Abstract
LAGLIDADG homing endonucleases (LHEs) cleave 18–24 bp DNA sequences and are promising enzymes for applications requiring sequence-specific DNA cleavage amongst genome-sized DNA backgrounds. Here, we report a method for cell surface display of LHEs, which facilitates analysis of their DNA binding and cleavage properties by flow cytometry. Cells expressing surface LHEs can be stained with fluorescently conjugated double-stranded oligonucleotides (dsOligos) containing their respective target sequences. The signal is absolutely sequence specific and undetectable with dsOligos carrying single base-pair substitutions. LHE–dsOligo interactions facilitate rapid enrichment and viable recovery of rare LHE expressing cells by both fluorescence-activated cell sorting (FACS) and magnetic cell sorting (MACS). Additionally, dsOligos conjugated with unique fluorophores at opposite termini can be tethered to the cell surface and used to detect DNA cleavage. Recapitulation of DNA binding and cleavage by surface-displayed LHEs provides a high-throughput approach to library screening that should facilitate rapid identification and analysis of enzymes with novel sequence specificities.
INTRODUCTION
Homing endonucleases of the LAGLIDADG family (LHEs) form homodimers or pseudosymmetric monomers that generally recognize DNA sequences 18–24 bp in length (1). Their molecular structures are built around two conserved alpha helices that contain a LAGLIDADG consensus sequence, which forms the center of the interface between enzyme subunits or domains (2). The final acidic residues from the LAGLIDADG helix form part of each domain's active site that cleaves one strand of the double-stranded DNA target sequence. The DNA-binding interface of each domain is made up of a four-stranded antiparallel beta-sheet that is supported by a series of framework alpha helices, which form the core of the domain. Unlike restriction endonucleases which form densely packed and highly saturated DNA–protein interfaces, the DNA-binding interface of LHEs make fewer hydrogen bonds per target sequence base pair (3). These structural properties account for the ability of LHEs to withstand moderate variability in target sequence recognition (4–7), a characteristic which has been essential in maintaining their genetic mobility and horizontal proliferation (8) and which make LHEs ideal substrates for engineering altered DNA-binding interfaces with novel endonucleolytic specificities (9–13). The combination of high target sequence specificity and adaptable DNA-binding interfaces make LHEs attractive tools for genome engineering applications, which require the introduction of a double-stranded break at a precise genomic location (13–16).
Since only a limited number of native LHEs have been identified (1), attempts to use existing LHEs as scaffolds for creating novel enzymes able to target desired sequences have been widespread (4,5,17–20). Recent efforts have employed a variety of approaches that individually (5,20) or successively (12) utilize computational redesign with substrate cleavage screening or two-plasmid selection systems (21,22). While these methodologies have shown promise, they are limited in their screening throughput because they require the generation of combinatorial endonuclease mutant libraries and the variant endonucleases must be well tolerated by the host's genomic DNA. An additional limitation is that the intracellular cleavage system must be redesigned and generated for each sequence targeted for selection. We sought to develop a system where LHE proteins could be rapidly screened to identify and isolate variants with new DNA target specificities. Here, we demonstrate that LHEs can be expressed on the plasma membrane of a lymphocyte cell line by targeting the expression of an LHE-CD80 transmembrane fusion protein to the secretory pathway. Surface-expressed LHEs faithfully recapitulate the properties of the native enzymes in solution, as assessed by flow cytometric analysis of both the binding and cleavage of fluorescently conjugated dsOligos. Furthermore, sequence-specific LHE interactions with dsOligos in conditions, which prohibit substrate cleavage allow for their physical isolation by multiple cell separation methods. The rapid analysis of LHE–DNA interactions on the cell surface with concurrent sorting options should facilitate a significant acceleration in the isolation of novel endonuclease variants with unique DNA target specificities.
Methods
Plasmid construction and generation of stable LHE expressing DT40 clones
Vectors-containing cDNA for both LHEs were PCR amplified using following primers: I-AniI For SfiI and I-AniI Rev SalI; H-DreI For SfiI and H-DreI Rev SalI and cloned into the pLHCX-phOx expression vector (23,24) by SfiI and SalI digestion to replace phOx coding sequence. To place the NeoR gene in frame in the I-AniI construct, the NeoR cDNA including the HSV polyA sequence was amplified using CD80-NeoR For and NeoR Rev ClaI, while the existing I-AniI-CD80 expression construct (including the 5′ SP and HA epitope) was amplified by primers SP For Hind3 and CD80-NeoR Rev. The entire fusion molecule was generated by fusion PCR as described previously (26), and subcloned back into the pLHCX plasmid by HindIII and ClaI digestion. Mutation of residues K21, T27 for I-AniIm generation was achieved by site-directed mutagenesis (Stratagene QuikChange II, no. 200523-5) using I-AniI K21 T27 SDM For and I-AniI K21 T27 SDM Rev, and the L223 mutation arose by PCR error. For transfection of DT40 cells, 30 μg of linearized plasmid DNA was electroporated into 107 DT40 cells (IgM-negative where indicated) using a Gene Pulser XCell (Bio-Rad, Hercules, CA, USA) in a final volume of 400 μl of serum-free RPMI media employing the exponential protocol: 550 V, 25 μF, ∞ resistance with a 4 mm cuvette gap. After 24 h of culture in drug-free media, cells were plated by limiting dilution in media containing 2 mg/ml G418 (Invitrogen, Carlsbad, CA, USA, no. 11811-098) for 10–14 days. Wells containing single G418-resistant clones were expanded and screened by flow cytometry for HA surface expression.
