Abstract
Although the vital role of the androgen receptor (AR) has been well demonstrated in primary prostate cancers, its role in the androgen-insensitive prostate cancers still remains unclear. Here, we used a small hairpin RNA approach to directly assess AR activity in prostate cancer cells. Reduction of AR expression in the two androgen-sensitive prostate cancer cell lines, LNCaP and LAPC4, significantly decreased AR-mediated transcription and cell growth. Intriguingly, in two androgen-insensitive prostate cell lines, LNCaP-C42B4 and CWR22Rv1, knockdown of AR expression showed a more pronounced effect on AR-induced transcription and cell growth than androgen depletion. Using cDNA microarrays, we also compared the transcriptional profiles induced by either androgen depletion or AR knockdown. Although a significant number of transcripts appear to be regulated by both androgen depletion and AR knockdown, we observed a subset of transcripts affected only by androgen depletion but not by AR knockdown, and vice versa. Finally, we demonstrated a direct role for AR in promoting tumor formation and growth in a xenograft model. Taken together, our results elucidate an important role for the AR in androgen-insensitive prostate cancer cells, and suggest that AR can be used as a therapeutic target for androgen-insensitive prostate cancers.
INTRODUCTION
The androgen-signaling pathway plays a critical role in the regulation of prostate cancer cell growth and survival. Consequently, androgen ablation has been used as an effective treatment for the majority of advanced prostate cancers (1–3). Androgens exert their biological effects mainly through androgen receptor (AR), a member of the steroid hormone receptor superfamily (4). The AR is expressed in normal prostate epithelial cells, in virtually all primary prostate cancer cells, and in most refractory prostate cancer cells (4–6). Although the mechanisms by which prostate cancer cells become androgen insensitive (AI) are currently unclear, it is believed that the tumor cells must either bypass or adapt the AR-mediated cell growth pathway in order to survive in a low-androgen microenvironment during androgen ablation therapy (3,7). Several lines of evidence suggest that the AR-signaling pathway remains active in AI prostate cancer. Mutated AR proteins have been identified in a significant portion of AI prostate cancers. Some of the mutations identified in the AR-ligand-binding domain can result in activation of the receptor by other steroid hormones (8,9). Amplification of the AR gene also occurs in prostate cancer samples after androgen ablation therapy (10). Finally, multiple lines of evidence have shown that dysregulation of AR co-regulators can modify AR activity to compensate for lower androgen levels during androgen ablation therapies (11).
In this article, we directly assess AR-mediated transcription and cell growth of prostate cancer cells using a small hairpin RNA (shRNA) approach to attempt to address a longstanding unresolved question: does the AR still play a dominant role in AI prostate cancer cells? Using in vitro and in vivo model systems, we assessed AR-mediated transcription and cell growth in both androgen-sensitive and -insensitive prostate cancer cells through the knockdown of AR expression. Our data provide additional evidence that the AR continues to play a critical role in transcriptional regulation and cell growth in AI prostate cancer cells. Our findings suggest that the AR remains a viable therapeutic target in AI prostate cancers.
MATERIALS AND METHODS
Cells and tissue culture
The LNCaP cell line and its subline C4-2B4 were maintained in T-medium containing 5% fetal bovine serum (FBS; HyClone, Denver, CO). LAPC4 and CWR22Rv1 cells were maintained in phenol-red free or regular RPMI 1640 medium containing 10% FBS, respectively.
Plasmids
The PSA promoter/reporter plasmid (pPSA7Kb-Luc) was kindly provided by Dr Jan Trapman (Department of Pathology, Erasmus University, Rotterdam, the Netherlands) (12). The pARE-luc reporter was obtained from Dr Chawnshang Chang (13). The MMTV pA3LUC reporter was a gift from Dr Richard Pestell (Albert Einstein Medical College, New York, NY) (8). The cytomegalovirus-driven β-galactosidase (β-gal) reporter (pcDNA3-β-gal) was generated in our laboratory previously (14). The AR shRNA constructs were created by inserting double-stranded oligonucleotides corresponding to the human AR cDNA sequences 5′-GGACACTTGAACTGCCGTCT-3′ [amino acids (aa) 335–342], 5′-GGACATGCGTTTGGAGACTG-3′ (aa: 535–542), and 5′-GGTGTCACTATGGAGCTCTC-3′ (aa 568–575) downstream of U6 promoter in the pBS/U6 vector (15).
Adenovirus and lentivirus production
The pBS/U6-AR shRNA constructs were released by restriction-enzyme digestions and cloned into the pAdTrack shuttle vector (16). The plasmids were then cleaved with PmeI, and transformed into BJ5183 cells that contain pAdEasy-1 vector. Adenoviral vectors were amplified in DH5α cells, and propagated in HEK293 cells. Viral titers were determined using plaque assays.
To make the AR shRNA lentiviral constructs, the pBS/U6-AR shRNA constructs were digested and the DNA fragments containing U6 promoter and AR sequences were subcloned into the pLenti-super vector (17). Lentiviruses of AR shRNA were produced in 293T cells as described previously (18).
Immunofluorescence
Cells infected with adenoviruses or lentiviruses were cultured in 8-well Lab Tek chamber slides (Nalge Nunc International, Naperville, IL). Three days post-infection, cells were fixed for 30 min with 3% formaldehyde in phosphate-buffered saline (PBS), permeabilized with 95% ethanol at –20°C for 10 min, blocked in 10% normal goat serum for 1 h, and then incubated with the antibody against the AR and H1 (Santa Cruz Biotechnology, Santa Cruz, CA) overnight at 4°C. Slides were washed three times with PBS followed by incubation with appropriate secondary antibodies (Molecular Probes, Eugene, OR), and analyzed with Zeiss LSM 510 confocal, two-photon laser scanning microscope.
Western blotting
Whole-cell lysates were prepared from transfected or infected cells by extraction in lysis buffer containing 50 mM Tris (pH 8), 150 mM NaCl, 1% NP-40, 0.1% SDS, 10 mM NaF, 1 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin, 1 μg/ml aprotinin and 1 mM dithiothreitol. Proteins were resolved by 10% SDS-PAGE, transferred onto nitrocellulose membranes, probed with appropriate antibodies and developed using the ECL kit (Amersham Biosciences, Piscataway, NJ).
