Abstract
Neuroendocrine tumors (carcinoids) are a heterogeneous group of neoplasms arising from the diffuse neuroendocrine system. Genetic changes underlying their tumorigenesis are primarily unknown. We used comparative genomic hybridization to screen 32 well-differentiated neuroendocrine tumors (21 gastrointestinal and 11 bronchial) and three associated metastases for genomic alterations. There were striking differences of genomic imbalances between the two subgroups of neuroendocrine tumors. Losses of chromosome 18q and 18p were shown in eight (38%) and seven (33%), respectively, out of 21 gastrointestinal tumors and in none of the 11 bronchial tumors. Conversely, deletions of 11q occurred in four of 11 (36%) bronchial tumors but only in one gastrointestinal tumor. These comparative genomic hybridization findings were confirmed by interphase cytogenetics. Our data indicate that neuroendocrine tumors of the two subgroups develop via different molecular pathways. Inactivation of one or several tumor suppressor genes on chromosome 18 may be important for the biological behavior of gastrointestinal tumors, whereas gene inactivation on 11q seems to be associated with tumor development of the bronchi.
Neuroendocrine tumors (NETs) are a heterogeneous group of neoplasms arising from the diffuse neuroendocrine system, which include a wide histopathological spectrum ranging from classical carcinoids with slow growth and relatively good prognosis to highly malignant undifferentiated neuroendocrine carcinomas. 1 According to Godwin, 2 ∼85% of all NETs occur in the gastrointestinal tract, 10% in the lung, and the rest in various organs such as the larynx, thymus, kidney, ovary, and skin. The most frequent location in the gastrointestinal tract is the appendix (40 to 50%), followed by the jejunum and ileum (20 to 30%), and rectum and colon (10%). Bronchial NETs, which are the most common type of NETs in the lung, are rare lung malignancies, accounting for 2% of all lung tumors. 3 Although NETs generally exhibit a characteristic growth pattern with common histological and immunohistochemical features, there are considerable differences in the clinical behavior and in responsiveness to therapy among different subgroups. Appendiceal NETs have a very low aggressive behavior with a metastasis frequency ranging from 1.4 to 8.8%, whereas other NETs have a relative high metastatic potential. 1,3-7 Mixed exocrine-endocrine tumors are extremely rare neuroendocrine lesions that share histological features of both endocrine and glandular differentiation. There is confusion regarding the classification and nomenclature of these neoplasms, reflected in a variety of their names, such as adenocarcinoid, goblet cell carcinoid, crypt cell carcinoid, mucous carcinoid, and mixed carcinoid-adenocarcinoma. Some of these tumors, especially the so-called mixed carcinoid-adenocarcinoma, are more aggressive than classical carcinoids, 8,9 suggesting that the biological behavior of this type of tumors may be dictated by the differentiation of the exocrine cell compartment.
Despite recent advances in the diagnosis, localization, and treatment of NETs, no etiological factors are proven to be associated with these tumors and little is known about the molecular genetic changes underlying their tumorigenesis. Several genetic alterations associated with bronchial NETs have been reported, including loss of heterozygosity of 11q 10 and the Rb gene, 11 as well as mutations of p53. 12 Recently, genome-wide surveys of lung NETs (typical and atypical carcinoids) revealed frequent 11q deletions. 13,14 In contrast to lung NETs, molecular and cytogenetic data for gastrointestinal NETs are very limited. So far, only one study on genomic imbalances in eight gastrointestinal NETs including five ileal, two duodenal, and one gastric carcinoids has been reported. 15 A few molecular studies have also been performed to elucidate the potential role of the tumor suppressor genes APC, DCC, and p53 16,17 as well as the oncogenes c-myc, bcl-2, c-erbB-2, and c-jun in the development of gastrointestinal NETs. 18 The importance of these genes, however, remains to be clarified. With regard to mixed exocrine-endocrine tumors of the gastrointestinal tract, a recent study by Ramnani et al, 17 which is thus far the sole molecular genetic investigation in this type of neoplasms, showed the involvement of p53 mutations in a small proportion of these tumors.
In the present study, we used comparative genomic hybridization (CGH), which allows screening of the entire genome of a tumor for all relative DNA copy-number changes, to identify genomic imbalances in 35 NETs comprising 21 well-differentiated primary gastrointestinal (including seven mixed exocrine-endocrine tumors), 11 well-differentiated primary bronchial and three associated metastases. Our goals were: 1) to screen for frequently altered chromosomal regions in these tumors; and 2) to elucidate the molecular similarities and diversities of these tumor subgroups, which would provide an explanation for their differences in clinical outcome. Interphase cytogenetics for chromosomes 18 and 11q were additionally performed to independently confirm some of the CGH results.