I-AniI For SfiI: GGCCCAGCCGGCCATGGGCAGCAGCCATCATCATC
I-AniI Rev SalI: GTCGACATAATTTGAAGGTATTTTTATTTTTTCTG
H-DreI For SfiI: GGCCCAGCCGGCCATGCATAATAATGAGAATGTT
H-DreI Rev SalI: GTCGACCGGGGACGATTTCTTTTTTTCACT
CD80-NeoR For: CAGACCGTCTTCCTTGGATCGGCCATTGAACAAG
NeoR Rev ClaI: ATCGATGAACAAACGACCCAACACCCGTGCG
SP For Hind3: AAGCTTATGGAGACAGACACACTCCTGCTATGGG
CD80-NeoR Rev: CTTGTTCAATGGCCGATCCAAGGAAGACGGTCTG
I-AniI K21 T27 SDM For: CAGCATCACCAACAAGGGTAAGTACCTACAGTATGAGCTGGGTATCGAG
I-AniI K21 T27 SDM Rev: CTCGATACCCAGCTCATACTGTAGGTACTTACCCTTGTTGGTGATGCTG
Western blotting and glycosylation analysis by PNGase F treatment
Here, 7.5 × 106 cells of the indicated cell lines were washed once in ice-cold PBS containing 0.1% BSA and lysed for 30 min at 4°C in lysis buffer (25 mM Tris-Cl pH 7.4, 140 mM NaCl, 2 mM EDTA, 1% NP-40, 0.05% sodium deoxycholate, 0.005% SDS and protease inhibitors). The crude cell lysates were clarified by centrifugation and 50 μg of total protein from post-nuclear cell lysates were used for incubation with PNGase F (New England Biolabs, Beverly, MA, USA, no. P0704S) for 2 h according to manufacturer's guidelines. Samples were analyzed by western blotting using anti-HA (Cell Signaling Technology, Danvers, MA, USA, no. 2367) and anti-β-actin Ab (Sigma-Aldrich, St. Louis, MO, USA, no. A1978) followed by HRP-conjugated anti-mouse-IgG (Amersham Biosciences, Piscataway, NJ, USA, no. NA931V).
Flow cytometry
Standard antibody staining was done in PBS-containing 0.2% BSA using the following antibodies: mouse monoclonal anti-HA (Cell Signaling Technology, no. 2367) followed by PE-conjugated goat anti-mouse IgG1 (Southern Biotech, Birmingham, AL, USA, no. 1070-09S); FITC-conjugated anti-chicken IgM (Bethyl Laboratories Inc., Montgomery, TX, USA, no. A30-102F). Preparation of dsOligos and subsequent staining was performed as follows: complementary 5′-biotinylated and non-biotinylated DNA oligonucleotides (Figure 2) were annealed by incubation at 94°C for 5 min and allowed to cool slowly to room temperature, sterilized by ethanol precipitation and resuspended to a stock concentration of 1.6 μM. Cells were first incubated at 4°C for 30 min in our standard dsOligo blocking and staining buffer containing 135 mM NaCl, 5 mM KCl, 10 mM CaCl2, 5.6 mM Glucose, 10 mM HEPES, 0.2% BSA and 1 μg/ml sonicated salmon sperm DNA, pH 7.4. Concurrent with this incubation, annealed dsOligos were complexed with SAv-PE (BD Biosciences, Palo Alto, CA, USA, no. 554061, Mw 300,000) at 1:1 molar ratio in the same buffer. The dsOligo-BT: SAv-PE complexes were used to stain the cells at a final concentration of 10–50 nM for 30–40 min at 4°C. Cells were washed twice with ice-cold buffer prior to analysis. Antibody and dsOligo-stained cells were analyzed by flow cytometry using the Beckton Dickenson FACSCalibur or LSRII instruments (BD Biosciences). 10 000–100 000 live cells were acquired per sample and the resulting raw data were processed using FlowJo software (FlowJo Ashland, OR, USA, LLC).
Figure 2.
Fluorescently conjugated dsOligos bind cell surface LHEs in a manner, which is sequence specific and easily resolved by flow cytometry. (a) H-DreI is an engineered enzyme composed of domains derived from the I-CreI and I-DmoI LHEs. Its 23-bp recognition site (dsDre4, boxed) is therefore a complex of the natural target sequences bound by I-CreI (green) and I-DmoI (purple). The 19-bp I-AniI recognition site (dsAni1, boxed) was placed between stretches of five GC base pairs designed to enhance the formation and stability of the double-stranded complex. Single base-pair changes (dsDre46T, dsDre410T, dsAni1−6A and dsAni1−9A) are indicated by red boxes and the cleavage sites by red arrows. The alternative I-AniI target sequence (dsAni2) containing two base-pair changes are shown in blue boxes. Conjugations with biotin at the 5′ termini are depicted, and Alexa Fluor 647 conjugated oligonucleotides for dsAni1 and dsAni1−9A were used in the flow cytometry cleavage assay. (b) Verification of efficient annealing of the complementary oligonucleotides run on a 3% agarose gel, with individual oligos (+S and −S) run as controls. (c) Flow cytometry analysis of clones stained with fluorescent dsOligos. Staining of I-AniI and H-DreI expressing clones in the presence of 10 mM Ca2+ are shown, with shaded and open histograms representing SAv-PE-only controls and dsOligo-BT: SAv-PE stained cells, respectively. The dsOligos used for each stain are indicated in the upper right corner of the histograms.
Fluorescence-activated cell sorting (FACS)
LHE expressing clones were mixed at the indicated ratios immediately prior to staining. The cells were stained using the above protocol with the indicated dsOligo complexes (SAv-Q655 from Invitrogen, no. Q10121MP). The PE- or Q655-positive populations of live-gated doublet-excluded cells were sorted using the BD Aria cell sorter. Sorted populations were cultured for 5–7 days and labeled with either dsOligos or anti-IgM for flow cytometry analysis. The above process was iterated for subsequent rounds of enrichment.
Magnetic cell sorting (MACS)
Cells were mixed at the indicated ratios (∼5–10 × 107 cells per sample) and labeled for 30 min at 4°C with 100 nM dsAni1 in the same buffer used for flow cytometry. After washing, the mixed population was incubated with 20–50 μl SAv-coated magnetic beads (Miltenyi Biotec, Auburn, CA, USA, no. 130-048-101) in a final volume of 0.5–1.0 ml for 20 min at 4°C. The samples were washed twice and resuspended at a concentration of 2 × 107 cells/ml prior to loading onto the AutoMACS cell separator. The ‘posselds’ double column separation program was run and the positive fraction was washed and placed immediately in culture. Cells were analyzed by staining separately with anti-IgM and dsAni1 as described above.
