Abstract
Chondrosarcomas are malignant cartilaginous tumors arising centrally in bone (central chondrosarcoma), or secondarily within the cartilaginous cap of osteochondroma (peripheral chondrosarcoma). We previously used DNA flow cytometry to demonstrate that near-haploidy is relatively frequent in peripheral chondrosarcomas. We performed fluorescence in situ hybridization (FISH) to interphase nuclei using centromeric probes, a genome wide loss of heterozygosity (LOH) analysis, and comparative genomic hybridization on five peripheral chondrosarcomas. We demonstrated near-haploidy in two low-grade tumors with only one copy and LOH of most chromosomes. Few chromosomes are disomic, with retention of heterozygosity and overrepresentation at comparative genomic hybridization. One tumor contains both a near-haploid clone with chromosomes in monosomic and disomic state, and an exactly duplicated clone. Two high-grade tumors clearly demonstrate polyploidization because most chromosomes show LOH and two copies at FISH, whereas few chromosomes have four copies with retention of heterozygosity. Using DNA from a relative, we demonstrate that chromosome loss is random regardless of parental origin. Using FISH on paraffin slides, we exclude near-haploidy to result from meiosis-like division in binucleated cells, characteristic for chondrosarcoma. In conclusion, our results indicate that near-haploidy characterizes the progression from osteochondroma toward low-grade chondrosarcoma. Moreover, further progression toward high-grade chondrosarcoma is characterized by polyploidization.
Chondrosarcoma, the second most common primary malignant bone tumor after osteosarcoma, is a malignant cartilage-forming tumor occurring predominantly in adults. The majority of chondrosarcomas develops de novo and is located centrally within the medullary cavity (central chondrosarcoma). In contrast, ∼15% arise within the cartilage cap of a long-standing osteochondroma (peripheral chondrosarcoma), mostly in patients suffering from the hereditary multiple exostoses syndrome. Hereditary multiple exostoses syndrome is a familial disorder with an autosomal dominant mode of inheritance. 1-4 Malignant transformation is estimated to occur in 1 to 5% of osteochondromas.
The development of peripheral chondrosarcoma results in genetic instability characterized by a high percentage of loss of heterozygosity (LOH) and a wide variation in DNA ploidy. 5 LOH in peripheral chondrosarcoma most frequently involves the TP53 (80%), RB1 (71%), and EXT1 (65%) loci. 5 In contrast, peridiploidy and a low percentage of LOH in central tumors indicates that a different oncogenic molecular mechanism may be operative. 5
We have detected hypodiploidy, which is uncommon for human solid tumors, in 29% of peripheral chondrosarcomas and not in central chondrosarcomas. 5 Others mention hypodiploidy in 8 to 31% of chondrosarcomas without further subclassification. 6-9 Near-haploidy, which is a very rare phenomenon in human solid tumors in general, is found especially in low-grade peripheral chondrosarcomas (DNA index, 0.56 and 0.70). 5
Most of the near-haploid solid tumors documented cytogenetically (carcinomas as well as sarcomas) show polyploidization of the near-haploid stem line. 10-12 In our original series we could demonstrate a hypodiploid fraction (DNA index, 0.76) as well as a polyploid fraction (DNA index, 1.56) in one tumor. 5 This led us to postulate that high-grade peripheral chondrosarcomas with high percentages of LOH may have evolved from polyploidization of near-haploid clones.
Our goal was to further investigate near-haploidy in low-grade peripheral chondrosarcoma and to confirm polyploidization in high-grade peripheral chondrosarcoma. We present an estimate of chromosome copy numbers studied by fluorescence in situ hybridization (FISH) to interphase nuclei, relative copy numbers studied by comparative genomic hybridization (CGH) and genome-wide LOH studied by microsatellite analysis for five peripheral chondrosarcomas.
Materials and Methods
Clinicopathological Data
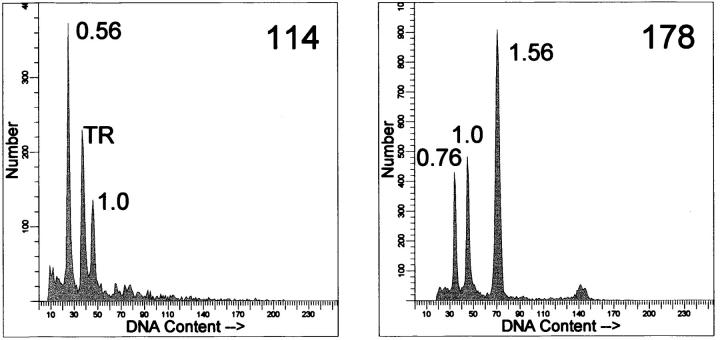
We previously documented 19 peripheral and 12 central chondrosarcomas, for which fresh-frozen tumor tissue and normal DNA were available, that were studied by LOH analysis and DNA flow cytometry. 5 This study rendered five peripheral tumors with either near-haploidy or an unusually high LOH percentage that were selected for the present study. Clinicopathological data are shown in Table 1 ▶ . For four cases primary tumors were used. Tumor DNA of case 215 is derived from the first local recurrence, because no suitable fresh-frozen material was available from the primary tumor. DNA indices were defined by DNA flow cytometry as described previously. 5 Examples of DNA histograms are shown in Figure 1 ▶ . Histological grading was performed according to Evans et al. 13
Table 1.
Clinicopathological Data of the Five Patients
| Case | DNA index* | Grade | Age† | Sex | Location | Follow-up (months) | HME¶ |
|---|---|---|---|---|---|---|---|
| 114 | 0.56 | I | 42 | M | Right proximal femur | 51 NED‡ | Yes |
| 408 | 0.70 | II | 38 | M | Costa | 17 NED and 29 alive | No |
| 178 | 0.76+ 1.56 | II | 50 | F | Costa | 121 NED | Yes |
| 262 | 1.22 | III | 61 | F | Right scapula | 51, had 3 recur§ at 27, 40, and 47 months, and now lung metastases | yes |
| 215 | 1.32 | III | 40 | F | Left scapula | 47 NED, had 4 recur§ at 6, 15, 21, and 28 months | No |
*DNA index measured by DNA flow cytometry as described previously.5
†Age: age at diagnosis.