Transfection, luciferase and β-gal assays
Cells were infected with adenovirus for 6 h and then transfected with different plasmids using LipofectAMINE 2000 (Invitrogen, Carlsbad, CA). Dihydrotestosterone (DHT) was added into cells 24 h after infection if required. Luciferase and β-gal activities were measured in total cell lysates after 18–24 h. The relative light units (RLUs) from individual transfections were normalized by β-gal activity in the same samples. Individual transfection experiments were done in triplicate, and the results are reported as the means ± standard deviations (SD) from representative experiments.
Northern blotting and RT-PCR
Total RNA was isolated from cells infected with control or AR shRNA adenoviruses in the presence or absence of DHT using RNAwiz extraction reagent (Ambion, Austin, TX). Five micrograms of RNA were fractionated on a 1% agarose-formaldehyde gel, transferred to Hybond-N nylon membranes (Amersham Biosciences, Piscataway, NJ), and hybridized with a DNA fragment derived from either the human prostate-specific antigen (PSA) cDNA (aa: 1–261) or from the human kallikrein 2 (KLK2) cDNA (aa: 1–224). The blots were stripped and rehybridized with a human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) fragment (aa: 104–168).
Two micrograms of total RNA isolated from LNCaP or LAPC4 cells infected with either the adenoviral vector as a control or AR shRNA adenoviruses in the presence or absence of DHT was reverse transcribed using oligo dT and random primers, and then amplified with appropriate primers for PSA (5′-ACCATGTGGGTCCCGGTTGT-3′ and 5′-GAGTTGATAGGGGTGCTCAGG-3′), KLK2 (5′-CTGTGTCAGCATGTGGGACCT-3′ and 5′-CCATGATGTGATACCTTGAAGCA-3′), and GAPDH cDNAs (5′-CCATGGAGAAGGCTGGGG-3′ and 5′-CAAAGTTGTCATGGATGACC-3′), respectively.
Proliferation and colony formation assays
Cells were infected with adenoviruses for 3 h and seeded on 96-well plates. The growth rate of cells was measured every other day by the MTS assay following the manufacturer's protocol (Promega, Madison, WI). For the colony formation assay, infected cells were seeded on 24-well plates at ∼400 cells/well and incubated at 37°C for 14 days. Colonies were then fixed with 2% formaldehyde and stained with 0.2% crystal violet (Sigma, St. Louis, MO). The experiments were conducted in triplicate and repeated more than three times.
cDNA microarray hybridizations
Fluorescently labeled cDNA probes were prepared from 50 to 70 µg of total RNA isolated from CS-treated or AR-knockdown LNCaP cells (Cy5 labeled) and Universal Human Reference RNA (Stratagene, La Jolla, CA) (Cy3-labeled) by reverse transcription with a 17-mer Oligo dT primer (QIAGEN, Valencia, CA) as described previously (19). Labeled probes prepared from LNCaP cell RNA and reference RNA were mixed and hybridized overnight to spotted cDNA microarrays with 42,941 elements (Stanford Functional Genomics Facility). Microarray slides were then washed and scanned with a GenePix 4000B scanner (Axon Instruments, Inc., Union City, CA).
Data processing and analysis
Fluorescence intensities for each fluoroprobe were analyzed with GenePix Pro3.0 software (Axon Instruments). Spots of poor quality were removed from further analysis by visual inspection. Data files containing fluorescence ratios were entered into the Stanford Microarray Database where biological data were associated with fluorescence ratios, and genes were selected for further analysis (20). Only spots with a signal intensity >150% above background in either Cy5 or Cy3 channels were used in the subsequent analysis. We arbitrarily selected transcripts whose expression level decreased at least 1.5-fold after CS treatment or AR knockdown compared with controls.
Xenografts
LAPC4 cells were transduced with the AR shRNA and control GFP lentiviruses at a multiplicity of infection (MOI) of 3 for 24 h, and then harvested, resuspended in PBS, and mixed with equal volume of Matrigel ECM (Becton Dickinson, Bedford, MA). Here, 100 μl of cell suspension (1 × 107 cells/ml) infected with either control or AR shRNA lentiviruses was injected subcutaneously into opposite lateral flanks of 6–8-week-old athymic male mice (Harlan Sprague Dawley, Inc., Indianapolis, IN). Mice were monitored twice weekly. Tumors were measured in two dimensions with calipers, and tumor volume (mm3) was calculated with the formula V = (length × width2)/2. All the animal experiments were done in accordance with NIH animal use guidelines and the protocol approved by the University Committee on Animal Resources at Stanford University.
Immunohistochemical staining
Tumor specimens were fixed in 10% neutral-buffered formalin and embedded in paraffin. Serial sections (5 μm thick) were cut on a microtome and mounted on glass slides. Sections were deparaffinized in xylene and hydrated in graded ethanol solutions and distilled water. Antigen retrieval was performed by microwave processing at full power for 5 min, followed by half power for 20 min in 10 μM citrate buffer, pH 6.0. Endogenous peroxidase activity was blocked with 3% hydrogen peroxide in methanol for 30 min followed by washing in PBS pH 7.4. The sections were then incubated with the antibody against the AR (Santa Cruz Biotechnology, Santa Cruz, CA), PSA (Dako, Carpinteria, CA), green fluorescent protein (GFP) (Upstate, Charlottesville, VA), caspase 3 (Cell Signaling Technology, Danvers, MA) or PCNA (Santa Cruz Biotechnology, Santa Cruz, CA) overnight at 4°C. Biotinylated goat anti-rabbit or goat anti-mouse secondary antibodies (Vector Labs, Burlingame, CA) were then applied. Slides were treated with horseradish peroxidase streptavidin (Vector Labs) and developed using the diaminobenzidine tetrahydrochloride (DAB) substrate kit (Vector Labs). All sections were counterstained with hematoxylin.
Statistical analysis
Tumor volumes were represented as mean ± SD. Relative fold activation/suppression was represented as mean ± SD. Student's t test was used for the statistical analysis. Probabilities of P < 0.05 were considered significant.
