h, spDmc1-mediated D-loop formation stopped at 1, 2.5, 5, 10, 20, 30 and 60 min, respectively; lane i, no protein (60 min reaction); lanes j–p, reactions with spDmc1 and spH2M1 stopped at 1, 2.5, 5, 10, 20, 30 and 60 min, respectively. (E–F) Quantification of (A), lanes b–g and (B), lanes c–h, respectively. (G) Quantification of (C), diamonds represent lanes c–i and squares correspond to lanes j–p. (H) Quantification of (D), diamonds signify lanes j–p and squares correspond to lanes b–h.
Figure 5.
(A) Effect of the order of addition on S. pombe Dmc1-Hop2-Mnd1 D-loop formation in Mg2+ or Ca2+ buffer. Reactions with Dmc1 (4 μM) and Hop2-Mnd1 (1 μM) were performed for 5 min (lanes a–d) or 30 min in MgCl2 (2.5 mM) (lanes e–h) or 5 min (lanes i–l) or 30 min (lanes m–p) in CaCl2 buffer. (2.5 mM). (B) SpH2M1 does not stimulate spRad51. Indicated concentrations of spRad51 and spH2M1 were used (lanes a–f) and mammalian Dmc1 and Hop2-Mnd1 reaction is shown for comparison (lane g). (C) SpH2M1 interacts with both spDmc1 and spRad51. Purified His-tagged spH2M1 was incubated with affinity beads containing crosslinked BSA (lane a), spRad51 (lane b), or spDmc1 (lane c). Eluted material was probed by western blotting using anti-His antibody.
hRad51 and hDmc1 strand exchange is stimulated by mouse Hop2 and Hop2-Mnd1
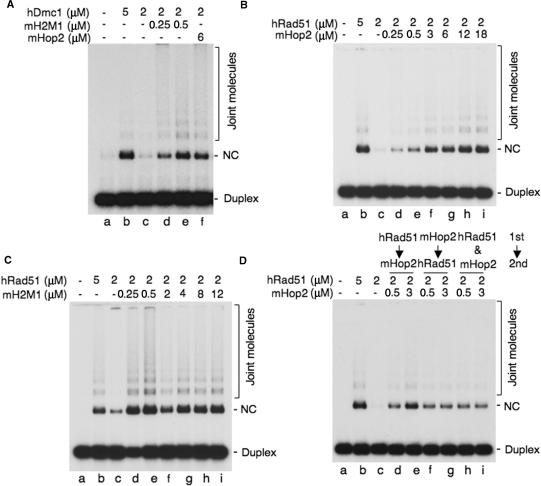
We were curious to verify whether this specificity was conserved during evolution. Using mouse Hop2 and Hop2-Mnd1, we next investigated its ability to promote hRad51 and hDmc1 strand exchange in vitro in the presence of larger substrates using a circular single-stranded DNA (4300 nucleotides) and homologous linear duplex DNA (400 bp). Although the proteins used in this assay come from different species, it is important to note that the human and mouse Rad51 and Dmc1 recombinases are homologous at 98% at the amino acid level. When hDmc1 was used at subsaturating concentrations (Figure 6A, lane c), we observed a strong stimulation by both mHop2-Mnd1 (lanes d–e) and mHop2 (lane f). hRad51 strand exchange was also enhanced by increasing concentrations of mHop2 (Figure 6B, lanes d–i) or mHop2-Mnd1 (Figure 6C, lanes d–i). Stimulation was also observed either when hRad51 or mHop2 was added to the ssDNA first, or when the proteins were premixed and added to the ssDNA (Figure 6D, lanes d–i). Enhancement of hRad51 or hDmc1 strand exchange by mHop2 or mHop2-Mnd1 required the presence of ATP. Strand transfer products were not observed when ATP was replaced by the non-hydrolysable form AMP-PNP, when Mg2+ was omitted or when heterologous single-stranded DNA was used (data not shown). To ascertain whether mHop2 or mHop2-Mnd1 was able to stimulate hRad51 or hDmc1 activity specifically, and not any recombinase, we tested whether mHop2 was able to stimulate E. coli RecA. Very low level of strand exchange was observed in the presence of both proteins (data nor shown). Altogether, we conclude that mouse Hop2 or Hop2-Mnd1 can work in concert specifically with hRad51 or hDmc1 and not a distant homolog such as RecA.