‡NED: No evidence of disease.
§Recur: local recurrence. All local recurrences, as well as the metastases were surgically treated.
¶HME: hereditary multiple exostoses.
Figure 1.
Examples of flow cytometric DNA histograms from nuclear suspensions. 5 DNA indices are indicated. Left: case 114, demonstrating near-haploidy. TR, trout red blood cells serving as internal standard. Right: case 178 is shown, demonstrating two clones, a hypodiploid clone of 0.76 and a polyploidized clone of 1.56. The trout erythrocytes peak overlaps with the hypodiploid peak and is therefore not shown.
FISH
Isolation of interphase nuclei from fresh-frozen tumor tissue was performed as described. 14 Tumor percentages were estimated on subsequent hematoxylin and eosin-stained slides. The chromosome-specific centromeric repeats used as probes are listed in Table 2 ▶ . They were labeled by standard nick-translation (Boehringer Mannheim, Almere, The Netherlands) with digoxigenin-11-dUTP or biotin-16-dUTP (Roche, Basel, Switzerland). Hybridization was performed as described. 14 Hybridization of biotin-labeled probes was detected in a three-step reaction with streptavidin-Texas Red, biotin-labeled goat anti-streptavidin and a second streptavidin-Texas Red (Vector Laboratories, Burlingame, CA). The hybridization of digoxigenin-labeled probes was detected in a three-step reaction with mouse anti-dioxin, fluorescein isothiocyanate-labeled rabbit anti-mouse (Sigma-Aldrich Chemie, Zwijndrecht, The Netherlands) and fluorescein isothiocyanate-labeled goat anti-rabbit (Vector Laboratories). Slides were counterstained with 4,6-diamidino-2-phenylindole. Slides with isolated interphase nuclei from placental tissue served as positive controls and were used to determine hybridization efficiency. For each hybridization 200 nuclei were scored. Deformed nuclei were excluded. To study chromosome division in binucleated cells in chondrosarcoma and to investigate whether meiosis or mitosis precedes the formation of binucleated cells, for a male patient (case 408), FISH using a chromosome X centromere probe was performed on paraffin sections as described. 15
Table 2.
Estimated Chromosome Copy Number Based on Combined Information of Number of Centromeric Signals Found Using FISH at Interphase Nuclei and Tumor Percentage (Shown in Italics)
| Chromosome | Probe | Repetitive family | Reference no. | Case | ||||
|---|---|---|---|---|---|---|---|---|
| 114 | 408 | 178 | 262 | 215 | ||||
| tum%: | 95% | <50% | 80% | 75% | 70% | |||
| 1 | pUC1.7 | Satellite III | 63 | 1 | 1* | 2 and 4 | 2 | 2 |
| 3 | pα3.5 | Alphoid satellite | 64 | NS | NS | NS | 2 | 2 |
| 6 | p308 | Alphoid satellite | 65 | 1 | 1* | 1 and 2 | 2 | 2 |
| 7 | p7t1(αp7) | Alphoid satellite | 66 | 1 | 2 | 2 and 4 | 2 | 3* |
| 8 | D8Z2 (αp8) | Alphoid satellite | 67 | 1 | 1* | 2 and 4 | 2 | 3* |
| 9 | pHUR98 (sp9) | Satellite III | 68 | 1 | 2 | 2 and 4 | 2 | 2 |
| 10 | D10Z1 (αp10) | Alphoid satellite | – | 1 | 1* | 1 and 2 | 2 | 2 |
| 11 | pLC11A | Alphoid satellite | 69 | 1 | 1* | 1 and 2 | 2 | 2 |
| 12 | pα12H8 (αp12) | Alphoid satellite | 70 | 2 | 2 | 2 and 4 | 4 | 2 |
| 15 | D15Z1 | Satellite III | 71 | 2 | 2 | 2 and 4 | 2 | 4* |
| 16 | pHUR195 | Satellite II | 68 | 1 | 1* | 2 and 4 | 2 | 2 |
| 17 | p17H8 | Alphoid satellite | 72 | 1 | 1* | 2 and 4 | 2 | 3* |
| 18 | L1.84 (#18) | Alphoid satellite | 73 | 1 | 2 | 2 and 4 | 2 | 2 |
| 20 | p3.4 | Alphoid satellite | – | 1 | 2 | NS | 4 | NS |
| X | pBAMX5 (αpX) | Alphoid satellite | 74 | 1 | 1 | 1 and 2 | 4 | 1* |
| Y | DYZ3 | Alphoid satellite | 75 | 1 | 0 | 0 | 0 | 0 |
*Many nuclei show two copies of the chromosomes, because of a high percentage of nontumor nuclei. NS: nonsuccessful hybridization.
Microsatellite Analysis
Normal and tumor DNA were isolated as described. 5 A single cell suspension from case 408 was used to sort the aneuploid fraction on a fluorescence-activated cell sorter (FACS Vantage cell sorter; Becton Dickinson, Mountain View, CA) to enrich for the tumor cell fraction. DNA was isolated as described. 5,16 Thirty-nine markers selected from the Marshfield screening set, v6 (http://www.marshmed.org/genetics), distributed over the chromosomes were used to investigate heterozygosity (Table 3) ▶ . Polymorphic markers are described in the Genome Database (http://gdbwww.gdb.org). PCR and gel analyses on an automated ABI377 DNA sequencer were performed as described. 17 LOH was scored when the quotient of the ratios of both alleles of normal and tumor was larger than or equal to 1.7. 18 Ratios between 1.3 and 1.7 were regarded inconclusive. 19 To investigate whether chromosome loss was randomly distributed or paternally or maternally derived, DNA isolated from the father of patient 178, obtained after informed consent, was included.
Table 3.