Materials and Methods
Patients and Samples
Tumor samples available from 34 patients with NETs were either deep frozen or fixed in 4% formalin and embedded in paraffin. The histopathological classification was based on the recent proposed criteria. 1,19-21 Among the 35 tumors collected, there were 21 primary gastrointestinal NETs (including seven mixed exocrine-endocrine tumors), 11 primary bronchial NETs, and three associated metastases (Table 1) ▶ . The gastrointestinal NETs examined were well-differentiated primary tumors, including four appendiceal, nine ileal, two colonic, four duodenal, and two gastric tumors. The mixed exocrine-endocrine tumors were all of the goblet-cell type. Pancreatic endocrine tumors (islet cell tumors) were excluded from the study. All of the 11 bronchial tumors were so-called typical (well-differentiated) carcinoids. 20 Other types of lung NETs, such as atypical carcinoid, were excluded. The average diameter of the tumors was 4 cm (range, 0.7 to 12 cm) for gastrointestinal and 2.6 cm (range, 1 to 4.8 cm) for bronchial NETs. For some tumors, two or three different regions were investigated to evaluate intratumoral genetic heterogeneity. The three metastatic lesions, present in three patients with NET, were located in the liver, paraortal lymph nodes, and mesenteric lymph nodes, respectively. For the two formers, the exact tumor sites of the primaries in the gastrointestinal tract were not known. The latter had arisen from a colonic NET.
Table 1.
Clinical and Genetic Data of 32 Well-Differentiated Neuroendocrine Tumors and Three Associated Metastases
| No. | Age/ sex | Diagnosis | Metastasis | Size (cm) | CGH finding | FISH finding | |||
|---|---|---|---|---|---|---|---|---|---|
| Gain | Loss | 18c | 11q13 | 11q13.4-q21 | |||||
| 1 | 73 /m | BNET | Liver, spleen | 4.5 | +17q24-qter | −6q14-qter, −11q13-qter | L | ||
| 2 | 68 /f | BNET | nk | 3 | +7p,+7q, +17p,+17cen-q21 | −1p,−11q13-qter | L | L | |
| 3 | 34 /f | BNET | nk | 4.8 | +5p,+5q24-qter, +14q24-qter | ndc | ndc | ||
| 4 | 72 /f | BNET | nk | 3.2 | +9q,+16q,+20q | ndc | ndc | ||
| 5 | 30 /f | BNET | nk | 2.5 | ndc | ndc | |||
| 6 | 44 /m | BNET | nk | 1.4 | +5p,+5q,+8p,+8q, +16q | ndc | ndc | ||
| 7 | 47 /f | BNET | nk | 1.3 | +9q34 | −15q,−22q | ndc | ndc | |
| 8 | 45 /f | BNET | nk | 2 | −11cen-p14, −11q13-qter | ndc | L | ||
| 9 | 45 /m | BNET | nk | 2 | −11p15,−11q13.2-qter | ndc | L | ||
| 10 | 45 /f | BNET | nk | 1 | ndc | ndc | |||
| 11 | 55 /m | BNET | nk | 3 | ndc | ndc | |||
| 12 | 42 /f | GINET | Local LN | nk | −18p,−18q | ndc | |||
| 13 | 68 /f | GINET | Local LN | 1 | −16q13-q24,−18p, −18q | L | |||
| 14# | 63 /f | GINET Area 1 | Local LN | 2 | L* | ||||
| Area 2 | 2 | −18p,−18q | L | ||||||
| 15 | 26 /f | GINET | Liver | 5 | +9q34,+17p,+17q | −1q22-qter | |||
| 16 | 79 /m | GINET | Local LN | 1 | −18p,−18q | L | |||
| 17 | 89 /f | GINET | Local LN | 4 | −18p,−18q | L | |||
| 18 | 70 /f | GINET | nk | +10p,+10q | −18q22-qter | ||||
| 19# | 70 /m | GINET | LN | 2 | |||||
| 20 | 38 /f | GINET | Local LN | 0.7 | |||||
| 21 | 16 /f | GINET | 3.5 | +9q34 | ndc | ||||
| 22 | 19 /f | GINET | Local LN | 2 | |||||
| 23 | 53 /m | GINET | 10 | +17p,+17q | |||||
| 24 | 50 /m | GINET | Liver and LN | 2.5 | +1q,+7p,+7q,+15q,+10q22-qter,+20q | −3q21-qter, −y | |||
| 25 | 55 /m | GINET | LN | 12 | +3p,+3q,+9p13-pter, +9q21-qter | −10p,−10q,−13q,−16p,−16q,−17p,−18p,−18q, −21q,−22q | |||
| 26 | 68 /m | GINETα | Liver | 1.5 | |||||
| 27 | 73 /f | GINETα | Local LN | 2 | |||||
| 28 | 27 /m | GINETα | nk | nk | |||||
| 29# | 70 /m | GINETα | nk | ndc | |||||
| 30 | 68 /f | GINETα | nk | ||||||
| 31 | 65 /f | GINETα | Local LN | 1 | −1q13.2-qter, −18p,−18q | L | |||
| 32 | 65 /m | GINETα primary | nk | ||||||
| metastasis | Stomach | nk | −3p,−3q | ||||||
| 33 | 35 /f | NET’s metastasis | Liver | 10 | +4p,+4q, +20q | −9p21-qter,−13q21-q22,−16q21-qter,−18p,−18q | L | ||
| 34 | 46 /m | NET’s metastasis | Paraortal LN | 4 | +5p, +5q,+7p,+7q | ||||
#Two or three different regions of the tumors were examined for genetic heterogeneity; m, male; f, female; BNET, bronchial neuroendocrine tumors; GINET, gastrointestinal neuroendocrine tumors; α, mixed endocrine-exocrine tumors; nk, not known; LN, lymph node; ndc, no detectable change; L, loss; 18c, centromere 18 probe; 11q13, MEN1 gene; 11q13.4-q21, cClll-270.