RESULTS
Knockdown of AR expression in androgen-sensitive prostate cells
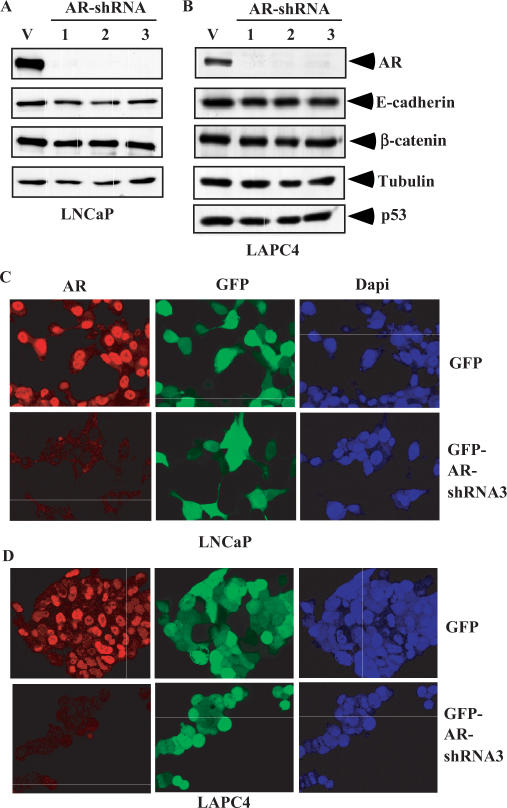
Multiple lines of evidence have shown that AR activity is required for the growth and survival of prostate cancer cells. To directly test the role of AR in transcriptional regulation and cell growth, we created three adenoviral-based AR shRNA constructs and tested them in prostate cancer cells. Infection of three AR shRNA adenoviruses into two AR-positive prostate cancer cell lines, LNCaP and LAPC4, showed an obvious reduction of AR protein expression (Figure 1A and B). The knockdown effect appeared to be sequence specific since the expression of other proteins in the infected cells was unchanged. Among these AR shRNA adenoviruses, AR shRNA virus 3 appeared the most effective at the silencing of AR expression in both LNCaP and LAPC4 cells when we infected cells with different MOI (data not shown). Immunofluorescent microscopy confirmed the knockdown of AR protein expression by the AR shRNA in prostate cancer cells. Since the adenoviral constructs used in this study express GFP, we monitored the infection efficiency directly by examining GFP expression. Here, ∼90% of cells infected with either the control or AR shRNA adenoviruses appeared GFP positive after 72 h of transduction (middle panels, Figure 1C and D). The AR proteins were mainly localized in the nuclei of both LNCaP and LAPC4 cells. Notably, the levels of AR proteins were much lower in the cells infected with the AR shRNA viruses than with the GFP viruses (Left panels, Figure 1C and D). These data demonstrate that the AR shRNA adenoviruses efficiently and specifically knockdown the expression of endogenous AR proteins in AR-positive prostate cancer cells.
Figure 1.
Down-regulation of endogenous AR expression by AR shRNA in prostate cancer cells. (A) LNCaP cells were infected with either the GFP adenovirus or the different AR shRNA adenovirus at an MOI of 40. Whole-cell lysates were prepared after 48 h of viral infection, and then analyzed by western blotting. Specific antibodies used to detect protein expression are labeled in the figure. (B) Identical experiments performed in LAPC4 cells. (C) LNCaP cells were infected with either the GFP adenovirus or AR shRNA3 adenovirus at an MOI of 40. Cells were fixed and immunostained 72 h after viral infection. Representative confocal laser scanning microscopy images of cells are shown. (D) Identical experiments performed in LAPC4 cells.
Knockdown of AR-mediated transactivation by AR shRNA adenovirus
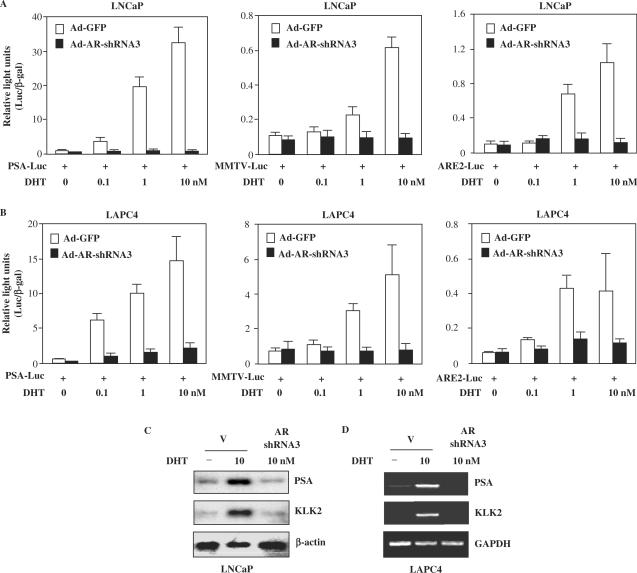
Next we examined the effects of AR knockdown on androgen-induced transcription. The human PSA is an AR-regulated target that has been widely used as a prostate-specific tumor marker (12). To determine if the knockdown of AR could affect the activation of the PSA promoter, transient transfections were carried out with a luciferase reporter driven by the 7-kb PSA promoter in both LNCaP and LAPC4 cells (34). Ligand-dependent induction of PSA promoter/reporter activities was observed in both cell lines (left panels, Figure 2A and B). The expression of AR shRNA adenovirus 3 significantly reduced the activities of endogenous AR on PSA promoter/reporter in both LNCaP and LAPC4 cells. In addition, the basal levels of AR activity were also decreased in the cells infected with the AR shRNA viruses in the absence of DHT compared to those infected with the control viruses. We repeated the transient transfection experiments with the other two androgen-inducible promoter/reporters, MMTV-LTR (21) and ARE luciferase (14). Both LNCaP and LAPC4 cells transfected with the AR shRNA plasmids showed greatly reduced activity with both promoters/reporters (middle and right panels, Figure 2A and B). These results demonstrate that the AR shRNA viruses specifically silence the transactivation potential of endogenous AR proteins.
Figure 2.