Flow cytometry assay for dsOligo cleavage
Complementary 5′-biotin and 5′-Alexa Fluor647 conjugated (Invitrogen) DNA oligonucleotides were annealed as described above. The buffer used for all steps of the cleavage assay contained 10 mM NaCl, 90 mM KCl, 10 mM HEPES, 5.6 mM Glucose, 0.2% BSA, 1 μg/ml salmon sperm DNA and pH 8.5. Approximately 1 × 106 cells were first incubated at 4°C with biotinylated mouse anti-HA Ab (Abcam, Cambridge, UK, no. AB27987-100) at a dilution 1:300 for 30–40 min. After washing, the cells were stained with 30–50 nM 647-dsOligo-BT: SAv-PE for 30 min on ice. For cleavage 10 mM MgCl2 was added to the buffer and the reaction was carried out at 42°C for the designated time points. The cells were washed in Mg2+-free buffer and analyzed by flow cytometry.
In-vitro LHE cleavage assay and fluorescence gel imaging
Reaction conditions were identical to those described in the flow cytometry cleavage assay except that 30 nM recombinant I-AniI was used in place of cells for the in vitro assays. For the in vitro assay with bead-complexed oligos, 647-dsOligo-BT: SAv-bead complexes were formed by incubating 50 nM dsOligo with 20 μl SAv-conjugated Dynabeads for 30 min at room temperature. The unbound 647-dsOligo-BT was removed by extensive washing in cleavage assay buffer, followed by incubation with 30 nM recombinant I-AniI for 1 h at 42°C. Oligonucleotide fragments were purified by phenol extraction followed by ethanol precipitation. The purified samples were resuspended in Ficoll-based loading buffer and resolved by PAGE. The gels were scanned using the Typhoon 9410 system (GE Healthcare, Piscataway, NJ, USA) with excitation by the 633 nM laser. Images were acquired with detector PMT voltages at both optimal (between 450 and 600 V) and maximal (between 700 and 850 V) settings to observe all fluorescent species. Images were processed with Adobe Photoshop using linear adjustments and all detectible bands in each lane are visible.
RESULTS
Expression of homing endonucleases on the plasma membrane surface
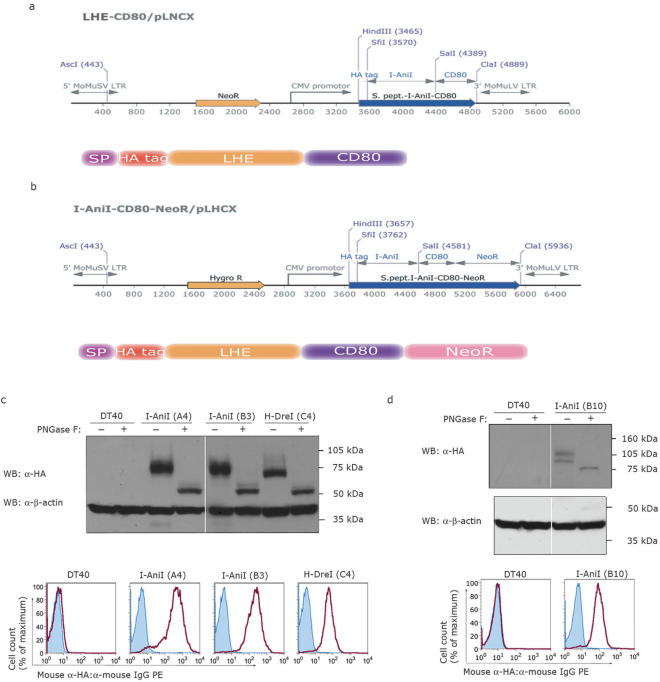
LHEs are normally expressed in the cytosol and targeted to DNA-containing organelles post-translationally. Cell surface display requires both the cotranslational targeting to the secretory pathway and fusion to an appropriate transmembrane domain. The strategy we chose was the one previously used to support surface display of antibody fragments (23,24). LHE genes were inserted between the coding sequences of the N-terminal murine immunoglobulin signal peptide (SP) and the transmembrane region of the murine CD80 molecule (Figure 1a). Two different LHE-coding sequences were integrated into the CMV promoter-driven surface expression constructs: I-AniI, an endonuclease encoded in the mitochondrial genome of Aspergillus nidulans (25); and H-DreI (Hybrid-Dmo/CreI, formerly called ‘E-DreI’), an engineered endonuclease containing an N-terminal domain derived from I-DmoI LHE (Desulfurococcus mobilis) and a C-terminal domain derived from I-CreI (Chlamydomonas reinhardtii) (10). These constructs included a hemagglutinin (HA) epitope tag downstream of the SP to facilitate biochemical and flow cytometric detection. Transfection of the linearized constructs into DT40 cells resulted in the isolation of clonal lines with high levels of I-AniI and H-DreI surface expression (Figure 1c).
Figure 1.
Vector schematics and validation of efficient LHE fusion protein expression in DT40 chicken B-cells. (a) LHE cDNAs were placed in-frame between a murine immunoglobulin-derived N-terminal signal peptide (SP) and the transmembrane spanning region of the murine CD80 molecule at the C-terminus. G418 resistance was conferred by a NeoR gene driven by an independent promoter. (b) The SP-HA-LHE-CD80 cassette was placed in-frame with the NeoR gene to allow coupled expression from a single promoter. Both constructs include an HA epitope tag at the N-terminus of the LHE and transcription is driven by the CMV promoter. (c) Western blot and flow cytometry analysis from clones expressing I-AniI (A4 and B3) and H-DreI (C4) and (d) from clone B10 expressing I-AniI as a fusion with C-terminal NeoR. Treatment with PNGase F is indicated above each lane. The corresponding clones were analyzed by flow cytometry for surface HA detection.
Intracellularly expressed LHEs are not exposed to glycosyltransferase enzymes, however this is an important consideration when their expression is directed to the cell surface. Primary sequence analysis revealed that LHE fusion proteins do contain potential N-glycosylation motifs (N-X-S/T where X ≠ P or D). To evaluate their N-glycosylation status, we incubated lysates of LHE expressing cells with the enzyme peptide-N-glycosidase F (PNGaseF). The N-glycosylation status was estimated by observing changes in band mobility during electrophoresis, which demonstrated that PNGaseF-treated LHE fusion proteins migrated faster and with less variability compared with the untreated controls (Figure 1c and d). These results indicate that the membrane-anchored molecules were indeed N-glycosylated, consistent with their surface expression through the secretory pathway.