*Deletion of chromosome 1 was also detected, suggesting monosomy.
DNA Preparation for CGH
Isolation of genomic DNA from frozen tumor samples was performed using the D-5000 Puregene DNA Isolation Kit (Gentra Systems Inc., Minneapolis, MN). Approximately 2 mm 3 of frozen tumor material was homogenized and DNA extraction performed according to the manufacturer’s recommendations. DNA extraction from paraffin-embedded tumors was done as previously described. 22 Direct fluorescence labeling of DNA was performed by nick translation using a commercial kit (BioNick kit; Life Technologies, Gaithersburg, MD).
CGH Analysis
CGH was performed as previously described. 23 The hybridization mixture consisted of 200 to 400 ng of spectrum-green-labeled tumor DNA, 200 ng of spectrum-red-labeled normal reference DNA, and 10 μg of unlabeled human Cot-1 DNA dissolved in 10 μl of hybridization buffer (50% formamide, 10% dextran sulfate, 2× standard saline citrate, pH 7.0). Hybridization took place throughout 3 days at 37°C to sex-matched normal metaphase spreads (Vysis, Downer Grove, IL). Digital images were collected from six to seven metaphases using a cooled Photometrics charge-couple device camera (Microimager 1400; Xillix Technologies, Vancouver, British Columbia, Canada). The VYSIS software program was used to calculate average green-to-red ratio profiles for each chromosome. At least five observations per autosome and three observations per sex chromosome were included in each analysis.
Thresholds used for definition of DNA sequence copy number gains and losses were based on the results of CGH analyses of normal tissues. A gain of DNA sequences was assumed at chromosomal regions where the hybridization resulted in a green-to-red ratio of ≥1.20. Overrepresentations were considered amplifications when the fluorescence ratio values exceeded ≥1.5 in a subregion of a chromosome arm. A loss of DNA sequences was presumed at chromosomal regions where the tumor to normal ratio was ≤0.80. Because some false-positive results were found in normal tissues at chromosomes 1p, 16p, 19, and 22, gains at these G-C-rich regions were excluded from all analyses.
Fluorescence in Situ Hybridization (FISH) Analysis
A combination of two centromere probes specific for chromosomes 1 and 18 was used to analyze interphase cytogenetics in 10 selected tumors, of which eight exhibited losses of chromosome 18 identified by CGH, aiming to independently confirm the CGH results of chromosome 18 abnormalities. Touch preparations were made from five frozen tumor samples. Paraffin-embedded sections were used for the other five tumors.
For FISH analysis on touch preparations, centromere probes specific for chromosomes 1 and 18 were labeled using spectrum-green dUTP and spectrum-red dUTP (Vysis), respectively. Hybridization, posthybridization washes, and detection of the hybridized signals were performed as previously described. 23
For paraffin-embedded sections, the centromere probe of chromosome 18 was labeled using digoxigenin-11-dUTP (Roche, Basel, Switzerland). Interphase analysis on paraffin-embedded sections was performed according to published protocols 24 with some modifications. Briefly, sections were deparaffinized in xylol (3 times for 10 minutes each) and washed twice for 5 minutes each in 100% methanol. Slides were immersed in 85% formic acid/0.3% H2O2 for 10 minutes at room temperature, and then soaked in prewarmed 1 mol/L sodium thiocyanate (NaSCN) at 80°C for another 10 minutes. Subsequently, sections were treated with 4 mg/ml of pepsin in 0.02 N HCl for 30 minutes at 37°C. Ten to 15 μl of hybridization solution containing 10 to 15 ng probes were applied on each section. Hybridization took place overnight in a humidified chamber at 37°C. Detection of centromere signal for chromosome 18 was achieved using sheep anti-digoxigenin-rhodamine (Roche).
Two cosmid probes containing the chromosomal region of 11q13 (MEN1) and 11q13.4-q21 (cCl11-270), respectively, were used in a combination with a centromere 11 probe to analyze the 11 bronchial NETs, of which four showed 11q losses and seven no detectable alterations as revealed by CGH. The cosmid probes were labeled with spectrum-green dUTP, whereas the centromere 11 probe was labeled using biotin-16-dUTP (Vector, Vector Laboratories, Burlingame, CA). Detection of hybridized cosmid probes was performed using rabbit anti-fluorescein isothiocyanate (DAKO, Glostrup, Denmark) and swine anti-rabbit Ig fluorescein isothiocyanate (DAKO). Biotin-labeled chromosome 11-specific centromere probe was detected by using avidin-tetramethylrhodamine B isothiocyanate (Vector) and biotinylated goat anti-avidin. 25
At least 100 interphase nuclei with strong hybridization signals were scored for each tumor. Normal frozen specimen or connective tissue in the vicinity of tumors served as control that exhibited two copies of centromere or 11q signals in ≥95% of nuclei. A loss was assumed if >30% of nuclei exhibited only one signal of centromere, 11q13 or 11q13.4-q21.