Figure 6.
Mouse Hop2 or Hop2-Mnd1 stimulates hDmc1 or hRad51 strand exchange. (A) Stimulation of hDmc1 strand exchange by mHop2 or mH2M1 using long DNA substrates (lanes a–f). Single-stranded DNA (15 μM) was incubated with the indicated concentrations of hDmc1 and mHop2 or mH2M1 proteins for 5 min at 37°C followed by addition of 32P-end labeled double-stranded DNA (1.38 μM) and incubation at 37°C for 90 min. hDmc1 was added before mHop2 or mHop2-Mnd1 to the ssDNA. (B) Stimulation of hRad51 strand exchange by the indicated amounts of mouse Hop2 (lanes a–i) and (C) mH2M1 (lanes a–i). The reactions were carried out as described in (A). (D) Effect of the order of addition of the proteins on hRad51-mHop2 DNA strand exchange. Lane a, no protein; lane b, hRad51 (5 μM); hRad51 (2 μM, lane c) was added to ssDNA first followed by addition of the mHop2 protein (0.5 and 3 μM, lanes d and e); mHop2 was added to ssDNA first followed by addition of the hRad51 protein (lanes f and g); or both of the proteins were added to ssDNA at the same time (lanes h and i). The reactions were then carried out as described in (A). Joint molecules and Nicked circles (NC) are indicated.
Visualization of fission yeast Hop2-Mnd1 on DNA
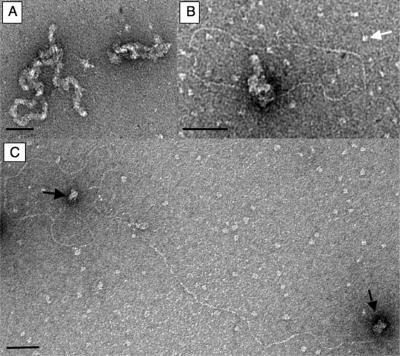
When complexes formed between spHop2-Mnd1 and DNA were visualized by electron microscopy, we observed large protein complexes bound to single-strand DNA (Figure 7A, right). We also observed less frequently irregular filament-like structures possibly due to many molecules bound to the ssDNA (Figure 7A, left). Consistent with gel-retardation assays, spHop2-Mnd1 had high affinity for ssDNA since unbound protein was not distinguished. Because meiotic recombination is thought to be initiated by DNA that contains single-stranded tails, the possibility that spHop2-Mnd1 may bind specifically to tailed linear duplex molecules was investigated. Following uranyl acetate staining, duplex DNA is well visible while single-stranded DNA, if not bound by proteins, is collapsed to very small blobs or bushy structures. Consistent with a specific interaction with ssDNA, spHop2-Mnd1 (5 μM) selectively bound the ssDNA of linear duplex DNA molecules with ssDNA at both ends (Figure 7B). Very interestingly, the two ends of the tails were often found entangled in one and other, suggestive of homology search. Unbound spHop2-Mnd1 was also discerned (white arrow). In order to confirm specific interactions with ssDNA, we also observed binding of ssDNA ends at lower concentrations of spHop2-Mnd1 (Figure 7C). About 70% of the molecules observed had spHop2-Hop2 at both extremities. These results are consistent with specific roles for spHop2-Mnd1 in strand invasion during meiotic homologous recombination.
Figure 7.
Electron microscopic visualization of fission yeast Hop2-Mnd1. (A) Electron microscopic visualization of spH2M1 (1.65 μM) bound to ϕX174 single-stranded DNA (5 μM). (B) Close up view of spH2M1 (0.75 μM) bound to linear duplex with a single-stranded tail at both ends (5 μM). The white arrows designate unbound protein. (C) spH2M1 (5 μM) bound to linear duplex with a single-stranded tail at both ends (5 μM). The black arrows designate bound spH2M1 on ssDNA. The magnification bars represents 50 nm.
DISCUSSION
The DNA strand exchange activity of both Rad51 and Dmc1 is critical for proper meiotic DNA double-strand break repair in lower eukaryotes. Many factors have been found to stimulate Rad51, including RPA, Rad52 and Rad54. On the other hand, human Dmc1 is stimulated by the human Rad54B, which could be an ortholog of budding yeast Rdh54 and Tid1 (40). Much attention is now turned toward the Hop2 and Mnd1 proteins because of their important roles in meiotic recombination.