As one intended application of surface expressed LHEs is identification of desired LHE variants from large libraries generated by random or targeted mutagenesis, a tight linkage between surface LHE expression and a selection marker is desirable as a means to enrich for variants which are efficiently expressed. For this purpose, we evaluated a strategy involving fusion of a neomycin resistance (NeoR) gene in frame with the C-terminus of the CD80 transmembrane domain (Figure 1b) (26), such that the NeoR activity is positioned on the cytosolic face of vesicles and the plasma membrane after expression. Transfection of LHE-CD80-NeoR constructs and application of neomycin selection allowed the isolation of multiple DT40 clones with stable surface expression of HA immunoreactivity from a single promoter (Figure 1d).
Surface-expressed LHEs are efficiently labeled with fluorescently conjugated dsOligos and detected by flow cytometry
We next tested the ability of the surface LHEs to bind annealed oligonucleotides representing their natural target specificities using flow cytometry. HEs are enzymatically active in the presence of Mg2+ ions, which are present in the active site (27). When Mg2+ ions are replaced with Ca2+ ions, LHEs retain their DNA-binding properties, while the cleavage of target DNA sequence is abolished (27,28). We therefore used a buffer containing 10 mM Ca2+ for cell-surface staining of LHE expressing clones using fluorescently labeled dsOligos. In an effort to minimize the effects of variations in dissociation kinetics of different LHEs, we used a single-step staining protocol with pre-formed complexes of biotinylated dsOligos (dsOligo-BT, Figure 2b) with phycoerythrin-conjugated streptavidin (SAv-PE). Since streptavidin contains four high affinity biotin-binding subunits, we created complexes (dsOligo-BT: SAv-PE) at a 1:1 molar ratio to maximize the fluorescent signal per target sequence. Staining I-AniI and H-DreI expressing clones with dsOligos of their respective natural target sequences generated clearly labeled populations despite their apparent N-glycosylation (Figure 2c). This analysis indicates that glycosylation does not confound surface analysis of these particular LHEs.
To rule out the possibility that our expression and detection system leads to degenerate DNA substrate recognition, we stained I-AniI and H-DreI expressing clones with dsOligos containing modifications to their respective target sequences. As expected, we observed no detectable staining when dsAni1 or dsDre4 were used to stain non-corresponding LHE expressing clones (Figure. 2c). To achieve a precise characterization of staining specificity, we designed dsOligos bearing single base-pair differences from the known target sequence (dsAni1−9A, dsAni1−6A, dsDre46T, dsDre410T, Figure 2a). These substitutions were chosen to interrupt direct contacts within the I-AniI and H-DreI DNA–protein interfaces (10,25). Remarkably, these single base-pair changes resulted in little or no detectable staining above non-specific background levels (Figure 3), consistent with the predicted destabilization of the binding interactions with their respective LHEs. Conversely, we have generated NeoR-linked clones with mutant I-AniI enzymes (generally denoted as I-AniIm) expressed stably on the cell surface (Figure 4b). Two I-AniIm clones were used in our experiments and were predicted to have either core structural changes or designed to have lost specific contacts at the DNA-binding interface. Although we have not validated the structural consequences of these mutations, the failure of the mutant enzymes to bind dsAni1 indicates that structural alterations, which do not inhibit LHE expression have DNA-binding consequences that are resolvable by our approach. We further extended our analysis to a unique target sequence variation against which wild-type I-AniI is known to maintain its cleavage activity (dsAni2, unpublished data, Figure 2a). This second I-AniI target sequence readily stained clones expressing I-AniI, further supporting the correlation of dsOligo-based interrogation of LHEs on the cell surface with biochemical cleavage data (Figure 3a, bottom panels). These data show that surface-expressed LHEs reliably discriminate closely related dsOligo sequences in a manner which both parallels their reported target sequence cleavage specificities and is sensitive to mutations in the DNA binding and core regions of the enzyme.
Figure 3.
LHEs expressed on the cell surface reliably discriminate dsOligos containing single base-pair differences from their natural target sequences. (a) I-AniI and (b) H-DreI expressing clones were stained with dsOligo-BT: SAv-PE complexes containing the natural target sequences (dsAni1 and dsDre4) or containing single base-pair changes (dsAni1−6A and dsAni1−9A; dsDre46T and dsDre410T). Known target sequence degeneracy for I-AniI is also recapitulated by dsOligo staining and analysis by flow cytometry. The cells expressing I-AniI were efficiently stained with dsAni2 corresponding to an alternative I-AniI target sequence known to be cleaved with an efficiency that is similar to the natural target sequence.
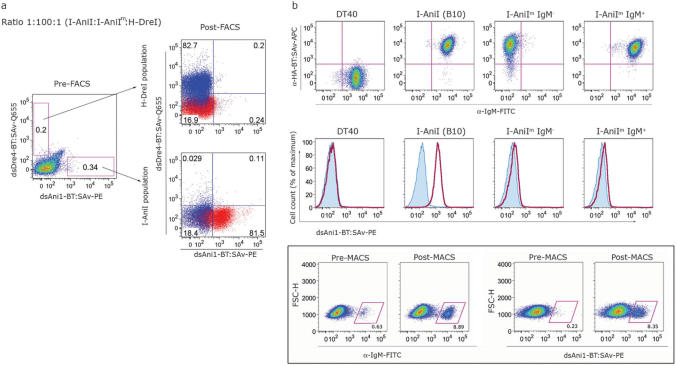
Figure 4.