Statistics
Contingency table analysis and Student’ t-test were used to compare the number of alterations and the frequency of individual changes between tumors of different types.
Results
Genomic Alterations Detected by CGH
DNA copy number changes were observed in 12 of 21 primary gastrointestinal NETs and eight of 11 primary bronchial NETs (Table 1) ▶ . The average number of alterations per tumor was 2.2 ± 3.3 for gastrointestinal NETs and 2.5 ± 2 for bronchial NETs. In gastrointestinal NETs, 75% of the tumors with metastasis exhibited genomic alterations. All three of the metastatic tumors examined also showed genomic aberrations. These findings indicate that genomic imbalances are associated with NET progression. However, such an association could not be determined for the bronchial tumor group, because of lack of the follow-up data on metastasis. Comparing the mean number of total genomic changes of the mixed exocrine-endocrine tumors (0.5 ± 1.3) with that of the remaining gastrointestinal NETs (2.9 ± 3.7) showed an obvious difference which, however, did not reach statistical significance (P = 0.1467). One tumor (tumor 14) exhibited intratumoral heterogeneity.
Regions of Frequent Genomic Aberrations
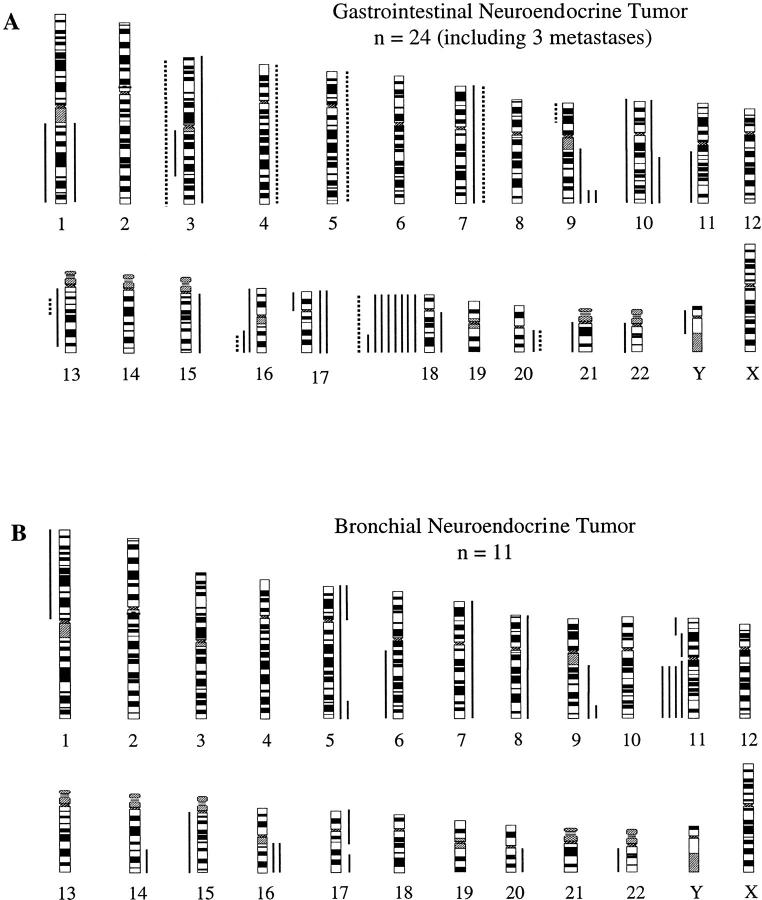
The chromosomal regions with DNA copy number alterations (losses and gains) identified by CGH are illustrated in Figure 1 ▶ . The most common DNA copy number changes were losses of chromosome 18q (38%) and 18p (33%) in gastrointestinal NETs and losses of 11q in 36% bronchial NETs (Table 2) ▶ . Other areas with less frequent alterations included +9q34 (14% each), −3q, −16q, and +10q (each 10%), −9p, −11q, −13q, +7p, +7q, and +20q (each 5%) in gastrointestinal NETs, as well as −11p, +5p, +5q, +9q, and +16q (each 18%) in bronchial NETs. Among all of the alteration differences between gastrointestinal and bronchial NETs, only −11p and +16q reached statistical significance. No high-level gains (amplifications) were observed in this study.
Figure 1.
Summary of all DNA copy number alterations detected by CGH in 21 gastrointestinal NETs and three associated metastases (A) and 11 bronchial NETs (B). The vertical green lines on the right of the chromosome ideograms indicate gains, the red on the left losses of the corresponding chromosomal regions. The solid and dotted lines in (A) represent genomic alterations detected in primary gastrointestinal NETs and associated metastases, respectively. Each line represents one alteration.
Table 2.