Down-regulation of AR expression reduces AR transactivation in prostate cancer cells. (A) LNCaP cells infected with either the GFP adenovirus (control) or AR shRNA3 adenovirus at an MOI of 40 for 6 h were transiently transfected with PSA7 kb-Luc (PSA-Luc), reporter, MMTV-Luc or ARE2-Luc reporter and pcDNA3-β-gal in T-medium with 5% CS-FBS for 24 h, and then incubated in different amounts of DHT for another 24 h. Luciferase and β-gal activities were measured and reported as RLU. (B) Transient transfection experiments carried out in LAPC4 cells infected with the AR shRNA3 or control adenoviruses at an MOI of 40. (C) LNCaP cells infected with control or AR shRNA3 adenovirus in the presence or absence of 10 nM DHT. Total RNA was isolated and analyzed by northern blot using radiolabeled probes for PSA, KLK2 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). (D) LAPC4 cells were infected with control adenovirus or AR shRNA3 adenovirus in the presence or absence of 10 nM DHT. Total RNA was isolated, reverse transcribed and analyzed by PCR.
Knockdown of endogenous AR affects the expression of AR target genes
To further evaluate the AR shRNA-mediated knockdown effect in a physiologically relevant cellular context, we examined the expression of PSA and kallikrein 2 (KLK2), two AR downstream target genes, in LNCaP and LAPC4 cells. Here, ∼10-fold induction of PSA and KLK2 transcripts was observed in LNCaP cells in the presence of 10 nM of DHT (Figure 2C). Infection of the AR shRNA adenovirus significantly reduced the expression of PSA and KLK2 transcripts to basal levels. The expression of β-actin or GAPDH, used as a negative control, was unchanged in the same samples. RT-PCR demonstrated a similar reduction of PSA and KLK2 transcript levels in the AR shRNA adenovirus-infected LAPC4 cells (Figure 2D). These results were consistent with the transactivation assays and provide additional evidence for the specificity of the AR shRNA adenoviruses in silencing AR-mediated transcription.
Androgen-induced transcription is mediated primarily through the AR
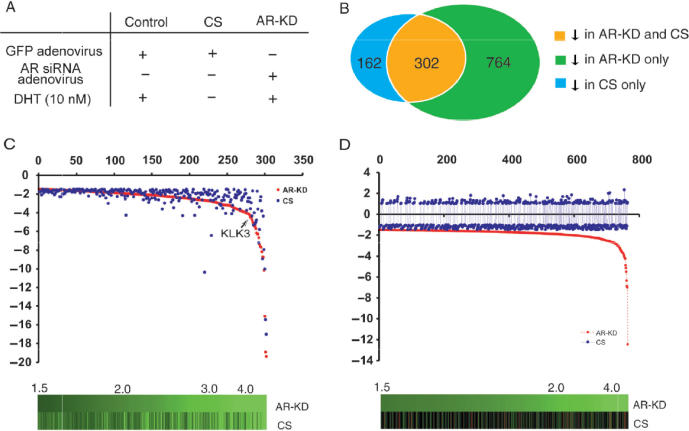
The AR and other ligand-dependent nuclear hormone receptors possess identifiable activation domains that confer transactivation potential when fused to a heterologous DNA-binding domain (22). However, one important feature of the AR and other nuclear hormone receptors, which distinguishes them from other transcription factors, is that the transcriptional activities of the receptors can only be induced by specific ligands through binding to the ligand-binding domains of the receptors (23). Therefore, androgen-induction has been widely used to assess AR-mediated transcription. Although androgen-induced transcription is mainly mediated through the AR, it is possible that other signaling pathways could be involved in androgen induced transcription. To attempt to distinguish between the direct effects of ligand and those of the AR protein, we compared effects of androgen depletion and AR knockdown on global gene expression profiles. RNA samples isolated from LNCaP cells that were either cultured in the absence of DHT for overnight or infected with the AR shRNA lentiviruses were analyzed using spotted cDNA microarrays (Figure 3A). Here, 302 genes were down-regulated in both groups, likely reflecting that these genes are regulated by androgens through the AR. Among them, 36 genes showed >4-fold decrease, including PSA (KLK3), a well-known AR target gene. Interestingly, there were 162 genes found to be down-regulated uniquely in samples isolated from the cells cultured in the absence of DHT. In the cells infected with the AR shRNA lentiviruses, there were 764 genes that were uniquely down-regulated. Complete gene expression datasets are available at http://www.stanford.edu/~hongjuan/AR. Although the precise mechanisms by which androgens or AR independently regulate gene expression are not clear, the difference in the transcriptional profiles from cells treated either with androgen ablation or AR knockdown suggests that alternative non-AR-dependent pathways may be involved in androgen-induced transcription and that AR might activate transcription for some genes in the absence of DHT.
Figure 3.
Identification of genes down-regulated by androgen deprivation and AR knockdown using cDNA microarray. (A) Treatment scheme for microarray experiment. (B) The number of genes with at least 1.5-fold decrease in expression in response to androgen deprivation, AR knockdown or both. (C) Fold change distribution of genes that showed decreased expression (>1.5 fold) in response to androgen deprivation or AR knockdown. Brightness of green in the figures below the graphs corresponds to the degree of decreased expression (fold change). (D) Genes whose expression decreased after AR knockdown (1.5-fold) but were not affected by androgen deprivation.