Fluorescent and magnetic strategies facilitate sequence specific sorting of cells expressing surface LHEs. (a) Three populations of cells expressing different LHEs (I-AniI, I-AniIm and H-DreI) were mixed at a 1:100:1 ratio and double stained with dsAni1-BT: SAv-PE and dsDre4-BT: SAv-Q655, followed by FACS. The resulting sorted populations were cultured for 5–7 days prior to analysis and subsequent rounds of sorting. In post-sort analyses, cells stained with dsAni1 and dsDre4 are shown in red and blue, respectively. (b) Enrichment of low frequency dsOligo-binding cells by MACS. IgM-negative DT40 cells expressing I-AniIm (top row, third panel) were used as a background population into which IgM-positive B10 cells were added at a frequency of 0.1%. IgM-positive I-AniIm cells were included at 0.5% to control for potential background dsOligo binding caused by surface immunoglobulin expression, leading to a total of 0.6% IgM-positive cells in the input population, the majority of which do not stain with dsAni1. This mixed population was stained and sorted using AutoMACS (see Methods section for details). The positive fraction was grown out and analyzed for IgM expression. Staining with dsAni1 confirmed that the enriched IgM-positive population primarily expressed wild-type I-AniI.
Multi-parameter fluorescence-activated cell sorting (FACS) of cells labeled with dsOligos
We next evaluated whether our labeling method is suitable for sequence-dependent physical separation of LHE expressing cells by flow cytometry. We mixed three DT40 clones expressing different LHEs: clone B3 expressing I-AniI; clone C4 expressing H-DreI; and an I-AniIm clone carrying a mutation proximal to the LAGLIDADG dimerization alpha-helix was utilized as the background population. The cells were mixed at a ratio of 1:100:1 for B3:I-AniIm:C4 clones, respectively, and the mixed population was then stained with dsAni1-BT: SAv-PE and a quantum dot-conjugated dsDre4-BT: SAv-Q655. The dsAni1-specific and dsDre4-specific populations were isolated concurrently using fluorescence-activated cell sorting (FACS) and analyzed for their relative target specificities (Figure 4a). We achieved a significant enrichment of both I-AniI and H-DreI positive populations to 80% after the first round of sorting, and essentially no cross-contamination of the purified I-AniI or H-DreI populations was detected. The capacity of dsOligo-dependent cell sorting was further explored by assessing the enrichment of low frequency I-AniI expressing cells from a background of I-AniIm expressing cells, for which two iterative rounds of FACS sorting enriched an initial 0.01% population to 33% (Supplementary Figure 1). These data demonstrate that FACS sorting using fluorescently conjugated dsOligos is a highly effective method for the viable recovery of LHE expressing cells based on their DNA target specificity, and that rare clones with desired specificities may be isolated and enriched from large background populations.
Magnetic cell sorting rapidly isolates LHE expressing cells labeled with biotin-conjugated dsOligos
As an alternative to FACS, we tested the utility of magnetic cell sorting (MACS) for isolation of low-frequency LHE expressing cells (Figure 4b). The principle advantage of MACS is its ability to process extremely large sample sizes in short time periods (screening rates greater than 105 cells per second were routinely used in our protocols), thereby providing a convenient mechanism to sample large libraries of LHE clones. We employed an IgM-negative background population expressing high levels of an I-AniIm clone containing a mutated DNA-binding interface that was designed to eliminate direct contacts with one side of the asymmetric wild-type target sequence. Consistent low-level staining with dsAni1 indicates that low-affinity interactions with the wild-type target sequence are retained (Figure 4b, middle panels). The IgM-positive B10 clone expressing wild-type I-AniI was added at a frequency of 0.1%. The use of IgM as a surrogate marker for wild-type I-AniI expression allows for more accurate discrimination of low-percentage populations after dsOligo-dependent sorting due to a higher signal to noise ratio compared with dsOligo staining. To control for potential low-affinity interactions of dsOligos with IgM on the cell surface we included IgM-positive cells expressing I-AniIm in the initial sample at a frequency of ∼0.5%. The mixed population was labeled with dsAni1-BT in the presence 10 mM Ca2+, followed by incubation with SAv-coated magnetic beads. Binding and non-binding fractions were isolated using a double-column positive selection protocol on an AutoMACS cell sorter. Our initial experiments indicate that 0.1% starting populations can be consistently enriched to by two orders of magnitude after a single round of MACS with sample sizes as large as 108 cells, despite residual low-affinity interactions with the bulk of cells expressing a mutated enzyme. Importantly, the enriched IgM-positive population was entirely composed of dsAni1-binding cells expressing wild-type I-AniI and not the IgM-positive fraction expressing I-AniIm (Figure 4b, lower panels). Importantly, these results establish that high-level expression of surface molecules with the potential for both spurious (IgM) and specific (I-AniIm) low-affinity interactions with DNA substrates do not compromise the specificity of dsOligo-dependent enrichment by MACS.
Flow cytometry-based cleavage assay for surface-expressed LHEs
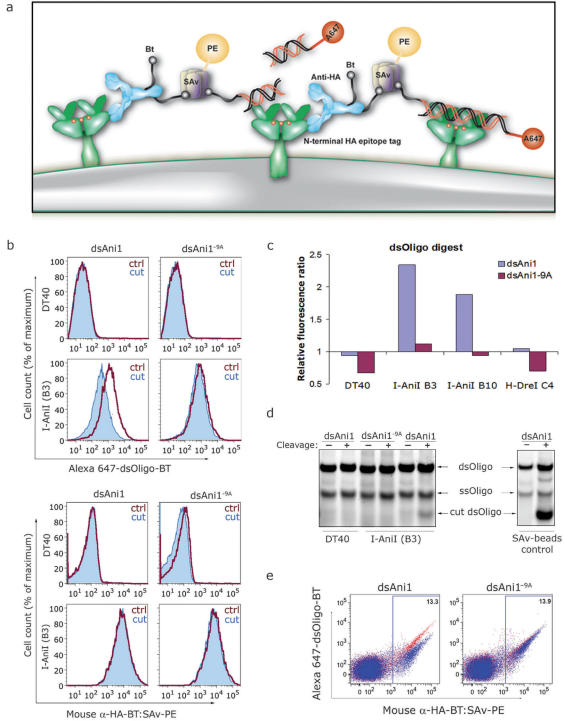
To evaluate whether surface LHEs retained sequence-specific endonuclease activity, we designed LHE target sequences with two distinct fluorophores at opposite termini. Each oligo was modified at its 5′ terminus with either Alexa Fluor 647 or biotin during synthesis and were annealed to obtain dually conjugated dsOligos (647-dsOligo-BT, Figure 2a) which were mixed with SAv-PE at a 1:1 molar ratio to obtain a bifluorescent 647-dsAni1-BT: SAv-PE staining reagent. Cells were first labeled with a biotin-conjugated anti-HA monoclonal antibody (α-HA-BT) followed by the addition of pre-formed 647-dsAni1-BT: SAv-PE complexes which should contain an average of three remaining BT-binding sites per SAv tetramer. This staining protocol serves to tether the 647-dsAni1-BT: SAv-PE to the cell surface independent of any specific LHE–dsOligo interaction, yet still placing the dsOligo within the LHE's immediate environment (Figure 5a). We reasoned that if the tethered 647-dsOligo-BT can be cleaved by the surface LHE, the cells would lose the fluorescence signal contribution from Alexa Fluor 647 yet retain signal from the tightly bound bridging SAv-PE.