Thirty-nine Microsatellite Markers Used for the LOH Analysis
| Marker | Cytogenetic location | Marker | Cytogenetic location |
|---|---|---|---|
| D01S1595 | 1p21.2 | D09S922 | 9q21.32 |
| D01S518 | 1p31.1–1p31.3 | D09S938 | 9q22.31–9q31.1 |
| D01S1612 | 1q36.31 | D10S1435 | 10p15.3 |
| D01S1663 | 1q32.1–1q32.2 | D11S1999 | 11p15.3 |
| D01S551 | 1q31.1 | D12S1052 | 12q14.3–12q15 |
| D01S547 | 1q43 | D13S787 | 13p11.2–13q11 |
| D02S1363 | 2q37.1–2q37.2 | D13S285 | 13q34 |
| D02S441 | 2q13.3 | D14S749 | 14q31.3–14q32.11 |
| D03S2460 | 3q13.31 | D15S816 | 15q26.1 |
| D04S2368 | 4q32.1 | D16S2622 | 16p13.3–16q24.3 |
| D05S2494 | 5p13.3 | D17S1303 | 17p12 |
| D05S2501 | 5q21.1–5q21.2 | D17S784 | 17p13.3–17q25.3 |
| D06S1053 | 6p12.1–6q11.2 | D17S1843 | 17q12–17q21 |
| D06S1277 | 6q26 | D18S851 | 18q21.1–18q22.1 |
| D07S1824 | 7q34 | D19S714 | 19p13.3–19q13.43 |
| D07S550 | 7q36.3 | D20S477 | 20p13–20q13.33 |
| D07S2846 | 7q32–7qter | D21S1446 | 21p13–21q22.3 |
| D07S820 | 7q21.11–7q21.2 | D22S689 | 22q12.1 |
| D07S1804 | 7q31–7q32 | DXS6789 | Xq22.1 |
| D08S136 | 8p22–8p11.1 |
Microsatellite markers were selected from the Marshfield Screening Set, v6 (http://www.marshmed.org/genetics). Cytogenetic locations were retrieved from the genome database (http://gdbwww.gdb.org).
CGH
Tumor DNA was isolated as described 5 and tumor percentage was estimated on subsequent hematoxylin and eosin-stained slides. The CGH procedure was based on the protocol described by Kallioniemi et al, 20 with few modifications as described previously. 21,22 Briefly, test DNA was directly labeled with fluorescein isothiocyanate-dUTP and reference DNA was labeled with lissamine-dUTP (Dupont-New England Nuclear, Boston, MA), both by nick translation. Nick-translated fragment sizes ranged from 400 to 2,000 bp. Two hundred ng of each labeled DNA and 10 μg of Cot-1 DNA were hybridized to normal metaphases and incubated at 37°C for 4 days. Posthybridization washes were performed with 2× standard saline citrate at 37°C, followed by 0.1× standard saline citrate at 60°C. Slides were counterstained with 4,6-diamidino-2-phenylindole in an antifade solution. Digital images were analyzed using QUIPS XL software from Vysis (Downers Grove, IL). Underrepresentation of DNA sequences was defined as chromosomal regions where the average green-to-red ratio and its 95% confidence interval were <0.9 whereas overrepresentation was defined as >1.1. These threshold values were based on measurements from a series of normal controls
Results
FISH
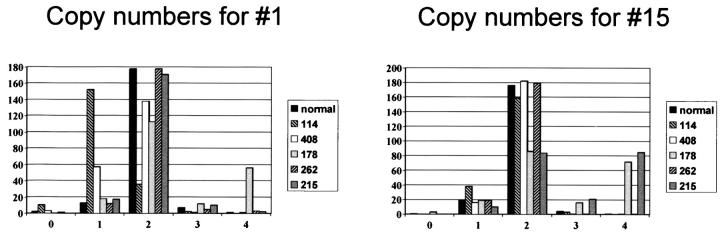
The estimated chromosome copy numbers based on combined information of number of centromeric signals found using FISH at interphase nuclei and the tumor percentage are presented for each case in Table 2 ▶ . As an example of the diversity found, the distribution of numbers of centromeric signals over 200 nuclei in the different tumor samples are shown for chromosomes 1 and 15 (Figure 2) ▶ . Near-haploidy is confirmed in case 114 and 408 because most of the chromosomes demonstrate monosomy. The estimation for case 408 is difficult because of the low tumor percentage estimated at maximal 50% using a hematoxylin and eosin cryostat section. This value is probably overestimated considering the distribution of copy numbers over 200 nuclei counted (Figure 2) ▶ . Contamination with nonneoplastic cells in chondrosarcoma is easily underestimated because of the often poor quality of cryostat sections of bony material and the low cellularity of cartilaginous tissue compared to highly cellular contaminating bone marrow. 5 Chromosomes 1, 6, 8, 10, 11, 16, and 17 are estimated at one copy per tumor cell, which is seen in 48 to 71 of 200 nuclei counted. This is elevated as compared to the placenta control (range, 7 to 15 of 200 nuclei with one spot) and to those chromosomes (7, 9, 12, 15, 18, and 20) that are clearly present in duplicate (range, 5 to 23 of 200 nuclei with one spot).
Figure 2.
The distribution of number of signals for centromeres of chromosome 1 and 15 over 200 counted nuclei in the different tumor samples compared to the normal placenta control. The tumor percentage of case 408 is estimated at maximal 50%. Chromosome 1 in case 408 is estimated at one copy per tumor cell, which is seen in only 57 of 200 nuclei counted. This is however elevated as compared to the placenta control.