S. pombe Hop2 and Mnd1 form a complex binding ssDNA preferentially
Sequence analysis revealed that Hop2 and Mnd1 possesses a coiled-coil motif (41,42). Fission yeast Hop2 coiled-coil region spans amino acids 128–159 whereas spMnd1 possess a long coiled-coil from amino acids 82 to 149. Using two-hybrid analysis we showed that spHop2 interacts with spMnd1. Domain mapping experiments revealed that the C-terminus of spHop2 (amino acids 111–216) interacts with the C-terminal portion of spMnd1 (amino acids 107–211). Hence, it is not the complete coiled-coiled region of spMnd1 that is required for the interaction with spHop2. These results are in accordance with a recent report showing that mouse Hop2 and Mnd1 interact through their coil–coil motif and that the C-terminus of the protein is important for the interaction (41). Gel filtration experiments revealed that spHop2-Mnd1 eluted in a broad profile, suggesting that it can form structures containing a variable number of molecules. However, our results suggest that purified spHop2-Mnd1 strictly forms a hetero-complex in solution. Indeed, we were unsuccessful in detecting soluble spHop2 or spMnd1 alone suggesting that these proteins are unstable without each other. In accordance with this, the overexpression of Mnd1 suppressed a temperature-sensitive mutant allele of Hop2 in budding yeast (19). Instability of Hop2 without Mnd1 was also observed with the human homologs (26). The 40 kDa complex observed by native electrophoresis thus corresponds to spHop2-Mnd1. The 66 kDa complex most likely corresponds to a dimer of Hop2 bound to Mnd1 (or a dimer of Mnd1 bound to Hop2) or spHop2-Mnd1 having a different conformation. It is most likely that one or both of these spHop2–Mnd1 complexes exist in vivo. Immunoprecipitation analyses from S. cerevisiae meiotic cell extracts revealed that Hop2-Myc and Mnd1-GFP interact (18). Moreover, in pat1 fission yeast strain, Hop2-GFP and Mnd1-HA show almost identical expression profiles during meiosis and both proteins interact (21). Co-immunoprecipitation, of mouse Hop2 and Mnd1 has been reported (24). We do not exclude the possibility of other spHop2–Mnd1 complexes in vivo. We observed spHop2–Hop2 and spMnd1–Mnd1 interactions by two-hybrid analysis. In this context, perhaps endogenous S. cerevisiae proteins could stabilize spHop2–Hop2 or spMnd1–Mnd1 complexes in vivo.
SpHop2-Mnd1 were purified with short histidine tags at their N-terminus. The tags should not affect the function of the heterodimer as tagging spMnd1 or Hop2 with large proteins such as GFP did not affect spore viability [(20,21) and Hiroshi Nojima and Takamune Saito, personal communication]. When observed by electron microscopy, purified spHop2-Mnd1 formed filament-like structures on single-strand DNA. The nature of this structure remains to be determined but it might be the result of random binding of spHop2-Mnd1 to the ssDNA so the whole DNA is covered. Our electron microscopic observations support the concept of specific binding of spHop2-Mnd1 to ssDNA ends on 3′-tailed DNA. The preferential interaction with ssDNA tails may have important biological consequences at the initiation of recombination during meiosis since double-strand breaks are processed into 3′-tailed DNA molecules. Presumably, the resected tails could act as a landing site for spHop2-Mnd1 leading to the nucleation of a presynaptic filament of spDmc1. In contrast, S. cerevisiae and human Hop2-Mnd1 was found to bind dsDNA preferentially (23,26), whereas mouse Hop2–Mnd1 binds DNA with no preference toward single- or double-strand DNA (41). This is an important issue since it influences the way we perceive the roles of these proteins in vivo. Very importantly, we found that the addition of calcium eliminates the order of addition requirement on DNA in spDmc1-Hop2-Mnd1 strand invasion assays. Dependence of meiosis on Ca2+ has been observed during meiosis I, when homologous recombination occurs, and it has been shown that calcium stimulates strand exchange by the human Rad51 (43) and human Dmc1 proteins (44). Hence, although the preferential binding of Hop2–Mnd1 proteins on DNA differs between species, calcium might be a key component in the regulation of their interactions with DNA in vivo by altering preferential binding.