Figure 5.
LHE mediated cleavage of cell surface-tethered dsOligo substrates conjugated with distinct fluorophores at opposite termini. (a) Schematic diagram for assaying surface LHE cleavage of α-HA-BT tethered dually fluorescent dsOligos and the release of Alexa Fluor 647 following addition of Mg2+ (red dots). (b) DT40 and B3 cells were stained with α-HA-BT followed by 647-dsOligo-BT: SAv-PE pre-formed complexes to tether the dsOligos to the surface LHE via the HA epitope. Cells with surface-tethered dsAni1 or dsAni1−9A substrates were incubated at 42°C for 20 min with (filled histograms) or without (open histograms) Mg2+ and analyzed by flow cytometry. Although the fluorescence data was collected simultaneously, the fluorescence from Alexa Fluor 647 and PE are represented separately in the top and bottom panel sets, respectively, to demonstrate specific loss of the untethered fluorophore signal. (c) To quantify the extent of dsOligo cleavage by I-AniI, we calculated a ratio of the mean PE to Alexa Fluor 647 fluorescence intensities. Blue columns indicate changes in the PE:647 fluorescence ratio for dsAni1 cleavage whereas purple columns show relative ratio shifts for the dsAniI−9A substrate. (d) DT40 cells and I-AniI expressing cells (B3) were stained as described in (b) and incubated at 42°C for 30 min in the presence (+) or absence (−) of Mg2+ (left panel). Also, 647-dsOligos-BT were bound to SAv-conjugated magnetic beads and incubated with recombinant I-AniI for 1 h at 42°C (right panel). DNA fragments were purified from supernatants and analyzed by PAGE followed by fluorescence imaging (see Methods section). (e) DT40 cells and I-AniI expressing cells (B3) were mixed at 10:1 ratio, labeled as described in (b) and incubated at 42°C for 20 min with (blue) or without (red) Mg2+ followed by flow cytometry analysis.
As both antibody binding and SAv: BT interactions are independent of divalent cation contribution, we used a Ca2+ and Mg2+-free buffer to stain I-AniI expressing cells with α-HA-BT followed by 647-dsAni1-BT: SAv-PE. The cells were then spiked with 10 mM Mg2+ and placed at 42°C in order to restore optimal cleavage conditions (29) (without Mg2+ for control samples). Using bifluorescent dsAni1, we were able to readily assay sequence-specific endonuclease activity by clones expressing wild-type I-AniI by monitoring changes in the fluorescence signals from each fluorophore (Figure 5b). Time-course experiments were performed to observe the relative disappearance of Alexa Fluor 647 fluorescence, which indicated that the signal progressively decreased during the first 20 min of incubation (data not shown). Given our data demonstrating the strict sequence specificity of the surface-expressed LHE DNA-binding interaction, we utilized bifluorescent dsAni1−9A as a stringent control for the specificity of the cleavage reaction. Consistent with the clear differences in the binding data for these dsOligos, we observed no relative fluorescent signal changes for dsAni1−9A under optimal cleavage conditions, confirming that dsAni1−9A was not cleaved by the surface LHEs. We calculated the PE:647 fluorescence ratios and their relative changes with each dsOligo species as an indicator of the relative substrate cleavage. This quantification clearly demonstrates a substantial increase in the PE:647 ratio only where the bifluorescent dsOligo matched the natural target sequence for I-AniI (Figure 5c). One possible interpretation of this result is that the sequence-specific reduction of the Alexa Fluor 647 signal was due to fluorophore quenching following LHE binding and not necessarily from cleavage and release of the fragment. We therefore verified the presence of the cleaved fragment in the supernatants of cleavage experiments (Figure 5d). Importantly, the cells used for the cleavage reactions were analyzed by flow cytometry to confirm specific loss of the Alexa Fluor 647 signal (as in Figure 5b). Control cleavage assays were performed in vitro using recombinant I-AniI to confirm that 647-dsAni1-BT alone or complexed with SAv-coated beads was readily accessible and efficiently cleaved by the purified enzyme. In both experiments, we identified co-migrating fluorescent fragments of smaller molecular weight compared to full-length double-stranded and residual single-stranded oligonucleotides. Smaller fragments were not detected in controls with dsAni1−9A or where the cleavage reaction was performed in the absence of either Mg2+ or I-AniI.
We next performed an experiment to confirm that the tethered dsOligos were being cleaved by LHEs on the very cells to which they were tethered. This is an important validation because cleavage caused by LHEs from adjacent cells might confound future attempts at FACS sorting by fluorescent signal loss following dsOligo cleavage. Using a mixed population of DT40 cells and I-AniI expressing (B3) cells at a 10:1 ratio where contacts between individual I-AniI expressing cells are decreased, we continued to observe sequence-specific reduction of Alexa Fluor 647 fluorescence to a similar extent as in a pure I-AniI-positive population (Figure 5e). We propose that individual dsOligos are primarily bound and digested by LHEs autonomously on the cell surface. These results demonstrate that under optimal reaction conditions the surface-expressed LHEs are catalytically active and functionally recapitulate their highly sequence-specific nuclease activity.