Knockdown of AR expression inhibits androgen-induced cell growth
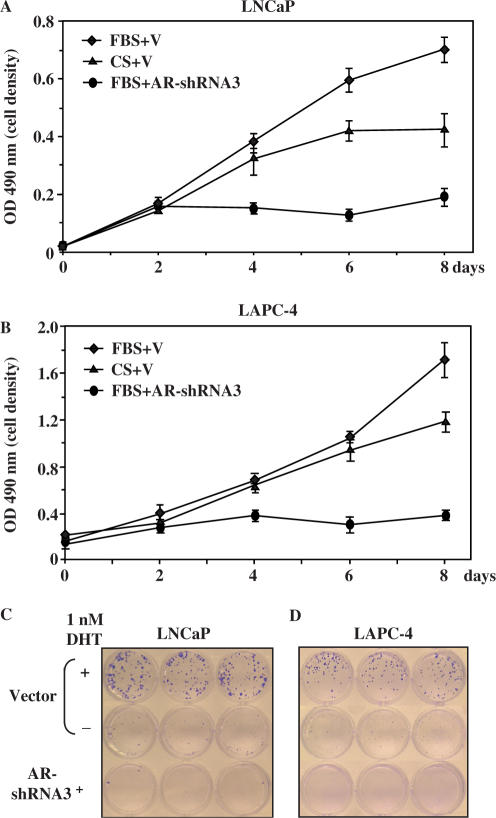
Multiple lines of evidence have shown that the depletion of androgens significantly suppresses the growth of prostate cancer cells, implying an essential role of androgens in prostate cancer cell growth (1,24). To further confirm that the growth-promoting effects of androgens in prostate cells are directly mediated through the AR, we examined the androgen-induced cell growth in LNCaP cells with the AR shRNA viruses. As shown in Figure 4A, cells infected with the control virus grew faster in the full medium than in the CS-FBS medium, which is consistent with the previous reports showing the growth-promoting role of androgens in LNCaP cells. The cells infected with the AR shRNA virus grew more slowly than the cells infected with the control virus, suggesting that the knockdown of AR expression inhibits the growth of LNCaP cells. In addition, infection with the AR shRNA virus significantly inhibited growth of LNCaP cells in colony formation assays (Figure 4C). Both the MTS and colony formation assay demonstrated similar growth inhibition by the AR shRNA adenovirus in LAPC4 cells, which contain a wild-type AR protein (25) (Figure 4B and D).
Figure 4.
Down-regulation of AR expression inhibits the growth of androgen-sensitive prostate cancer cells. (A) LNCaP cells were seeded into 96-well plates in media with or without DHT after 3 h adenovirus infection at an MOI of 10. Cell growth was measured every other day by MTS assay. The data represent the mean ± SD of three independent experiments. (B) Identical experiments performed in LAPC4 cells. (C) LNCaP cells were seeded into 24-well plates at 400 cells/well after 3 h adenovirus infection at an MOI of 10. Cells were cultured with the media in the presence or absence of DHT for 14 days and colonies were fixed and stained with crystal violet. (D) Similar experiments performed in LAPC4 cells.
Knockdown of AR expression affects AR-mediated transcription and cell growth in AI prostate cancer cells
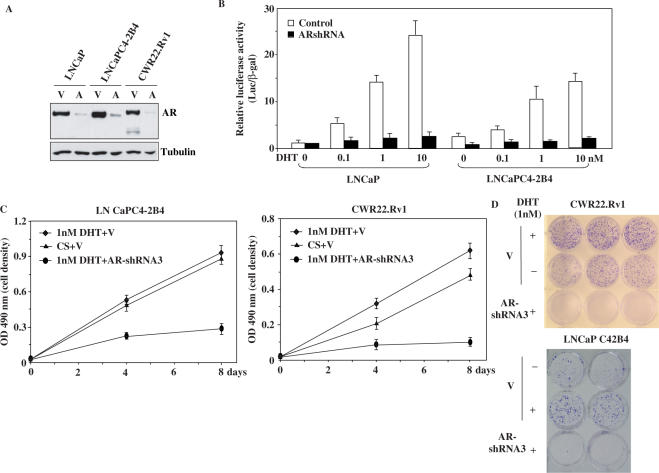
Compared to LNCaP and LAPC4, LNCaP C4-2B4 and CWR22Rv1 show blunted androgen-induced transcription and cell growth in response to androgens, even though both cell lines express the AR (26–28). Infection of the AR shRNA virus significantly decreased the expression of different forms of AR proteins in both LNCaP C4-2B4 and CWR22Rv1 cells (Figure 5A). Transient transfection of these cells with the 7-kb PSA promoter/reporter was carried out to assess the effects of the AR shRNA on AR-regulated transcription. In the absence of DHT, the activity of the PSA promoter/reporter was slightly higher in LNCaP C4-2B4 than in its parent line, LNCaP (Figure 5B). However, LNCaP cells were more responsive to DHT as compared to C4-2B4 cells, shown in the higher luciferase activities (Figure 5B), consistent with results reported previously (29). Intriguingly, knockdown of AR expression in C4-2B4 cells reduced PSA-luciferase activity to levels comparable to LNCaP cells, suggesting that the AR still plays a critical role in the regulation of PSA transcription in these AI prostate cancer cells.
Figure 5.
Down-regulation of AR expression affects AR-mediated transcription and cell growth in androgen-insensitive prostate cancer cells. (A) LNCaP, LNCaP C4-2B4 and CWR22-Rv1 cells were infected with control or AR shRNA3 lentivirus, and incubated in medium with 10 μg/ml blastcidin for selection. Whole-cell lysates were prepared at Day 7, and analyzed by western blotting with AR and tubulin antibodies. (B) LNCaP and LNCaP C4-2B4 cells were infected with either control or AR shRNA3 adenovirus at an MOI of 20 for 6 h, and transfected with PSA7 kb-luc reporter (PSA-Luc) and pcDNA3-β-gal. Cells were incubated in T-medium with 5% CS-FBS for 24 h, and then induced with different amounts of DHT for another 24 h. Luciferase and β-gal activities were measured and reported as RLU. (C) LNCaP C4-2B4 and CWR22-Rv1 cells were infected with control or AR shRNA3 lentivirus and incubated with medium containing 10 μg/ml of blastcidin for 7 days. Cell growth was measured by the MTS assay. The data represent the mean ± SD of three independent experiments. (D) CWR22-Rv1 and LNCaP C4-2B4 cells were infected with the control or AR shRNA3 lentivirus, and cultured with 10 μg/ml blastcidin for 7 days. Cells resistant to the selection were re-plated into 12-well plates and cultured for 14 days. Colonies were fixed and stained with crystal violet.
To further investigate the role of AR in LNCaP C4-2B4 and CWR22-Rv1 cells, we infected the cells with the AR shRNA and control lentiviruses and then cultured them in medium either with FBS or CS-FBS. As shown in Figure 5C, both cell lines infected with the control virus showed similar growth patterns in the presence or absence of androgens, indicating that androgens are not essential for the growth of these cells. However, cells infected with the AR shRNA viruses and cultured in the presence of androgens grew more slowly than the cells infected with the control virus (Figure 5C). Similarly, in the colony formation assays, depletion of androgens slightly reduced the growth of LNCaP C4-2B4 and CWR22-Rv1 cells in comparison with LNCaP and LAPC4 cells (Figure 4C and D). However, the knockdown of AR expression in these cells profoundly inhibited cell growth, resulting in no colony formation after a 12-day incubation (Figure 5D). Taken together, these data indicate an essential role for AR in the transcription and cell growth of AI prostate cancer cells.