Genomic Aberrations of Neuroendocrine Tumors
| Locus of alterations | Bronchial NETs (n = 11) | Gastrointestinal NETs (n = 21) | P value* |
|---|---|---|---|
| −3q | 0 | 2 (10%) | NS |
| −9p | 0 | 1 (5%) | NS |
| −11p | 2 (18%) | 0 | 0.0436 |
| −11q | 4 (36%) | 1 (5%) | 0.0194 |
| −13q | 0 | 1 (5%) | NS |
| −16q | 0 | 2 (10%) | NS |
| −18p | 0 | 7 (33%) | 0.0303 |
| −18q | 0 | 8 (38%) | 0.0181 |
| +5p | 2 (18%) | 0 | NS |
| +5q | 2 (18%) | 0 | NS |
| +7p | 1 (9%) | 1 (5%) | NS |
| +7q | 1 (9%) | 1 (5%) | NS |
| +9q | 2 (18%) | 3 (14%) | NS |
| +10q | 0 | 2 (10%) | NS |
| +16q | 2 (18%) | 0 | 0.0436 |
| +20q | 1 (9%) | 1 (5%) | NS |
*Contingency table analysis. NS, not significant.
Losses of Chromosomes 18 and 11q Confirmed by FISH
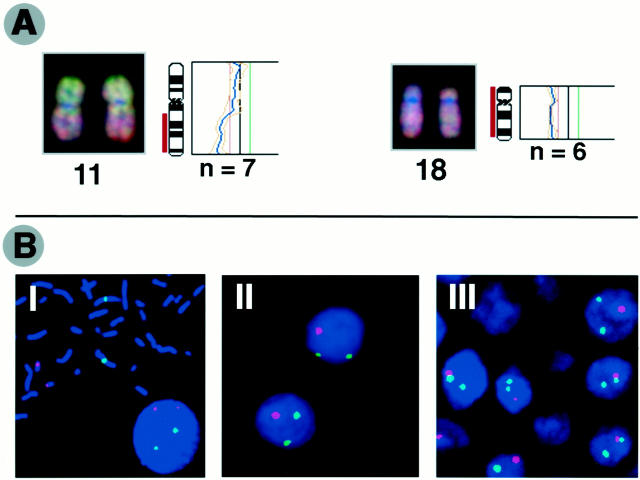
FISH analyses confirmed losses of chromosome 18 identified by CGH in seven of eight gastrointestinal NETs. In the other two NETs, which exhibited no CGH detectable alterations of chromosome 18, FISH also showed no alterations of this chromosome. CGH results of bronchial NETs regarding 11q losses were confirmed using the FISH method, too. FISH showed deletions of 11q13.4-q21 in all four tumors with CGH-detected 11q losses, and deletions of 11q13 only in two of the four tumors (Table 1) ▶ . In the seven remaining bronchial NETs, FISH demonstrated no alterations of 11q, confirming the CGH data. Taken together, both techniques provide highly comparable results of chromosome 18 losses in nine of 10 gastrointestinal NETs. Representative examples of CGH images and corresponding profiles as well as interphase cytogenetics are illustrated in Figure 2, A and B ▶ ).
Figure 2.
A: Representative examples of CGH digital images and corresponding profiles illustrating losses of chromosome11q and 18. Tumor DNA was labeled using green dUTP and normal reference DNA with red dUTP. The color ratio values 0.8, 1.2, and 1.5 were used as thresholds for chromosomal losses, gains, and amplifications, respectively. B: Representative pictures of FISH analysis. I: Signals of centromere probes specific for chromosomes 1 (green) and 18 (red) on metaphase from normal human lymphocytes. II: Deletions of chromosome 18 detected on paraffin-embedded section. III: Losses of chromosome 18 on interphase touch preparation. For the FISH analysis on paraffin-embedded sections, the centromere probe of chromosome 18 was labeled using digoxigenin-11-dUTP and then detected using sheep anti-digoxigenin-rhodamine (red). The centromere probe specific for chromosome 1 was directly labeled using spectrum-green dUTP. In the interphase analysis on touch preparations, centromere probes specific for chromosomes 1 and 18 were labeled using spectrum-green dUTP and spectrum-red dUTP, respectively.
Discussion
CGH allows a rapid detection of DNA sequence copy number changes anywhere in the entire genome, providing an overview of genomic imbalances in a given tumor. The present CGH results show different patterns of genomic alterations in gastrointestinal and bronchial NETs, thereby pinpointing different genetic events responsible for the initiation and development of the two subgroups of NETs.
The CGH results of the present study on gastrointestinal NETs demonstrate frequent losses of chromosome 18, which represent characteristic genomic imbalances involved in this type of tumors. Previous molecular studies have shown that loss of heterozygosity on chromosome 18q is also a frequent event in gastrointestinal adenocarcinomas, 26-28 suggesting an important role of 18q in the tumorigenesis and progression of these tumors. Several tumor suppressor genes are known to be located on chromosome 18, including DCC (deleted in colorectal carcinoma), DPC4 (deleted in pancreas carcinomas at locus 4), and Smad2. These genes have been implicated to be associated with tumorigenesis through inactivation in a variety of human tumors such as colorectal cancer. 29-31 Our present CGH data indicate that all three of the genes may be involved in gastrointestinal NETs. Additional studies are under way to further clarify the role of these tumor suppressor genes in NET tumorigenesis and progression of the gastrointestinal tract. Recently, Terris et al 15 detected extensive genomic imbalances in eight gastrointestinal NETs. Characteristic genomic changes, however, were difficult to recognize from the small series of tumors. The tumors they examined were probably high-grade neuroendocrine neoplasms rather than typical carcinoids as examined in this study, which would also explain for the discrepancies of their results from our data.