Case 178 with two aneuploid clones (DNA index, 0.76 + 1.56) demonstrates either one and two copy numbers (chromosomes 6, 10, and 11), or two and four copy numbers (chromosomes 1, 7, 8, 9, 12, 15, 16, 17, and 18). To further demonstrate the presence of two populations in case 178 we performed hybridization simultaneously with a biotin-labeled probe for a chromosome present in two and four copies (chromosome 8 or 18) and a digoxigenin-labeled probe for a chromosome present in one and two copies (chromosome 10 or 11). Indeed we demonstrate that 52 of 100 nuclei contain two copies of chromosome 8 and one copy of chromosome 11, whereas 23 nuclei contain four copies of chromosome 8 and two copies of chromosome 11 (Figure 3) ▶ . The remaining nuclei are derived from normal cells, demonstrating two copies of each chromosome. Combining chromosome 11 and 18 probes gives similar results. Combining chromosome 10 and 11 demonstrate 54 of 100 nuclei with only one copy of each chromosome whereas 29 nuclei demonstrate two copies of both chromosomes.
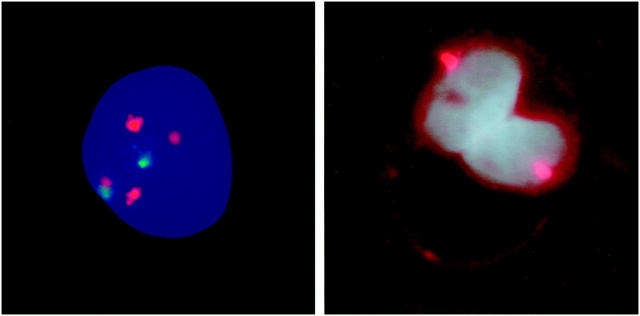
Figure 3.
Left: FISH micrograph showing an example of case 178. Hybridization was simultaneously performed with a probe for chromosome 8 (present in either two or four copies) and chromosome 11 (present in either one or two copies). Indeed we could confirm the presence of two populations. An example of the smallest population, containing four copies of chromosome 8 and two copies of chromosome 11, is shown. Right: FISH on paraffin slides using a sex chromosome (X) centromere probe on a male patient (case 408) is demonstrated. One copy of the X centromere is demonstrated in both nuclei of binucleated cells, indicating that a normal mitosis precedes the formation of these cells, which are highly characteristic for chondrosarcoma. A meiosis-like division in binucleated cells is therefore most probably not the cause of near-haploidy in chondrosarcoma.
We performed FISH on paraffin slides of a male patient (case 408) using a sex chromosome (X) centromere probe to investigate whether a meiosis or a mitosis precedes the formation of binucleated cells in chondrosarcoma. Both nuclei of binucleated cells are shown to contain one copy of chromosome X (Figure 3) ▶ .
Microsatellite Analysis
Results of the LOH analysis are shown in Table 4 ▶ , together with FISH and CGH results. Case 114 shows LOH for most chromosomes tested. Chromosomes 12 and 15, that present two signals at FISH at interphase nuclei, demonstrate retention of heterozygosity. LOH is found at 7q36.3 and 7q21 whereas retention of heterozygosity is seen at 7q34. Only one copy of chromosome 7 is found at interphase FISH, indicating that in case 114 part of the second copy of chromosome 7, containing band 7q34, is translocated to another chromosome.
Table 4.
Results of FISH, LOH, and CGH Summarized for Each Case
| Case | FISH | LOH | CGH |
|---|---|---|---|
| 114 | 1 copy: 1, 6, 7, 8, 9, 10, 11, 16, 17, 18, 20, X, Y | LOH: 1, 3, 4, 5, 6, 7q21, 7q36, 8, 9, 11, 13p11-q11, 14, 17, 19, 22 | Over: 12, 15, 20, X |
| 2 copies:12, 15 | ROH: 7q34, 12, 13q34, 15, 21 | ||
| 408 | 0 copies: Y | LOH: 1, 3, 4, 6, 7q31-32, 8, 14, 16, 17 | |
| 1 copy: 1, 6, 8,10,11,16,17, X | ROH: 2, 7q32-qter, 9, 12, 15, 18, 19, 20 | ||
| 2 copies: 7, 9, 12, 15, 18, 20 | |||
| 178 | 1 or 2 copies: 6, 10, 11 X | LOH: 1, 4, 6, 8, 9, 10, 11, 13p11-q11, 14, 17, 22, X | Over: 2, 3, 5, 7q, 8, 12, 18 |
| 2 or 4 copies: 1, 7, 8, 9, 12, 15, 16, 17, 18 | ROH: 2, 3, 5, 7, 13q34, 15, 18, 19 | Under: 9p, 10, 11, 14, 17p, 22, X | |
| 262 | 2 copies: 1, 3, 6, 7, 8, 9, 10, 11, 15, 16, 17, 18 | LOH: 1, 2, 3, 4, 6, 7, 8, 9, 11, 13, 14, 17p12, 17q12-21, 18 | Unsuccessful hybridization |
| 4 copies: 12, 20, X | ROH: 12, 17p13-q25, 19, 20, 22, X | ||
| 215 | 1 copy: X | LOH: 1, 3, 4, 5, 6, 9, 10, 11, 14, 16, 17, 18, X | Normal |
| 2 copies: 1, 3, 6, 9, 10, 11, 12, 16, 18 | ROH: 19, 20, 21 | ||
| 3 copies: 7, 8, 17 | Inconclusive: 7, 8, 13 | ||
| 4 copies: 15 |
Near-haploidy is confirmed in case 114 and 408, demonstrating only one copy and LOH of most chromosomes. Few chromosomes are disomic, with retention of heterozygosity and over-representation at CGH. Case 178 contains both a near-haploid clone with chromosomes in monosomic and disomic state, and an exactly duplicated clone. Cases 262 and 215 clearly demonstrate polyploidization since most chromosomes show both LOH as well as 2 copies at FISH, while few chromosomes have 4 copies combined with retention of heterozygosity. Abbreviations: LOH, loss of heterozygosity, ROH, retention of heterozygosity.
Case 178 has LOH for chromosomes 4, 6, 10, 11, and X which demonstrate either one or two copies at interphase FISH. Some of the chromosomes with either two or four copies (chromosomes 7, 15, and 18) at FISH at interphase nuclei, have retention of heterozygosity, whereas others (chromosomes 1, 8, 9, and 17) show LOH, probably because of additional mitotic recombination.