DISCUSSION
We have generated a high-throughput flow cytometry based method for screening LAGLIDADG homing endonuclease DNA target specificity by expression and analysis of the normally intracellular endonucleases on the cell surface. Surface expression of LHEs followed by simple fluorescent staining protocols allowed us to verify cell surface display and DNA binding and cleavage specificity of LHEs with single cell resolution and high-throughput capacity. The precise recognition of their respective target sequence enabled multi-parameter FACS enrichment of specific LHE expressing cells. Low frequency clone enrichment by 3–4 orders of magnitude was accomplished from an initial 0.01% representation in a mixed population (Supplementary Figure 1). This was achieved with as little as two iterative selection rounds and we expect that this can be resolved for significantly lower frequencies and a greater number of fluorescent parameters with continued iteration of the process and/or with clone isolation by sorting into multi-well plates. Similarly, MACS achieved significant enrichment of I-AniI expressing cells from a low frequency mixed population following a single round of selection. Overall, the surface-expression approach is simple and appears to be readily applicable to multiple homing endonucleases of the LAGLIDADG family.
When compared to previously described methods used in selection experiments to identify LHEs with novel specificities, our approach has several useful properties. While transfection efficiencies are lower for mammalian cells, thus limiting transfected LHE library sizes, the potential exists to utilize somatic hypermutation to create and iteratively screen virtually limitless LHE libraries (30). In addition, LHEs expressed on the cell surface are physically separated from its host cell's genome. This allows for the possibility that iterative selections produce lower specificity endonucleases as intermediates to the generation of a high-specificity enzyme. Such intermediates would likely be toxic to a host organism if the endonuclease were expressed intracellularly where it would have access to host genomic DNA. Furthermore, this method allows rapid generation of quantitative information regarding binding and cleavage properties of novel LHE variants which otherwise would require in vitro expression and purification followed by electrophoretic analysis of gel shifts and cleavage. Finally, when combined with present generation flow cytometers, this approach offers significant flexibility in screening strategies. One may choose to screen a population of LHEs for multiple unique target sites in a single experiment. This approach would involve simultaneous staining with multiple dsOligos representing desired target sequences that are labeled with unique fluorophores. Alternatively, selective population gating could enrich for desired specificities while simultaneously excluding undesired cross-reactivity as an approach to refining target sequence stringency. With the capacity to analyze dsOligo substrate cleavage, one could also potentially screen for both binding and catalytic activities in sequential flow cytometry assays to ensure that isolated binding properties do not compromise catalysis. Overall, the capacity to recapitulate DNA binding and cleavage by LHEs on the cell surface provides a high-throughput approach to LHE library screening that should facilitate the identification and analysis of enzymes with novel sequence specificities for use in a variety of genome engineering applications.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Noel Blake for assistance with developing flow cytometry protocols, and Dr David Baker and Dr James J. Havranek for designing mutant I-AniI enzymes. P.V. is supported by Young Investigator Award from Children's Hospital & Regional Medical Center in Seattle and J.J. by the Cancer Research Institute (CRI) and the Natural Sciences and Engineering Research Council of Canada (NSERC). This work was supported by NIH grant R21AI064581 to A.M.S. Funding to pay the Open Access publication charges for this article was provided by Seattle Children's Hospital & Regional Medical Center in Seattle.
Conflicts of interest statement. None declared.
REFERENCES
- 1.Chevalier BS, Stoddard BL. Homing endonucleases: structural and functional insight into the catalysts of intron/intein mobility. Nucleic Acids Res. 2001;29:3757–3774. doi: 10.1093/nar/29.18.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heath PJ, Stephens KM, Monnat RJ, Jr, Stoddard BL. The structure of I-Crel, a group I intron-encoded homing endonuclease. Nat. Struct. Biol. 1997;4:468–476. doi: 10.1038/nsb0697-468. [DOI] [PubMed] [Google Scholar]
- 3.Galburt EA, Stoddard BL. Catalytic mechanisms of restriction and homing endonucleases. Biochemistry. 2002;41:13851–13860. doi: 10.1021/bi020467h. [DOI] [PubMed] [Google Scholar]
- 4.Jurica MS, Monnat RJ, Jr, Stoddard BL. DNA recognition and cleavage by the LAGLIDADG homing endonuclease I-CreI. Mol. Cell. 1998;2:469–476. doi: 10.1016/s1097-2765(00)80146-x. [DOI] [PubMed] [Google Scholar]
- 5.Chevalier B, Turmel M, Lemieux C, Monnat RJ, Jr, Stoddard BL. Flexible DNA target site recognition by divergent homing endonuclease isoschizomers I-CreI and I-MsoI. J. Mol. Biol. 2003;329:253–269. doi: 10.1016/s0022-2836(03)00447-9. [DOI] [PubMed] [Google Scholar]
- 6.Moure CM, Gimble FS, Quiocho FA. The crystal structure of the gene targeting homing endonuclease I-SceI reveals the origins of its target site specificity. J. Mol. Biol. 2003;334:685–695. doi: 10.1016/j.jmb.2003.09.068. [DOI] [PubMed] [Google Scholar]
- 7.Moure CM, Gimble FS, Quiocho FA. Crystal structure of the intein homing endonuclease PI-SceI bound to its recognition sequence. Nat. Struct. Biol. 2002;9:764–770. doi: 10.1038/nsb840. [DOI] [PubMed] [Google Scholar]