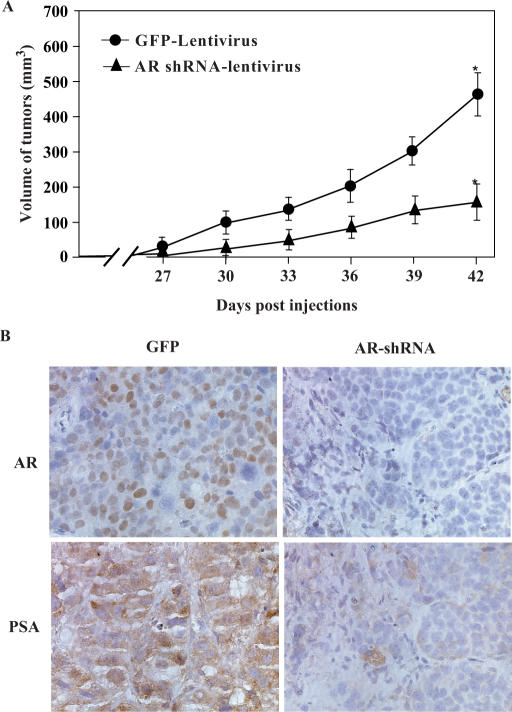
Reduction of AR expression delays tumor formation in a xenograft model
Next, we explored the AR's role in tumor formation using a xenograft model. LAPC4 cells infected with AR shRNA or GFP lentiviruses were implanted into male athymic nude mice. As shown in Figure 6A, LAPC4 cells that were infected with the AR shRNA lentiviruses developed palpable tumors later than the cells infected with GFP (control) viruses. By Day 42 post-infection, tumors formed by LAPC4 cells infected with the AR shRNA viruses were significantly smaller than their counterparts infected with control viruses (P < 0.05). Interestingly, by Day 42, staining of the tumor tissues from LAPC4 cells infected with the AR shRNA viruses showed that most tumor cells were AR positive (data not shown), whereas tumor samples isolated at D15 post-infection were largely AR negative (Figure 6B). While the reasons for this change are unclear, and could be due to loss of expression of the lentivirus, growth of a subpopulation of the cells lacking the lentivirus or escape from AR knockdown by other mechanisms. Regardless, the data suggest a critical role for AR in prostate cancer tumor growth in vivo.
Figure 6.
Reduction of AR expression inhibits tumor xenograft formation in athymic mice. (A) LAPC4 cells were transduced with the AR shRNA or GFP lentiviruses at a MOI of 3 for 24 h. Cells were harvested, resuspended in PBS and mixed with an equal volume of Matrigel ECM. Here, 100 μl of cell suspension (1 × 107 cells/ml) was injected subcutaneously in opposite lateral flanks of 6–8-week-old athymic male mice. Mice were monitored twice weekly. Tumors were measured in two dimensions with calipers, and tumor volume (mm3) was calculated with the formula V = (length × width2)/2. ‘Asterisk’ indicates a significant difference (P < 0.05) between the two groups of animals. (B) The tumor specimens isolated from xenograft animals at Day 15 were fixed in 10% neutral-buffered formalin and embedded in paraffin, and then analyzed by immunohistochemistry using anti-AR antibodies.
DISCUSSION
Androgen ablation, through either surgical or biochemical approaches, to reduce the level of serum testosterone or competitively repress AR function is frequently used in the treatment of prostate cancer patients (1,24). Initially, most tumors respond to androgen ablation, implying that the androgen-signaling pathway is required in the growth of prostate cancer cells in vivo. It has been shown that androgen-induced transcription and cell growth are mediated mainly through the AR (3,7,11). Thus, modification of AR activity directly affects the growth and progression of prostate cancer cells. Mice engineered to overexpress AR develop lesions similar to prostatic intraepithelial neoplasia (PIN), a putative prostate cancer precursor lesion (30). Increasing cellular levels of the AR not only enhances androgen-induced cell growth but increases the sensitivity of prostate cancer cells to androgens, allowing the tumor cells to grow in a low androgen environment (31). To understand whether the AR is critical in prostate carcinogenesis, we used an RNA interference approach to directly assess the AR's effect on androgen-induced transcription and cell growth (15). Three AR shRNA constructs that contain 21-mer sequences derived from different coding regions of the human AR all showed specific silencing of AR expression. Using these vectors, we evaluated the downstream consequences of AR knockdown in prostate cancer cells. We observed that the silencing of AR expression fully abolished androgen-induced transcription in three ARE-containing promoters/reporters in two AR-positive prostate cancer cell lines. In addition, the expression of two AR downstream target genes, PSA and KLK2, was also significantly reduced in cells infected with the AR shRNA viruses. These data are consistent with previous studies by others and confirm the critical role of AR in androgen-induced transcription. Moreover, in both cases, it appears that the repression of androgen-induced transcription is more pronounced by the AR knockdown than by the androgen ablation, implying that transcription of PSA and KLK2 in LNCaP and LAPC4 cells is solely regulated through the AR.
Using the AR shRNA constructs, we also directly assessed the effect of the AR on androgen-induced prostate cancer cell growth. Both colony formation and MTS assays showed that knockdown of the AR expression by the AR shRNA affects the growth of LNCaP and LAPC4 cells, even when androgens were supplied in the media. The effect of the AR shRNA adenoviruses on cell growth appears more potent than that induced by androgen depletion. Consistent with these findings, AR shRNA affected significantly more transcripts than androgen deprivation alone. Although the precise targets of AR, that are primarily responsible for prostate cancer cell growth are currently unknown, candidate target genes are likely to be directly involved in promoting prostate cancer initiation and progression. Our demonstration that knockdown of AR expression reduces both androgen-induced transcription and cell growth argues for an essential role for AR-mediated transcription in prostate cancer growth and progression in hormone naïve and hormone refractory tumors.