Strand invasion and strand exchange by S. pombe Hop2-Mnd1
Strand exchange with oligonucleotides can occur in an ATP-independent fashion and also by a DNA melting and strand separation activity (34). We show that SpHop2-Mnd1 does not possess DNA melting activity. We have shown previously that fission yeast Dmc1 and Rad51 can both promote strand exchange (31). At a concentration of one monomer per nucleotide, spDmc1 promotes weak strand exchange unless a single-strand binding protein such as SSB is included in the reaction (31). In support of this, RPA is required for meiotic recombination in budding yeast (45). Very interestingly, the addition of spHop2-Mnd1 markedly increases strand exchange activity of the spDmc1 protein. SpDmc1 strand invasion, but not spRad51, was stimulated by spHop2-Mnd1. Budding yeast Hop2-Mnd1 and mouse Hop2 can also stimulate Dmc1 specifically (23–25). However, it is not clear how this stimulation occurs. It may be caused by changes in the DNA structure resulting either from spHop2–Mnd1 binding. Second, although spHop2-Mnd1 interacts with both spRad51 and spDmc1, protein–protein interactions might only be beneficial for Dmc1 nucleoprotein filaments. Since spHop2-Mnd1, spDmc1 and spRad51 bind DNA, this might be difficult to assess experimentally.
D-loop products from Dmc1–spHop2–Mnd1 reactions were about 3-fold more abundant in calcium buffer than in magnesium buffer. Moreover, we observed that the D-loop reaction was not reversible in the presence of calcium. These results suggest that calcium might be an important regulator of spDmc1-Hop2-Mnd1 strand invasion in vivo. D-loop cycle was first described for RecA protein and was proposed to protect from illegitimate recombination (39). Only if the sequences are truly homologous, successive D-loop cycles at many sites will result in stable pairing. Ca2+ was shown to stimulate the strand exchange activity of human Rad51 protein by inhibiting the ATPase activity of Rad51 thus preventing the conversion of the Rad51-ATP-ssDNA filament to an inactive Rad51-ADP-ssDNA form (43). Hence, activation of DNA strand exchange appears to be correlated with calcium-triggered transition from inactive ring-shaped oligomers to active nucleoprotein filaments (40,46). It is possible that a similar mechanism might be involved in the S. pombe Dmc1–Hop2–Mnd1 reaction. Dmc1 filaments might be more stable in the presence of calcium and cycling of the D-loop reaction in Mg2+ buffer is observed only in the presence of ATP, but not when ATP is replaced by non-hydrolysable analog AMP-PNP (our unpublished data).
Knowing that fission yeast Hop2-Mnd1 enhances Dmc1 recombinase activity specifically, we investigated whether we could find the same stimulatory effect for mouse Hop2 or Hop2-Mnd1. As expected, the addition of mouse Hop2 or Hop2-Mnd1 markedly increases strand exchange activity of the human Dmc1 or Rad51 proteins. However, in order to achieve the same level of stimulation, a smaller amount of mHop2-Mnd1 was required compared to mHop2. Hence, mHop2-Mnd1 perhaps functions in a catalytic manner. It is interesting to note that mammalian Hop2 functions in transcription (47) and interacts with the DNA-binding domains of nuclear receptors (48). Our results suggest that Hop2, in the absence of Mnd1, can also function in DNA recombination. We believe that the stimulation observed involves a specific interplay between mHop2 or mHop2-Mnd1 and hRad51/Dmc1 for three reasons. First, we could not detect any stimulatory effect of mHop2 or mHop2/Mnd1 on RecA-promoted strand exchange. Second, the stimulatory activity was not severely affected by the order of addition of proteins on the single-stranded DNA, suggesting that there was no sequestration of ssDNA by mHop2 or mHop2-Mnd1 alone. This is important as sequestration of ssDNA would lead to a decrease in the available concentration of ssDNA by hDmc1 or hRad51 therefore increasing the local concentration of the recombinases per DNA molecule, which could explain the strand exchange activity. If it was the case, mHop2 or mHop2-Mnd1 would have also stimulated RecA. Third, hRad51 and hDmc1 stimulation by mHop2 or mHop2-Mnd1 was dependent on ATP hydrolysis, which suggests that the stimulation required the activity of both recombinases. Altogether, these results would argue that mHop2 is not a non-specific factor capable of stimulating any strand exchange reaction in general. Our results are in accordance with recent studies showing that human Hop2-Mnd1 stimulates hRad51 and hDmc1 strand exchange (26). In addition, we show that mHop2 can also stimulate hRad51 and hDmc1 and perhaps Mnd1 is the catalytic subunit.