Our CGH results on bronchial NETs show that these tumors are characterized by prevalent 11q deletions, a finding that is in accordance with previous studies. 13,14 The region of 11q13 has been known to harbor the tumor suppressor gene MEN1 that was recently cloned. 32 Loss of heterozygosity studies have demonstrated frequent allelic losses at or around the 11q13 locus involved in NETs of the lung associated or not with the multiple endocrine neoplasia type 1 (MEN1) syndrome. 10,33,34 Somatic mutations of the gene have also been found in sporadic lung carcinoids, albeit only in a subset of tumors. 35 In our FISH analyses, deletions were revealed at the region of cCl11-270 (11q13.4-qter) but not at the MEN1 locus in two tumors (tumors 8 and 9) which showed losses of 11q13-qter in the CGH examination. This finding suggests that, in addition to MEN1, one or more other possible tumor suppressor genes located distal from the MEN1 gene on 11q may be involved in the pathogenesis of bronchial NETs. Previous molecular studies have shown the involvement of the genes Rb and p53 in lung NETs. 11,12 In contrast, the present study did not demonstrate losses of 13q and 17p in the bronchial NETs, at which the Rb gene and p53 are located, respectively. However, our data could not rule out a possible role of the both genes in the bronchial NETs because small chromosomal alterations (<10 Mb) may be not detected by CGH.
CGH and FISH are two complementary powerful tools for detection and mapping of genetic alterations in tumors. Both methods used in the present study provide well-consistent results regarding losses of chromosomes 18 and 11q, which were most frequently observed in gastrointestinal and bronchial NETs, respectively. Attempting to evaluate genetic heterogeneity of a tumor, we investigated two or three different regions in three neoplasms. One tumor (tumor 14) exhibited a clearly intratumoral genetic heterogeneity, which may implicate the presence of two or more neoplastic cell populations in these tumors. In one tumor (tumor 12), losses of chromosome 18 were revealed by CGH, but not by FISH. These conflicting results are most likely also because of intratumoral genetic heterogeneity.
The number of genomic alterations detected by CGH could be used as a predictor of tumor progression or recurrence, as suggested by studies on a variety of tumor types. 23,36 In the present study, genomic alterations were observed in 75% of the advanced gastrointestinal NETs and in all of the three investigated metastases, thus supporting the predictive value of the number of genomic changes. Surprisingly, however, genomic alterations were only seen in two of seven mixed exocrine-endocrine tumors (goblet-cell carcinoids) examined, including one metastasis in the stomach. The average number of genomic changes found in these tumors was 0.5, which was much lower than that in the remaining gastrointestinal NETs (mean, 2.9). This finding would explain why goblet-cell carcinoids usually exhibit an indolent clinical course. Another explanation would be that the sensitivity of CGH in these tumors was compromised by the presence of high amounts of normal cells. Remarkably, however, losses of chromosome 18 were also found in one of the two goblet-cell carcinoids that exhibited genomic alterations. This implies that the tumorigenesis of these neoplasms may be driven by the same genetic events involved in the remaining gastrointestinal NETs.
In conclusion, our data indicate that NETs of the gastrointestinal tract and the lung develop via different molecular pathways. Inactivation of one or several tumor suppressor genes on chromosome 18 may be important for the biological behavior of gastrointestinal NET, whereas gene inactivation on 11q seems to be associated with bronchial NET development.
Acknowledgments
We thank Drs. Guido Sauter and Holger Moch, University of Basel, Switzerland for continuous support and helpful discussions, and Norbert Wey and Ida Schmieder for photographic and computer-assisted reproductions.
Footnotes
Address reprint requests to Dr. Paul Komminoth, Department of Pathology, University of Zurich, Schmelzbergstrasse 12, CH-8091 Zurich, Switzerland. E-mail: paul.komminoth@pty.usz.ch.
Supported by the Swiss National Science Foundation and the Swiss Cancer League.