Both cases 215 and 262 reveal LOH for most of the chromosomes found to be present in two copies by FISH at interphase nuclei. In case 262 retention of heterozygosity is associated with four copies at FISH analysis (chromosomes 12, 20, and X). In case 215 the presence of three copies of chromosomes 7 and 8 is reflected in inconclusive allelic imbalance ratios (1.5 to 1.63). These chromosomes were therefore most probably not involved in haploidization and lost one of four copies after polyploidization.
Using the blood of the father of patient 178, we have determined that for markers D1S1612, D1S1663, D6S127, and D10S1435 the paternal allele is involved in LOH, whereas for D8S136, D13S787, D14S749, D17S1303, D22S689, and DXS6789 the maternal allele is involved. This refutes a bias for loss of either paternal or maternal chromosomes in this tumor.
CGH
For case 114, overrepresentation is found for chromosomes 12, 15, 20, and X. Of chromosomes 12 and 15 two copies are found at FISH at interphase nuclei, combined with retention of heterozygosity. Because most chromosomes in case 114 are lost, CGH identifies one copy as balanced, and the retained chromosomes are seen as overrepresented.
The CGH data of case 178 show results typical of intermediate ploidy. Because CGH normalization is based on the average ploidy of the cell, the average in this case does not represent either one or two copies, but a value in between. Chromosome regions which are present in one copy are seen as underrepresented, whereas the regions with two copies are seen as overrepresented. 23 For chromosomes that present a flat profile in all its length, a perfect correlation is seen with FISH results using centromeric probes, which showed one signal for underrepresented chromosomes (chromosomes 6, 10, 11, and X) and two for overrepresented (chromosomes 7, 8, 12, 15, and 18). Some chromosomes did not present a flat CGH profile, and have a different copy number of their long and short arms (chromosomes 1, 9, 16, and 17). In these cases, it is assumed that structural chromosome rearrangements occurred, and the number of centromeric signals might represent either the copy number of the long or the short arm.
CGH of case 215 reveals no aberrations. This is not surprising, because the 3:2 ratio (chromosomes 7, 8, and 17) present in maximum 50% of the cells is close to the limit of sensitivity of CGH. 24 CGH results are summarized in Table 4 ▶ .
Discussion
Near-haploidy is a very rare phenomenon in solid tumors. It is more frequently found in hematological disorders, especially in acute lymphoblastic leukemia 25 and blast phase of chronic myelocytic leukemia. 26 In the literature we have found only 29 different solid tumors documented cytogenetically to contain near-haploid stem lines with <38 chromosomes, including both epithelial as well as mesenchymal neoplasms. 10-12,27-46 Three of these 29 solid tumors are conventional chondrosarcomas with 29 to 37 chromosomes. 46 In the present study we clearly demonstrate the presence of near-haploidy and polyploidization in five additional peripheral conventional chondrosarcomas. Near-haploidy is absent in central chondrosarcomas. 5 Compared to other solid tumors, near-haploidy is therefore a relatively frequent phenomenon in peripheral chondrosarcoma.
The mechanism leading to near-haploidy in general is unknown. Speculations include a meiosis-like division, 10,36 abnormal mitoses such as multipolar mitosis in tri- or tetraploid, mostly multinucleated cells, 10,26,28,47,48 extreme nondisjunction, 28 sequential nonrandom loss and telomeric associations leading to unstable dicentric chromosomes that will be eliminated during subsequent cell divisions. 28
Near-haploidy may suggest a preceding meiosis-like division, especially in the binucleated cells that are highly characteristic for chondrosarcoma. However, by studying segregation of the sex chromosomes in binucleated cells in case 408, we show that a normal mitosis precedes the formation of binucleated cells in this tumor. Furthermore, by studying DNA isolated from a relative of patient 178 we demonstrate that the chromosome loss in case 178 is random regardless of parental origin. Finally, FISH at interphase nuclei shows retention of both sex chromosomes in case 114. Thus, a meiosis-like division is most probably not the cause of near-haploidy in chondrosarcoma.
Therefore, a selective process of chromosomal loss during several rounds of mitosis most probably results in tumor cells that have an advantage in the tumorigenic process in chondrosarcoma. Chondrosarcomas demonstrating near-haploid clones are predominantly low grade (grade I and grade II, 2x) in our series and grade I (2x) and grade III in the literature. 46 ) Follow-up revealed prolonged disease-free survival for our patients, which is consistent with the favorable prognosis suggested for solid tumors with very low chromosome numbers. 12,49 Low-grade chondrosarcomas are well differentiated, have a remarkably slow growth rate, and as in normal cartilage the supply of nutrients by blood vessels is limited. 50,51 Tumor cells must be able to survive this low state of energy and may do so by decreasing their DNA content, thereby decreasing nucleic acid and protein (histone and other nuclear protein) synthesis.
Sixteen (55%) out of the 29 near-haploid solid tumors documented cytogenetically show polyploidization of the near-haploid stem line. We could still identify the near-haploid fraction (0.76) as well as the polyploid fraction (1.56) in case 178, and FISH data confirm the presence of a clone with chromosomes either in monosomic or disomic state, and a clone in which all copy numbers are exactly doubled. LOH and FISH data are not completely concordant, probably because of mitotic recombination instead of true chromosome loss leading to LOH at chromosomes 1, 8, 9, and 17. These chromosomes are described to be involved in LOH in chondrosarcoma. 5
We postulated that cases 262 and 215 with unusually high percentages of LOH in a previous study had evolved from polyploidization of near-haploid clones. 5 Indeed, of most chromosomes two copies are found displaying LOH, whereas few chromosomes demonstrate four copies at interphase FISH combined with retention of heterozygosity. Remarkably, both tumors are high-grade chondrosarcomas with multiple local recurrences and development of lung metastases during follow-up. This would suggest that progression from low grade to high grade, which is observed to occur in chondrosarcoma, 13,52 is characterized by polyploidization (Figure 4) ▶ . The first alteration, loss of many chromosomes leading to near-haploidy, can still be detected, either as a distinct clone (such as seen in case 178), or only as a high LOH percentage after polyploidization and total overgrowth of the near-haploid clone.