Roles of Hop2-Mnd1 in meiotic recombination
Genetic studies have established that fission yeast Hop2 is important during DSB-independent homolog pairing when cells are transferred to sporulation media (20,49). Our results also support roles for Hop2-Mnd1 later, during homologous pairing and strand exchange. After resection of a meiotic double-strand break, the 3′-tailed DNA invades an homologous double-strand DNA, a step known as homologous pairing without extensive heteroduplex formation. In meiosis, this occurs between homologs rather than sister chromatids. Then, the heteroduplex region is expanded by strand exchange to form joint molecules and Holliday junctions. The Holliday junctions are resolved to produce at least one crossover per chromosome. We observed that spHop2-Mnd1 can promote strand exchange with oligonucleotides and stimulate strand invasion by spDmc1. These activities demonstrate a key role for spHop2-Mdn1 in strand invasion. Also, mHop2-Mnd1, stimulates hRad51 and hDmc1 strand exchange, which means that these proteins can stabilize interactions between homologous sequences, as suggested recently with genetic studies in budding yeast (50). A subset of meiotic DSBs is also repaired by synthesis-dependent strand annealing (51). It is thought that following the initial strand invasion and repair synthesis, the invading strand containing newly synthesized DNA is displaced and reannealed to the other 3′ end. Annealing by spHop2-Mnd1 might be very important in this reaction. Indeed, spHop2-Mnd1 promoted strong annealing with long 400 bases single-strand DNAs. Moreover, this reaction persisted in the presence of a single-strand binding protein. Altogether, our results suggest important functions of Hop2–Mnd1 complex in the various steps of homologous recombination. These results are supported in vivo by the phenotypes of the hop2 knock-out mice which displays failure of DSB repair and sterility, although the Rad51 and Dmc1 recombinases are still present (16).
ACKNOWLEDGEMENTS
We are grateful to Isabelle Brodeur and Amélie Rodrigue for helpful comments on the manuscript. We thank Drs Ting-Fang Wang, Hiroshi Nojima and Takamune Saito for communication of unpublished results, Dr Doug Bishop for S. cerevisiae Dmc1 antibody, Synthia Sauvageau and Pierre Plante for technical help, anonymous reviewers for helpful comments and Jacques Dubochet for support and interest. AS was supported by Swiss National Science Foundation J.Y.M is a Canadian Institutes of Health New Investigator and this research is supported by funds from the National Cancer Institute of Canada and the Natural Sciences and Engineering Research Council of Canada. Funding to pay the Open Access publication charges for this article was provided by the National Cancer Institute of Canada.
Conflict of interest statement. None declared.
REFERENCES
- 1.Jinks-Robertson S, Petes TD. Chromosomal translocations generated by high-frequency meiotic recombination between repeated yeast genes. Genetics. 1986;114:731–752. doi: 10.1093/genetics/114.3.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goldman ASH, Lichten M. The efficiency of meiotic recombination between dispersed sequences in Saccharomyces cerevisiae depends upon their chromosomal location. Genetics. 1996;144:43–55. doi: 10.1093/genetics/144.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Masson J-Y, West SC. The Rad51 and Dmc1 recombinases: a non-identical twin relationship. Trends Biochem. Sci. 2001;26:131–136. doi: 10.1016/s0968-0004(00)01742-4. [DOI] [PubMed] [Google Scholar]
- 4.Keeney S. Mechanism and control of meiotic recombination initiation. Curr. Top Dev. Biol. 2001;52:1–53. doi: 10.1016/s0070-2153(01)52008-6. [DOI] [PubMed] [Google Scholar]