References
- 1.Capella C, Heitz PU, Höfler H, Solcia E, Klöppel G: Revised classification of neuroendocrine tumors of the lung, pancreas and gut. Digestion 1994, 55:11-23 [DOI] [PubMed] [Google Scholar]
- 2.Godwin JD: Carcinoid tumors: an analysis of 2,837 cases. Cancer 1975, 36:560-569 [DOI] [PubMed] [Google Scholar]
- 3.Dusmet M, McKneally MF: Bronchial and thymic carcinoid tumors: a review. Digestion 1994, 55:70-76 [DOI] [PubMed] [Google Scholar]
- 4.Burke AP, Thomas RM, Elsayed AM, Sobin LH: Carcinoids of the jejunum and ileum: an immunohistochemical and clinicopathologic study of 167 cases. Cancer 1997, 79:1086-1093 [PubMed] [Google Scholar]
- 5.Federspiel BH, Burke AP, Sobin LH, Shekitka KM: Rectal and colonic carcinoids: a clinicopathologic study of 84 cases. Cancer 1990, 65:135-140 [DOI] [PubMed] [Google Scholar]
- 6.Burke AP, Sobin LH, Federspiel BH, Shekitka KM, Helwig EB: Carcinoid tumors of the duodenum: a clinicopathologic study of 99 cases. Arch Pathol Lab Med 1990, 114:700-704 [PubMed] [Google Scholar]
- 7.Memon MA, Nelson H: Gastrointestinal carcinoid tumors: current management strategies. Dis Colon Rectum 1997, 40:1101-1118 [DOI] [PubMed] [Google Scholar]
- 8.Bak M, Asschenfeldt P: Adenocarcinoid of the vermiform appendix: a clinicopathologic study of 20 cases. Dis Colon Rectum 1988, 31:605-612 [DOI] [PubMed] [Google Scholar]
- 9.Warkel RL, Cooper PH, Helwig EB: Adenocarcinoid, a mucin-producing carcinoid tumor of the appendix: a study of 39 cases. Cancer 1978, 42:2781-2793 [DOI] [PubMed] [Google Scholar]
- 10.Jakobovitz O, Nass D, DeMarco L, Barbosa AJ, Simoni FB, Rechavi G, Friedman E: Carcinoid tumors frequently display genetic abnormalities involving chromosome 11. J Clin Endocrinol Metab 1996, 81:3164-3167 [DOI] [PubMed] [Google Scholar]
- 11.Gouyer V, Gazzeri S, Brambilla E, Bolon I, Moro D, Perron P, Benabid AL, Brambilla C: Loss of heterozygosity at the RB locus correlates with loss of RB protein in primary malignant neuroendocrine lung carcinomas. Int J Cancer 1994, 58:818-824 [DOI] [PubMed] [Google Scholar]
- 12.Lohmann DR, Fesseler B, Putz B, Reich U, Bohm J, Prauer H, Wunsch PH, Hofler H: Infrequent mutations of the p53 gene in pulmonary carcinoid tumors. Cancer Res 1993, 53:5797-5801 [PubMed] [Google Scholar]
- 13.Walch AK, Zitzelsberger HF, Aubele MM, Mattis AE, Bauchinger M, Candidus S, Prauer HW, Werner M, Hofler H: Typical and atypical carcinoid tumors of the lung are characterized by 11q deletions as detected by comparative genomic hybridization. Am J Pathol 1998, 153:1089-1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ullmann R, Schwendel A, Klemen H, Wolf G, Petersen I, Popper HH: Unbalanced chromosomal aberrations in neuroendocrine lung tumors as detected by comparative genomic hybridization. Hum Pathol 1998, 29:1145-1149 [DOI] [PubMed] [Google Scholar]
- 15.Terris B, Meddeb M, Marchio A, Danglot G, Flejou JF, Belghiti J, Ruszniewski P, Bernheim A: Comparative genomic hybridization analysis of sporadic neuroendocrine tumors of the digestive system. Genes Chromosom Cancer 1998, 22:50-56 [DOI] [PubMed] [Google Scholar]
- 16.Vortmeyer AO, Lubensky IA, Merino MJ, Wang CY, Pham T, Furth EE, Zhuang Z: Concordance of genetic alterations in poorly differentiated colorectal neuroendocrine carcinomas and associated adenocarcinomas. J Natl Cancer Inst 1997, 89:1448-1453 [DOI] [PubMed] [Google Scholar]
- 17.Ramnani DM, Wistuba II, Behrens C, Gazdar AF, Sobin LH, Albores S-J: K-ras and p53 mutations in the pathogenesis of classical and goblet cell carcinoids of the appendix. Cancer 1999, 86:14-21 [DOI] [PubMed] [Google Scholar]
- 18.Wang DG, Johnston CF, Buchanan KD: Oncogene expression in gastroenteropancreatic neuroendocrine tumors: implications for pathogenesis. Cancer 1997, 80:668-675 [PubMed] [Google Scholar]
- 19.Klöppel G, Heitz PU, Capella C, Solcia E: Pathology and nomenclature of human gastrointestinal neuroendocrine (carcinoid) tumors and related lesions. World J Surg 1996, 20:132-141 [DOI] [PubMed] [Google Scholar]
- 20.Travis WD, Colby TV, Corrin B, Shimosato Y, Brambilla E (Eds): Histological typing of lung and pleural tumors. World Health Organization International Histological Classification of Tumors, ed 3. New York, Springer, 1999