Figure 4.
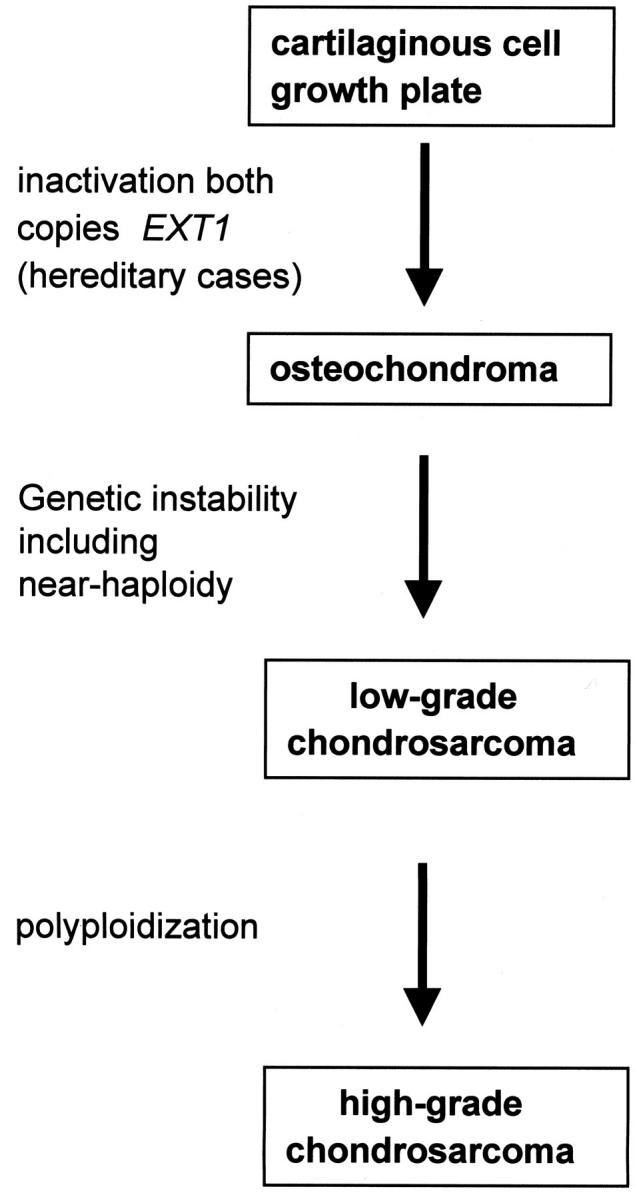
Multistep genetic model for peripheral cartilaginous tumorigenesis. First, inactivation of both copies of the EXT1 gene in cartilaginous cells of the growth plate is required for osteochondroma formation, as we previously showed by EXT1 germline mutations combined with loss of the remaining wild-type allele in hereditary osteochondromas. 56 If complete inactivation occurs in sporadic cases remains to be investigated. One or more additional genetic alterations, which may involve the EXT-genes, TP53 and RB1 as indicated by a high percentage of LOH at these loci, 5 are then required for peripheral chondrosarcomas to arise within its benign precursor. The process of malignant transformation is genetically represented by chromosomal instability as demonstrated by a high percentage of LOH involving various chromosomes and a broad range in DNA ploidy including near-haploidy. This may be caused by defective cell-cycle checkpoints. In osteochondromas, near-haploidy is absent 56 and can therefore be considered a progression marker toward a low-grade malignant phenotype. Further progression toward high-grade chondrosarcoma is characterized by polyploidization.
Loss of certain chromosomes (regions) may lead to inactivation of tumor suppressor genes, resulting in malignant transformation. Chromosomes 2 (81%), 3 (81%), 4 (81%), 6 (88%), 11 (80%), 13 (81%), 14 (77%), 16 (76%), and 17 (77%) were preferentially lost in the 29 near-haploid solid tumors documented in the literature. In the present study, chromosomes 6, 10, and 11 were monosomic in all five tumors. These data may suggest that important tumor suppressor genes are located at chromosomes 6 and 11. At chromosome 11p11-p12 the EXT2 gene, one of the genes involved in hereditary multiple exostoses, is located. 53 LOH is frequently demonstrated at 11p11-p12 in chondrosarcomas. 5,54,55 Although three patients had a hereditary multiple exostoses syndrome phenotype, no mutations were found in the EXT2 gene in these tumors as was documented in a previous study. 56 Chromosome 10 frequently demonstrates LOH in chondrosarcoma, especially at 10q11.2. 57
On the other hand the presence of two copies of certain chromosomes may be essential for growth of transformed cells. In the 29 near-haploid solid tumors retrieved from the literature, chromosome 7 is present in disomic state in 67% of near-haploid solid tumors. Moreover, of 27 hypodiploid solid tumors detected by DNA flow cytometry none demonstrated loss of chromosome 7. 58 In the present study, four cases demonstrate at least two copies of chromosome 7. In case 114 part of chromosome 7, including band 7q34, is translocated to another chromosome. In case 408, the 7q32-7qter region also shows retention of heterozygosity whereas 7q31-32 has LOH. Thus, at least two copies of the 7q32-7qter region is present in all five peripheral chondrosarcomas. This suggests that the presence of only one chromosomal copy of this region is deleterious for cells. One explanation may be that part of this region is either paternally or maternally inactivated. Remarkably a paternally imprinted gene PRG1/MEST was identified in this region. 59
In conclusion, we clearly demonstrate near-haploidy and polyploidization in a subset of peripheral chondrosarcoma. Near-haploidy is a relatively frequent phenomenon in peripheral chondrosarcoma as compared to other solid tumors. We demonstrate that in contrast to the evident loss of chromosomes in tumors demonstrating near-haploidy, after polyploidization with complete overgrowth of the near-haploid clone the initial loss of chromosomes can be recognized only by an unusually high LOH percentage involving several chromosomes. The frequency of near-haploidy in chondrosarcoma will therefore probably even be higher than expected based on DNA flow cytometric and cytogenetic data alone. We previously proposed a model for peripheral chondrosarcoma tumorigenesis, generally arising within benign sporadic or hereditary osteochondromas (Figure 4) ▶ . 5,56 Inactivation of both copies of an EXT gene was demonstrated to be required for (hereditary) osteochondroma development. 56 During malignant transformation additional genetic alterations are obtained, 5 probably causing defects in mitotic checkpoints leading to chromosomal instability which in turn may cause aneuploidy including near-haploidy. 60 In osteochondromas near-haploidy is absent 61,62 and near-haploidy can therefore be considered a progression marker toward a low-grade malignant phenotype. Our results suggest that further progression toward high-grade chondrosarcoma is characterized by polyploidization.