- 5.Baudat F, Manova K, Yuen JP, Jasin M, Keeney S. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol. Cell. 2000;6:989–998. doi: 10.1016/s1097-2765(00)00098-8. [DOI] [PubMed] [Google Scholar]
- 6.Romanienko PJ, Camerini-Otero RD. The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol. Cell. 2000;6:975–987. doi: 10.1016/s1097-2765(00)00097-6. [DOI] [PubMed] [Google Scholar]
- 7.Davis L, Smith GR. Meiotic recombination and chromosome segregation in Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA. 2001;98:8395–8402. doi: 10.1073/pnas.121005598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farah JA, Cromie G, Steiner WW, Smith GR. A novel recombination pathway initiated by the Mre11/Rad50/Nbs1 complex eliminates palindromes during meiosis in Schizosaccharomyces pombe. Genetics. 2005;169:1261–1274. doi: 10.1534/genetics.104.037515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsubouchi H, Roeder GS. The budding yeast mei5 and sae3 proteins act together with dmc1 during meiotic recombination. Genetics. 2004;168:1219–1230. doi: 10.1534/genetics.103.025700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sung P. Function of yeast Rad52 protein as a mediator between replication protein-A and the Rad51 recombinase. J. Biol. Chem. 1997;272:28194–28197. doi: 10.1074/jbc.272.45.28194. [DOI] [PubMed] [Google Scholar]
- 11.Sung P. Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein-A to promote DNA strand exchange by Rad51 recombinase. Genes Dev. 1997;11:1111–1121. doi: 10.1101/gad.11.9.1111. [DOI] [PubMed] [Google Scholar]
- 12.Petukhova G, Van Komen S, Vergano S, Klein H, Sung P. Yeast Rad54 promotes Rad51-dependent homologous DNA pairing via ATP hydrolysis-driven change in DNA double helix conformation. J. Biol. Chem. 1999;274:29453–29462. doi: 10.1074/jbc.274.41.29453. [DOI] [PubMed] [Google Scholar]
- 13.Petukhova G, Sung P, Klein H. Promotion of Rad51-dependent D-loop formation by yeast recombination factor Rdh54/Tid1. Genes Dev. 2000;14:2206–2215. doi: 10.1101/gad.826100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shinohara M, Gasior SL, Bishop DK, Shinohara A. Tid1/Rdh54 promotes colocalization of Rad51 and Dmc1 during meiotic recombination. Proc. Natl. Acad. Sci. USA. 2000;97:10814–10819. doi: 10.1073/pnas.97.20.10814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leu JY, Chua PR, Roeder GS. The meiosis-specific Hop2 protein of S. cerevisiae ensures synapsis between homologous chromosomes. Cell. 1998;94:375–386. doi: 10.1016/s0092-8674(00)81480-4. [DOI] [PubMed] [Google Scholar]
- 16.Petukhova GV, Romanienko PJ, Camerini-Otero RD. The Hop2 protein has a direct role in promoting interhomolog interactions during mouse meiosis. Dev. Cell. 2003;5:927–936. doi: 10.1016/s1534-5807(03)00369-1. [DOI] [PubMed] [Google Scholar]
- 17.Tsubouchi H, Ogawa H. A novel MRE11 mutation impairs processing of double-strand breaks of DNA during both mitosis and meiosis. Mol. Cell. Biol. 1998;18:260–268. doi: 10.1128/mcb.18.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsubouchi H, Roeder GS. The Mnd1 protein forms a complex with hop2 to promote homologous chromosome pairing and meiotic double-strand break repair. Mol. Cell Biol. 2002;22:3078–3088. doi: 10.1128/MCB.22.9.3078-3088.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerton JL, DeRisi JL. Mnd1p: an evolutionarily conserved protein required for meiotic recombination. Proc. Natl. Acad. Sci. USA. 2002;99:6895–6900. doi: 10.1073/pnas.102167899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nabeshima K, Kakihara Y, Hiraoka Y, Nojima H. A novel meiosis-specific protein of fission yeast, Meu13p, promotes homologous pairing independently of homologous recombination. Embo. J. 2001;20:3871–3881. doi: 10.1093/emboj/20.14.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saito TT, Tougan T, Kasama T, Okuzaki D, Nojima H. Mcp7, a meiosis-specific coiled-coil protein of fission yeast, associates with Meu13 and is required for meiotic recombination. Nucleic Acids Res. 2004;32:3325–3339. doi: 10.1093/nar/gkh654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsubouchi H, Roeder GS. The importance of genetic recombination for fidelity of chromosome pairing in meiosis. Dev. Cell. 2003;5:915–925. doi: 10.1016/s1534-5807(03)00357-5. [DOI] [PubMed] [Google Scholar]