Over time, clinical prostate cancers become unresponsive to androgen deprivation therapies (become AI) because of poorly understood molecular changes (3,7,11,32). Current hypotheses propose that prostate cancer cells either bypass the AR-signaling pathway altogether or adapt its function to a low androgen environment. Our data suggest that AR retains a critical role in prostate cancer growth in AI cancers. In two AI prostate cancer cell lines, we showed that the knockdown of AR expression still reduces AR-mediated transcription in reporter assays and the expression of endogenous AR target genes as examined by northern blotting and RT-PCR. Intriguingly, the silencing of AR expression also inhibits the growth of these cells in vitro and in vivo. Our data demonstrate an essential role for the AR in the regulation of transcription and cell growth in AI cells, implying that AI cells may still be ‘AR sensitive’.
Previous studies have shown that androgen-induced transcription is mainly mediated through the AR (33). Using the AR shRNA approach, we were able to readdress this question by comparing the transcript profiles induced by the AR knockdown and androgen depletion. We identified a group of genes that appear to be regulated by both androgen depletion and knockdown of AR protein, including the well known AR target gene, PSA. Interestingly, we also identified two subsets of genes that are only regulated by the either androgen depletion or AR knockdown. Although the precise mechanisms underlying the regulation of these two subsets of genes are not clear, these data suggest that androgen ligand and the AR might have effects on pathways outside the androgen ligand-induced, AR-signaling pathway. Further investigation of these pathways and regulatory mechanisms may enhance our current knowledge of androgen signaling and its role in the development of AI prostate cancer.
Finally, AR appears to play a direct role in prostate cancer growth in vivo. We demonstrated that reduction of endogenous AR expression in LAPC4 cells inhibits the onset and growth of tumors in nude mice. Intriguingly, at later stages of tumor growth, most cells in the tumors that developed from the AR shRNA-virus-infected LAPC4 cells were AR positive. This finding suggests that the tumors that grew out were derived from a subset of LAPC4 cells that had lost expression of the AR shRNA viruses or escaped AR inhibition, and underscores the importance of AR expression in LAPC4 tumor growth. Based on these data, we have begun to assess the potential therapeutic role of the AR shRNA in prostate cancer xenograft models.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Jane Lee and Yulan Yu for technical assistance and critical reading of this manuscript. This work was supported by National Institutes of Health Grants CA70297, CA87767 and DK61002, and the Department of Army Prostate Cancer grants DAMD17-03-1-0090 (to Z.S.), and CA123532-01 (to H.Z.). Funding to pay the Open Access publication charges for this article was provided by DK61002.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kyprianou N, Isaacs JT. Activation of programmed cell death in the rat ventral prostate after castration. Endocrinology. 1988;122:552–562. doi: 10.1210/endo-122-2-552. [DOI] [PubMed] [Google Scholar]
- 2.Culig Z, Klocker H, Bartsch G, Steiner H, Hobisch A. Androgen receptors in prostate cancer. J. Urol. 2003;170:1363–1369. doi: 10.1097/01.ju.0000075099.20662.7f. [DOI] [PubMed] [Google Scholar]
- 3.Gelmann EP. Molecular biology of the androgen receptor. J. Clin. Oncol. 2002;20:3001–3015. doi: 10.1200/JCO.2002.10.018. [DOI] [PubMed] [Google Scholar]
- 4.Jenster G. The role of the androgen receptor in the development and progression of prostate cancer. Semin. Oncol. 1999;26:407–421. [PubMed] [Google Scholar]
- 5.Culig Z, Hobisch A, Bartsch G, Klocker H. Androgen receptor – an update of mechanisms of action in prostate cancer. Urol. Res. 2000;28:211–219. doi: 10.1007/s002400000111. [DOI] [PubMed] [Google Scholar]
- 6.Koivisto P, Kolmer M, Visakorpi T, Kallioniemi OP. Androgen receptor gene and hormonal therapy failure of prostate cancer. Am. J. Pathol. 1998;152:1–9. [PMC free article] [PubMed] [Google Scholar]
- 7.Culig Z. Role of the androgen receptor axis in prostate cancer. Urology. 2003;62:21–26. doi: 10.1016/s0090-4295(03)00698-8. [DOI] [PubMed] [Google Scholar]
- 8.Taplin ME, Bubley GJ, Shuster TD, Frantz ME, Spooner AE, Ogata GK, Keer HN, Balk SP. Mutation of the androgen-receptor gene in metastatic androgen- independent prostate cancer. N. Engl. J. Med. 1995;332:1393–1398. doi: 10.1056/NEJM199505253322101. [DOI] [PubMed] [Google Scholar]
- 9.Gaddipati JP, McLeod DG, Heidenberg HB, Sesterhenn IA, Finger MJ, Moul JW, Srivastava S. Frequent detection of codon 877 mutation in the androgen receptor gene in advanced prostate cancers. Cancer Res. 1994;54:2861–2864. [PubMed] [Google Scholar]
- 10.Ruizeveld de Winter JA, Janssen PJ, Sleddens HM, Verleun-Mooijman MC, Trapman J, Brinkmann AO, Santerse AB, Schroder FH, van der Kwast TH. Androgen receptor status in localized and locally progressive hormone refractory human prostate cancer. Am. J. Pathol. 1994;144:735–746. [PMC free article] [PubMed] [Google Scholar]
- 11.Balk SP. Androgen receptor as a target in androgen-independent prostate cancer. Urology. 2002;60:132–138. doi: 10.1016/s0090-4295(02)01593-5. ; discussion 138–139. [DOI] [PubMed] [Google Scholar]