- 8.Burt A, Koufopanou V. Homing endonuclease genes: the rise and fall and rise again of a selfish element. Curr. Opin. Genet. Dev. 2004;14:609–615. doi: 10.1016/j.gde.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 9.Duan X, Gimble FS, Quiocho FA. Crystal structure of PI-SceI, a homing endonuclease with protein splicing activity. Cell. 1997;89:555–564. doi: 10.1016/s0092-8674(00)80237-8. [DOI] [PubMed] [Google Scholar]
- 10.Chevalier BS, Kortemme T, Chadsey MS, Baker D, Monnat RJ, Stoddard BL. Design, activity, and structure of a highly specific artificial endonuclease. Mol. Cell. 2002;10:895–905. doi: 10.1016/s1097-2765(02)00690-1. [DOI] [PubMed] [Google Scholar]
- 11.Epinat JC, Arnould S, Chames P, Rochaix P, Desfontaines D, Puzin C, Patin A, Zanghellini A, Paques F, et al. A novel engineered meganuclease induces homologous recombination in yeast and mammalian cells. Nucleic Acids Res. 2003;31:2952–2962. doi: 10.1093/nar/gkg375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arnould S, Chames P, Perez C, Lacroix E, Duclert A, Epinat JC, Stricher F, Petit AS, Patin A, et al. Engineering of large numbers of highly specific homing endonucleases that induce recombination on novel DNA targets. J. Mol. Biol. 2006;355:443–458. doi: 10.1016/j.jmb.2005.10.065. [DOI] [PubMed] [Google Scholar]
- 13.Steuer S, Pingoud V, Pingoud A, Wende W. Chimeras of the homing endonuclease PI-SceI and the homologous Candida tropicalis intein: a study to explore the possibility of exchanging DNA-binding modules to obtain highly specific endonucleases with altered specificity. Chembiochem. 2004;5:206–213. doi: 10.1002/cbic.200300718. [DOI] [PubMed] [Google Scholar]
- 14.Storici F, Durham CL, Gordenin DA, Resnick MA. Chromosomal site-specific double-strand breaks are efficiently targeted for repair by oligonucleotides in yeast. Proc. Natl Acad. Sci. USA. 2003;100:14994–14999. doi: 10.1073/pnas.2036296100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tzfira T, Frankman LR, Vaidya M, Citovsky V. Site-specific integration of Agrobacterium tumefaciens T-DNA via double-stranded intermediates. Plant Physiol. 2003;133:1011–1023. doi: 10.1104/pp.103.032128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller DG, Petek LM, Russell DW. Human gene targeting by adeno-associated virus vectors is enhanced by DNA double-strand breaks. Mol. Cell Biol. 2003;23:3550–3557. doi: 10.1128/MCB.23.10.3550-3557.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith J, Grizot S, Arnould S, Duclert A, Epinat JC, Chames P, Prieto J, Redondo P, Blanco FJ, et al. A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic Acids Res. 2006;34:e149. doi: 10.1093/nar/gkl720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sussman D, Chadsey M, Fauce S, Engel A, Bruett A, Monnat R, Jr, Stoddard BL, Seligman LM. Isolation and characterization of new homing endonuclease specificities at individual target site positions. J. Mol. Biol. 2004;342:31–41. doi: 10.1016/j.jmb.2004.07.031. [DOI] [PubMed] [Google Scholar]
- 19.Seligman LM, Chisholm KM, Chevalier BS, Chadsey MS, Edwards ST, Savage JH, Veillet AL. Mutations altering the cleavage specificity of a homing endonuclease. Nucleic Acids Res. 2002;30:3870–3879. doi: 10.1093/nar/gkf495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ashworth J, Havranek JJ, Duarte CM, Sussman D, Monnat RJ, Jr, Stoddard BL, Baker D. Computational redesign of endonuclease DNA binding and cleavage specificity. Nature. 2006;441:656–659. doi: 10.1038/nature04818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doyon JB, Pattanayak V, Meyer CB, Liu DR. Directed evolution and substrate specificity profile of homing endonuclease I-SceI. J. Am. Chem. Soc. 2006;128:2477–2484. doi: 10.1021/ja057519l. [DOI] [PubMed] [Google Scholar]
- 22.Chames P, Epinat JC, Guillier S, Patin A, Lacroix E, Paques F. In vivo selection of engineered homing endonucleases using double-strand break induced homologous recombination. Nucleic Acids Res. 2005;33:e178. doi: 10.1093/nar/gni175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chou WC, Liao KW, Lo YC, Jiang SY, Yeh MY, Roffler SR. Expression of chimeric monomer and dimer proteins on the plasma membrane of mammalian cells. Biotechnol. Bioeng. 1999;65:160–169. doi: 10.1002/(sici)1097-0290(19991020)65:2<160::aid-bit5>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 24.Liao KW, Chou WC, Lo YC, Roffler SR. Design of transgenes for efficient expression of active chimeric proteins on mammalian cells. Biotechnol. Bioeng. 2001;73:313–323. doi: 10.1002/bit.1064. [DOI] [PubMed] [Google Scholar]
- 25.Bolduc JM, Spiegel PC, Chatterjee P, Brady KL, Downing ME, Caprara MG, Waring RB, Stoddard BL. Structural and biochemical analyses of DNA and RNA binding by a bifunctional homing endonuclease and group I intron splicing factor. Genes Dev. 2003;17:2875–2888. doi: 10.1101/gad.1109003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mohler WA, Blau HM. Membrane-bound neomycin phosphotransferase confers drug-resistance in mammalian cells: a marker for high-efficiency targeting of genes encoding secreted and cell-surface proteins. Somat. Cell Mol. Genet. 1994;20:153–162. doi: 10.1007/BF02254756. [DOI] [PubMed] [Google Scholar]
- 27.Chevalier BS, Monnat RJ, Jr, Stoddard BL. The homing endonuclease I-CreI uses three metals, one of which is shared between the two active sites. Nat. Struct. Biol. 2001;8:312–316. doi: 10.1038/86181. [DOI] [PubMed] [Google Scholar]
- 28.Chevalier B, Sussman D, Otis C, Noel AJ, Turmel M, Lemieux C, Stephens K, Monnat RJ, Jr, et al. Metal-dependent DNA cleavage mechanism of the I-CreI LAGLIDADG homing endonuclease. Biochemistry. 2004;43:14015–14026. doi: 10.1021/bi048970c. [DOI] [PubMed] [Google Scholar]
- 29.Geese WJ, Kwon YK, Wen X, Waring RB. In vitro analysis of the relationship between endonuclease and maturase activities in the bi-functional group I intron-encoded protein, I-AniI. Eur. J. Biochem. 2003;270:1543–1554. doi: 10.1046/j.1432-1033.2003.03518.x. [DOI] [PubMed] [Google Scholar]
- 30.Wang L, Jackson WC, Steinbach PA, Tsien RY. Evolution of new nonantibody proteins via iterative somatic hypermutation. Proc. Natl Acad. Sci. USA. 2004;101:16745–16749. doi: 10.1073/pnas.0407752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.