- 23.Chen YK, Leng CH, Olivares H, Lee MH, Chang YC, Kung WM, Ti SC, Lo YH, Wang AH, Chang CS, et al. Heterodimeric complexes of Hop2 and Mnd1 function with Dmc1 to promote meiotic homolog juxtaposition and strand assimilation. Proc. Natl. Acad. Sci. USA. 2004;101:10572–10577. doi: 10.1073/pnas.0404195101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petukhova GV, Pezza RJ, Vanevski F, Ploquin M, Masson JY, Camerini-Otero RD. The Hop2 and Mnd1 proteins act in concert with Rad51 and Dmc1 in meiotic recombination. Nat. Struct. Mol. Biol. 2005;12:449–453. doi: 10.1038/nsmb923. [DOI] [PubMed] [Google Scholar]
- 25.Enomoto R, Kinebuchi T, Sato M, Yagi H, Shibata T, Kurumizaka H, Yokoyama S. Positive role of the mammalian TBPIP/HOP2 protein in DMC1-mediated homologous pairing. J. Biol. Chem. 2004;279:35263–35272. doi: 10.1074/jbc.M402481200. [DOI] [PubMed] [Google Scholar]
- 26.Enomoto R, Kinebuchi T, Sato M, Yagi H, Kurumizaka H, Yokoyama S. Stimulation of DNA strand exchange by the human TBPIP/Hop2-Mnd1 complex. J. Biol. Chem. 2006;281:5575–5581. doi: 10.1074/jbc.M506506200. [DOI] [PubMed] [Google Scholar]
- 27.Baumann P, West SC. The human Rad51 protein: polarity of strand transfer and stimulation by hRP-A. EMBO J. 1997;16:5198–5206. doi: 10.1093/emboj/16.17.5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McIlwraith MJ, Van Dyck E, Masson J-Y, Stasiak AZ, Stasiak A, West SC. Reconstitution of the strand invasion step of double-strand break repair using human Rad51, Rad52 and RPA proteins. J. Mol. Biol. 2000;304:151–164. doi: 10.1006/jmbi.2000.4180. [DOI] [PubMed] [Google Scholar]
- 29.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Meth. Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 30.Wang W, Malcolm BA. Two-stage PCR protocol allowing introduction of multiple mutations, deletions and insertions using QuikChange Site-Directed Mutagenesis. Biotechniques. 1999;26:680–682. doi: 10.2144/99264st03. [DOI] [PubMed] [Google Scholar]
- 31.Sauvageau S, Stasiak AZ, Banville I, Ploquin M, Stasiak A, Masson JY. Fission yeast rad51 and dmc1, two efficient DNA recombinases forming helical nucleoprotein filaments. Mol. Cell Biol. 2005;25:4377–4387. doi: 10.1128/MCB.25.11.4377-4387.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baumann P, Benson FE, Hajibagheri N, West SC. Purification of human Rad51 protein by selective spermidine precipitation. Mut. Res. DNA Repair. 1997;384:65–72. doi: 10.1016/s0921-8777(97)00028-1. [DOI] [PubMed] [Google Scholar]
- 33.Masson J-Y, Davies AA, Hajibagheri N, Van Dyck E, Benson FE, Stasiak AZ, Stasiak A, West SC. The meiosis-specific recombinase hDmc1 forms rings structures and interacts with hRad51. EMBO J. 1999;18:6552–6560. doi: 10.1093/emboj/18.22.6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lio YC, Mazin AV, Kowalczykowski SC, Chen DJ. Complex formation by the human Rad51B and Rad51C DNA repair proteins and their activities in vitro. J. Biol. Chem. 2003;278:2469–2478. doi: 10.1074/jbc.M211038200. [DOI] [PubMed] [Google Scholar]
- 35.Sogo J, Stasiak A, De Bernadin W, Losa R, Koller T. In: Electron Microscopy in Molecular Biology. Sommerville J, Scheer U, editors. Oxford: IRL Press; 1987. pp. 61–79. [Google Scholar]