- 12.Cleutjens KB, van Eekelen CC, van der Korput HA, Brinkman AO, Trapman J. Two androgen response regions cooperate in steroid hormone regulated activity of the prostate-specific antigen promoter. J. Biol. Chem. 1996;271:6379–6388. doi: 10.1074/jbc.271.11.6379. [DOI] [PubMed] [Google Scholar]
- 13.Yeh S, Chang C. Cloning and characterization of a specific coactivator, ARA70, for the androgen receptor in human prostate cells. Proc. Natl Acad. Sci. USA. 1996;93:5517–5521. doi: 10.1073/pnas.93.11.5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang F, Li X, Sharma M, Sasaki CY, Longo DL, Lim B, Sun Z. Linking beta-catenin to androgen signaling pathway. J. Biol. Chem. 2002;277:11336–11344. doi: 10.1074/jbc.M111962200. [DOI] [PubMed] [Google Scholar]
- 15.Sui G, Soohoo C, Affarel B, Gay F, Shi Y, Forrester WC. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc. Natl Acad. Sci. USA. 2002;99:5515–5520. doi: 10.1073/pnas.082117599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He TC, Zhou S, da Costa LT, Yu J, Kinzler KW, Vogelstein B. A simplified system for generating recombinant adenoviruses. Proc. Natl Acad. Sci. USA. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D, Naldini L. A third-generation lentivirus vector with a conditional packaging system. J. Virol. 1998;72:8463–8471. doi: 10.1128/jvi.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delenda C. Lentiviral vectors: optimization of packaging, transduction and gene expression. J. Gene. Med. 2004;6(Suppl. 1):S125–S138. doi: 10.1002/jgm.501. [DOI] [PubMed] [Google Scholar]
- 19.Zhao H, Whitfield ML, Xu T, Botstein D, Brooks JD. Diverse effects of methylseleninic acid on the transcriptional program of human prostate cancer cells. Mol. Biol. Cell. 2004;15:506–519. doi: 10.1091/mbc.E03-07-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sherlock G, Hernandez-Boussard T, Kasarskis A, Binkley G, Matese JC, Dwight SS, Kaloper M, Weng S, Jin H, et al. The Stanford Microarray Database. Nucleic Acids Res. 2001;29:152–155. doi: 10.1093/nar/29.1.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma M, Zarnegar M, Li X, Lim B, Sun Z. Androgen receptor interacts with a novel MYST protein, HBO1. J. Biol. Chem. 2000;275:35200–35208. doi: 10.1074/jbc.M004838200. [DOI] [PubMed] [Google Scholar]
- 22.Simental JA, Sar M, Wilson EM. Domain functions of the androgen receptor. J. Steroid Biochem. Mol. Biol. 1992;43:37–41. doi: 10.1016/0960-0760(92)90185-l. [DOI] [PubMed] [Google Scholar]
- 23.Zhou ZX, Wong CI, Sar M, Wilson EM. The androgen receptor: an overview. Recent Prog. Horm. Res. 1994;49:249–274. doi: 10.1016/b978-0-12-571149-4.50017-9. [DOI] [PubMed] [Google Scholar]
- 24.Isaacs JT, Kyprianou N. Biological basis for chemohormonal therapy for prostatic cancer. Cancer Treat. Res. 1989;46:177–193. doi: 10.1007/978-1-4613-1595-7_10. [DOI] [PubMed] [Google Scholar]
- 25.Craft N, Shostak Y, Carey M, Sawyers CL. A mechanism for hormone-independent prostate cancer through modulation of androgen receptor signaling by the HER-2/neu tyrosine kinase. Nat. Med. 1999;5:280–285. doi: 10.1038/6495. [DOI] [PubMed] [Google Scholar]
- 26.Wu TT, Sikes RA, Cui Q, Thalmann GN, Kao C, Murphy CF, Yang H, Zhau HE, Balian G, et al. Establishing human prostate cancer cell xenografts in bone: induction of osteoblastic reaction by prostate-specific antigen-producing tumors in athymic and SCID/bg mice using LNCaP and lineage-derived metastatic sublines. Int. J. Cancer. 1998;77:887–894. doi: 10.1002/(sici)1097-0215(19980911)77:6<887::aid-ijc15>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 27.Nagabhushan M, Miller CM, Pretlow TP, Giaconia JM, Edgehouse NL, Schwartz S, Kung HJ, de Vere White RW, Gumerlock PH, et al. CWR22: the first human prostate cancer xenograft with strongly androgen-dependent and relapsed strains both in vivo and in soft agar. Cancer Res. 1996;56:3042–3046. [PubMed] [Google Scholar]
- 28.Sramkoski RM, Pretlow TGII, Giaconia JM, Pretlow TP, Schwartz S, Sy MS, Marengo SR, Rhim JS, Zhang D, et al. A new human prostate carcinoma cell line, 22Rv1. In Vitro Cell Dev. Biol. Anim. 1999;35:403–409. doi: 10.1007/s11626-999-0115-4. [DOI] [PubMed] [Google Scholar]
- 29.Wu HC, Hsieh JT, Gleave ME, Brown NM, Pathak S, Chung LW. Derivation of androgen-independent human LNCaP prostatic cancer cell sublines: role of bone stromal cells. Int. J. Cancer. 1994;57:406–412. doi: 10.1002/ijc.2910570319. [DOI] [PubMed] [Google Scholar]
- 30.Stanbrough M, Leav I, Kwan PW, Bubley GJ, Balk SP. Prostatic intraepithelial neoplasia in mice expressing an androgen receptor transgene in prostate epithelium. Proc. Natl Acad. Sci. USA. 2001;98:10823–10828. doi: 10.1073/pnas.191235898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen CD, Welsbie DS, Tran C, Baek SH, Chen R, Vessella R, Rosenfeld MG, Sawyers CL. Molecular determinants of resistance to antiandrogen therapy. Nat. Med. 2004;10:33–39. doi: 10.1038/nm972. [DOI] [PubMed] [Google Scholar]
- 32.Abate-Shen C, Shen MM. Molecular genetics of prostate cancer. Genes Dev. 2000;14:2410–2434. doi: 10.1101/gad.819500. [DOI] [PubMed] [Google Scholar]
- 33.Heinlein CA, Chang C. The roles of androgen receptors and androgen-binding proteins in nongenomic androgen actions. Mol. Endocrinol. 2002;16:2181–2187. doi: 10.1210/me.2002-0070. [DOI] [PubMed] [Google Scholar]
- 34.Pang S, Dannull J, Kaboo R, Xie Y, Tso CL, Michel K, deKernion JB, Belldegrun AS. Identification of a positive regulatory element responsible for tissue-specific expression of prostate-specific antigen. Cancer Res. 1997;57:495–499. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.