Acknowledgments
We thank Prof. P. Pearson for helpful discussions; Prof. A. H. M. Taminiau for the clinical data; and K. Kleiverda, L. J. C. M. van den Broek, N. J. Kuipers-Dijkshoorn, and E. Geelen for expert technical assistance.
Footnotes
Address reprint requests to Judith V. M. G. Bovée, M.D., Department of Pathology, Leiden University Medical Center, P.O. Box 9600, L1-Q, 2300 RC Leiden, The Netherlands. E-mail: j.v.m.g.bovee@lumc.nl.
References
- 1.Schmale GA, Conrad EU, Raskind WH: The natural history of hereditary multiple exostoses. J Bone Joint Surg Am 1994, 76A:986-992 [DOI] [PubMed] [Google Scholar]
- 2.Wicklund LC, Pauli RM, Johnston D, Hecht JT: Natural history study of hereditary multiple exostoses. Am J Med Genet 1995, 55:43-46 [DOI] [PubMed] [Google Scholar]
- 3.Springfield DS, Gebhardt MC, McGuire MH: Chondrosarcoma: a review. J Bone Joint Surg Am 1996, 78A:141-149 [Google Scholar]
- 4.Mulder JD, Schütte HE, Kroon HM, Taconis WK: Radiologic Atlas of Bone Tumors. Edited by JD Mulder. Amsterdam, Elsevier, 1993
- 5.Bovee JVMG, Cleton-Jansen AM, Kuipers-Dijkshoorn N, Van den Broek LJCM, Taminiau AHM, Cornelisse CJ, Hogendoorn PCW: Loss of heterozygosity and DNA ploidy point to a diverging genetic mechanism in the origin of peripheral and central chondrosarcoma. Genes Chromosom Cancer 1999, 26:237-246 [PubMed] [Google Scholar]
- 6.Xiang JH, Spanier SS, Benson NA, Braylan RC: Flow cytometric analysis of DNA in bone and soft-tissue tumors using nuclear suspensions. Cancer 1987, 59:1951-1958 [DOI] [PubMed] [Google Scholar]
- 7.Mandahl N, Baldetorp B, Ferno M, Akerman M, Rydholm A, Heim S, Willen H, Killander D, Mitelman F: Comparative cytogenetic and DNA flow cytometric analysis of 150 bone and soft-tissue tumors. Int J Cancer 1993, 53:358-364 [DOI] [PubMed] [Google Scholar]
- 8.Heliö H, Karaharju E, Bohling T, Kivioja A, Nordling S: Chondrosarcoma of bone. A clinical and DNA flow cytometric study. Eur J Surg Oncol 1995, 21:408-413 [DOI] [PubMed] [Google Scholar]
- 9.Sandberg AA, Bridge JA (Eds): The Cytogenetics of Bone and Soft Tissue Tumors. Austin, RG Landes Company, 1994
- 10.Sreekantaiah C, Leong SPL, Davis JR, Sandberg AA: Cytogenetic and flow cytometric analysis of a clear cell chondrosarcoma. Cancer Genet Cytogenet 1991, 52:193-199 [DOI] [PubMed] [Google Scholar]
- 11.Dal Cin P, Sciot R, Fletcher CDM, Samson I, De Vos R, Mandahl N, Willen H, Larsson O, Van den Berghe H: Inflammatory leiomyosarcoma may be characterized by specific near-haploid chromosome changes. J Pathol 1998, 185:112-115 [DOI] [PubMed] [Google Scholar]
- 12.Atkin NB, Baker MC: A metastatic malignant melanoma with 24 chromosomes. Hum Genet 1981, 58:217-219 [DOI] [PubMed] [Google Scholar]
- 13.Evans HL, Ayala AG, Romsdahl MM: Prognostic factors in chondrosarcoma of bone. A clinicopathologic analysis with emphasis on histologic grading. Cancer 1977, 40:818-831 [DOI] [PubMed] [Google Scholar]
- 14.Vaandrager JW, Schuuring E, Raap T, Philippo K, Kleiverda K, Kluin P: Interphase FISH detection of BCL2 rearrangement in follicular lymphoma using breakpoint-flanking probes. Genes Chromosom Cancer 2000, 27:85-94 [PubMed] [Google Scholar]
- 15.Hoeve MA, Gisbertz IAM, Schouten HC, Schuuring E, Bot FJ, Hermans J, Hopman A, Kluin PM, Arends J, Krieken JHJMV: Gastric low-grade MALT lymphoma, high-grade MALT lymphoma and diffuse large B cell lymphoma show different frequencies of trisomy. Leukemia 1999, 13:799-807 [DOI] [PubMed] [Google Scholar]
- 16.Abeln ECA, Corver WE, Kuipers-Dijkshoorn N, Fleuren GJ, Cornelisse CJ: Molecular genetic analysis of flow sorted ovarian tumor cells: evidence for intra-tumor heterogeneity. Br J Cancer 1994, 70:255-262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wijmenga C, van den Heuvel LP, Strengman E, Luyten JA, van der Burgt IJ, de Groot R, Smeets DF, Draaisma JM, van Dongen JJ, De Abreu RA, Pearson PL, Sandkuijl LA, Weemaes CM: Localization of the ICF syndrome to chromosome 20 by homozygosity mapping. Am J Hum Genet 1998, 63:803-809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gruis NA, Abeln ECA, Bardoel AFJ, Devilee P, Frants RR, Cornelisse CJ: PCR-based microsatellite polymorphisms in the detection of loss of heterozygosity in fresh and archival tumour tissue. Br J Cancer 1993, 68:308-313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Devilee P, Van Vliet M, Bardoel AFJ, Kievits T, Kuipers-Dijkshoorn N, Pearson PL, Cornelisse CJ: Frequent somatic imbalance of marker alleles for chromosome 1 in human primary breast carcinoma. Cancer Res 1991, 51:1020-1025 [PubMed] [Google Scholar]