- 36.Pugh BF, Cox MM. Stable binding of RecA protein to duplex DNA. J. Biol. Chem. 1987;262:1326–1336. [PubMed] [Google Scholar]
- 37.Sugiyama T, Zaitseva EM, Kowalczykowski SC. A single-stranded DNA-binding protein is needed for efficient complex formation by the Saccharomyces cerevisiae Rad51 protein. J. Biol. Chem. 1997;272:7940–7945. doi: 10.1074/jbc.272.12.7940. [DOI] [PubMed] [Google Scholar]
- 38.Baumann P, West SC. Heteroduplex formation by human Rad51 protein: effects of DNA end-structure, hRP-A and hRad52. J. Mol. Biol. 1999;291:363–374. doi: 10.1006/jmbi.1999.2954. [DOI] [PubMed] [Google Scholar]
- 39.Shibata T, Ohtani T, Iwabuchi M, Ando T. D-loop cycle. A circular reaction sequence which comprises formation and dissociation of D-loops and inactivation and reactivation of superhelical closed circular DNA promoted by RecA protein of E.coli. J. Biol. Chem. 1982;257:13981–13986. [PubMed] [Google Scholar]
- 40.Sehorn MG, Sigurdsson S, Bussen W, Unger VM, Sung P. Human meiotic recombinase Dmc1 promotes ATP-dependent homologous DNA strand exchange. Nature. 2004;429:433–437. doi: 10.1038/nature02563. [DOI] [PubMed] [Google Scholar]
- 41.Pezza RJ, Petukhova GV, Ghirlando R, Camerini-Otero RD. Molecular activities of meiosis specific proteins Hop2, Mnd1 and the Hop2-Mnd1 complex. J. Biol. Chem. 2006;281:18426–18434. doi: 10.1074/jbc.M601073200. [DOI] [PubMed] [Google Scholar]
- 42.Burkhard P, Stetefeld J, Strelkov SV. Coiled coils: a highly versatile protein folding motif. Trends Cell Biol. 2001;11:82–88. doi: 10.1016/s0962-8924(00)01898-5. [DOI] [PubMed] [Google Scholar]
- 43.Bugreev DV, Mazin AV. Ca2+ activates human homologous recombination protein Rad51 by modulating its ATPase activity. Proc. Natl. Acad. Sci. USA. 2004;101:9988–9993. doi: 10.1073/pnas.0402105101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bugreev DV, Golub EI, Stasiak AZ, Stasiak A, Mazin AV. Activation of human meiosis-specific recombinase Dmc1 by Ca2+ J. Biol. Chem. 2005;280:26886–26895. doi: 10.1074/jbc.M502248200. [DOI] [PubMed] [Google Scholar]
- 45.Soustelle C, Vedel M, Kolodner R, Nicolas A. Replication protein A is required for meiotic recombination in Saccharomyces cerevisiae. Genetics. 2002;161:535–547. doi: 10.1093/genetics/161.2.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ristic D, Modesti M, van der Heijden T, van Noort J, Dekker C, Kanaar R, Wyman C. Human Rad51 filaments on double- and single-stranded DNA: correlating regular and irregular forms with recombination function. Nucleic Acids Res. 2005;33:3292–3302. doi: 10.1093/nar/gki640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tanaka T, Nakamura T, Takagi H, Sato M. Molecular cloning and characterization of a novel TBP-1 interacting protein (TBPIP): enhancement of TBP-1 action on Tat by TBPIP. Biochem. Biophys. Res. Commun. 1997;239:176–181. doi: 10.1006/bbrc.1997.7447. [DOI] [PubMed] [Google Scholar]
- 48.Ko L, Cardona GR, Henrion-Caude A, Chin WW. Identification and characterization of a tissue-specific coactivator, GT198, that interacts with the DNA-binding domains of nuclear receptors. Mol. Cell Biol. 2002;22:357–369. doi: 10.1128/MCB.22.1.357-369.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weiner BM, Kleckner N. Chromosome pairing via multiple interstitial interactions before and during meiosis in yeast. Cell. 1994;77:977–991. doi: 10.1016/0092-8674(94)90438-3. [DOI] [PubMed] [Google Scholar]
- 50.Henry JM, Camahort R, Rice DA, Florens L, Swanson SK, Washburn MP, Gerton JL. Mnd1/Hop2 facilitates interhomolog crossover formation in meiosis of budding yeast. Mol. Cell Biol. 2006;26:2913–2923. doi: 10.1128/MCB.26.8.2913-2923.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]