- 21.Solcia E, Klöppel G, Sobin LH (Eds): Histological typing of endocrine tumors. World Health Organization. International Histological Classification of Tumors, ed 2. New York, Springer, 2000
- 22.Zhao J, Richter J, Wagner U, Ackermann D, Schmid U, Roth B, Moch H, Mihatsch MJ, Gasser TC, Sauter G: Chromosomal imbalances in non-invasive papillary bladder neoplasms (pTa). Cancer Res 1999, 59:4658-4661 [PubMed] [Google Scholar]
- 23.Zhao J, Speel EJM, Muletta-Feurer S, Rütimann K, Saremaslani P, Roth J, Heith PU, Komminoth P: Analysis of genomic alterations in sporadic adrenocortical lesions: gain of chromosome 17 is an early event in adrenocortical tumorigenesis. Am J Pathol 1999, 155:1039-1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hopman AHN, Ramaekers FCS: Processing and staining of cell and tissue material for interphase cytogenetics. Curr Protocols Cytometry 1998, 8:5.1-22 [DOI] [PubMed] [Google Scholar]
- 25.Görtz B, Roth J, Speel EJ, Krahenmann A, de Krijger RR, Matias-Guiu X, Muletta-Feurer S, Rütimann K, Saremaslani P, Heitz PU, Komminoth P: MEN1 gene mutation analysis of sporadic adrenocortical lesions. Int J Cancer 1999, 80:373-379 [DOI] [PubMed] [Google Scholar]
- 26.Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM, Bos JL: Genetic alterations during colorectal-tumor development. N Engl J Med 1988, 319:525-532 [DOI] [PubMed] [Google Scholar]
- 27.Vogelstein B, Fearon ER, Kern SE, Hamilton SR, Preisinger AC, Nakamura Y, White R: Allelotype of colorectal carcinomas. Science 1989, 244:207-211 [DOI] [PubMed] [Google Scholar]
- 28.Jen J, Kim H, Piantadosi S, Liu ZF, Levitt RC, Sistonen P, Kinzler KW, Vogelstein B, Hamilton SR: Allelic loss of chromosome 18q and prognosis in colorectal cancer. N Engl J Med 1994, 331:213-221 [DOI] [PubMed] [Google Scholar]
- 29.Fearon ER, Cho KR, Nigro JM, Kern SE, Simons JW, Ruppert JM, Hamilton SR, Preisinger AC, Thomas G, Kinzler KW, Vogelstein B: Identification of a chromosome 18q gene that is altered in colorectal cancers. Science 1990, 247:49-56 [DOI] [PubMed] [Google Scholar]
- 30.MacGrogan D, Pegram M, Slamon D, Bookstein R: Comparative mutational analysis of DPC4 (Smad4) in prostatic and colorectal carcinomas. Oncogene 1997, 15:1111-1114 [DOI] [PubMed] [Google Scholar]
- 31.Eppert K, Scherer SW, Ozcelik H, Pirone R, Hoodless P, Kim H, Tsui LC, Bapat B, Gallinger S, Andrulis IL, Thomsen GH, Wrana JL, Attisano L: MADR2 maps to 18q21 and encodes a TGFbeta-regulated MAD-related protein that is functionally mutated in colorectal carcinoma. Cell 1996, 86:543-552 [DOI] [PubMed] [Google Scholar]
- 32.Chandrasekharappa SC, Guru SC, Manickam P, Olufemi SE, Collins FS, Emmert B-MR, Debelenko LV, Zhuang Z, Lubensky IA, Liotta LA, Crabtree JS, Wang Y, Roe BA, Weisemann J, Boguski MS, Agarwal SK, Kester MB, Kim YS, Heppner C, Dong Q, Spiegel AM, Burns AL, Marx SJ: Positional cloning of the gene for multiple endocrine neoplasia-type 1. Science 1997, 276:404-407 [DOI] [PubMed] [Google Scholar]
- 33.Dong Q, Debelenko LV, Chandrasekharappa SC, Emmert B-MR, Zhuang Z, Guru SC, Manickam P, Skarulis M, Lubensky IA, Liotta LA, Collins FS, Marx SJ, Spiegel AM: Loss of heterozygosity at 11q13: analysis of pituitary tumors, lung carcinoids, lipomas, and other uncommon tumors in subjects with familial multiple endocrine neoplasia type 1. J Clin Endocrinol Metab 1997, 82:1416-1420 [DOI] [PubMed] [Google Scholar]
- 34.D’Adda T, Keller G, Bordi C, Hofler H: Loss of heterozygosity in 11q13–14 regions in gastric neuroendocrine tumors not associated with multiple endocrine neoplasia type 1 syndrome. Lab Invest 1999, 79:671-677 [PubMed] [Google Scholar]
- 35.Görtz B, Roth J, Krahenmann A, de Krijger RR, Muletta-Feurer S, Rütimann K, Saremaslani P, Speel EJ, Heitz PU, Komminoth P: Mutations and allelic deletions of the MEN1 gene are associated with a subset of sporadic endocrine pancreatic and neuroendocrine tumors and not restricted to foregut neoplasms. Am J Pathol 1999, 154:429-436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sallinen SL, Sallinen P, Haapasalo H, Kononen J, Karhu R, Helen P, Isola J: Accumulation of genetic changes is associated with poor prognosis in grade II astrocytomas. Am J Pathol 1997, 151:1799-1807 [PMC free article] [PubMed] [Google Scholar]