- 20.Kallioniemi O, Kallioniemi A, Piper J, Isola J, Waldman FM, Gray JW, Pinkel D: Optimizing comparative genomic hybridization for analysis of DNA sequence copy number changes in solid tumors. Genes Chromosom Cancer 1994, 10:231-243 [DOI] [PubMed] [Google Scholar]
- 21.Rosenberg C, Van Gijlswijk RPM, Vos CBJ, Wiegant J, Cornelisse CJ, Tanke HJ, Raap AK: Comparative genomic hybridization with lissamine- and fluorescein-labelled nucleotides. Cytometry 1998, 32:337-341 [PubMed] [Google Scholar]
- 22.Bovee JVMG, Cleton-Jansen AM, Rosenberg C, Taminiau AHM, Cornelisse CJ, Hogendoorn PCW: Molecular genetic characterization of both components of a dedifferentiated chondrosarcoma, with implications for its histogenesis. J Pathol 1999, 189:454-462 [DOI] [PubMed] [Google Scholar]
- 23.Rosenberg C, Bakker-Schut T, Mostert MM, Tanke HJ, Raap AK, Oosterhuis JW, Looijenga LHJ: Comparative genomic hybridization in hypotriploid/hyperdiploid tumors. Cytometry 1997, 29:113-121 [DOI] [PubMed] [Google Scholar]
- 24.Du Manoir S, Schrock E, Bentz M, Speicher MR, Joos S, Ried T, Lichter P, Cremer T: Quantitative analysis of comparative genomic hybridization. Cytometry 1995, 19:27-41 [DOI] [PubMed] [Google Scholar]
- 25.Brodeur GM, Williams DL, Look AT, Bowman WP, Kalwinsky DK: Near-haploid acute lymphoblastic leukemia: a unique subgroup with a poor prognosis. Blood 1981, 58:14-19 [PubMed] [Google Scholar]
- 26.Verma RS, Macera MJ, Silver RT, Coleman M: Origin of near-haploidy in malignant hematopoietic cells. Leuk Res 1988, 12:941-950 [DOI] [PubMed] [Google Scholar]
- 27.Trent JM, Stanisic T, Olson S: Cytogenetic analysis of urologic malignancies: study of tumor colony forming cells and premature chromosome condensation. J Urol 1984, 131:146-151 [DOI] [PubMed] [Google Scholar]
- 28.Kovacs G, Soudah B, Hoene E: Binucleated cells in a human renal cell carcinoma with 34 chromosomes. Cancer Genet Cytogenet 1988, 31:211-215 [DOI] [PubMed] [Google Scholar]
- 29.Kristoffersson U, Olsson H, Kelly D, Akerman M, Mitelman F: Near-haploidy in a case of plasmocytoma. Cancer Genet Cytogenet 1986, 19:239-243 [DOI] [PubMed] [Google Scholar]
- 30.Petersen SE, Frederiksen P, Friedrich U: Cytogenetic analysis and flow cytometric DNA measurement of a human tumor with pronounced hypoploidy. Cancer Genet Cytogenet 1981, 4:1-9 [DOI] [PubMed] [Google Scholar]
- 31.Neumann E, Kalousek DK, Norman MG, Steinbok P, Cochrane DD, Goddard K: Cytogenetic analysis of 109 pediatric central nervous system tumors. Cancer Genet Cytogenet 1993, 71:40-49 [DOI] [PubMed] [Google Scholar]
- 32.Trent JM, Salmon SE: Karyotypic analysis of human ovarian carcinoma cells cloned in short term agar culture. Cancer Genet Cytogenet 1981, 3:279-291 [DOI] [PubMed] [Google Scholar]
- 33.Biegel JA, Womer RB, Emanuel BS: Complex karyotypes in a series of pediatric osteosarcomas. Cancer Genet Cytogenet 1989, 38:89-100 [DOI] [PubMed] [Google Scholar]
- 34.Jenkins RB, Kimmel DW, Moertel CA, Schultz CG, Scheithauer BW, Kelly PJ, Dewald GW: A cytogenetic study of 53 human gliomas. Cancer Genet Cytogenet 1989, 39:253-279 [DOI] [PubMed] [Google Scholar]
- 35.Ronne M, Poulsgard L, Elberg JJ: A case of meningioma with frequent relapses and a hyperhaploid stemline. Anticancer Res 1988, 8:545-548 [PubMed] [Google Scholar]
- 36.Sreekantaiah C, Crickard K, Crickard U, Yoonessi M, Sandberg AA: Three related near-haploid clones in a primary endometrioid carcinoma of the ovary. Gynecol Oncol 1990, 38:282-285 [DOI] [PubMed] [Google Scholar]
- 37.Orndal C, Mandahl N, Carlen B, Willen H, Wennerberg J, Heim S, Mitelman F: Near-haploid clones in a malignant fibrous histiocytoma. Cancer Genet Cytogenet 1992, 60:147-151 [DOI] [PubMed] [Google Scholar]
- 38.Aspberg F, Mertens F, Bauer HC, Lindholm J, Mitelman F, Mandahl N: Near-haploidy in two malignant fibrous histiocytomas. Cancer Genet Cytogenet 1995, 79:119-122 [DOI] [PubMed] [Google Scholar]
- 39.Flagiello D, Gerbault-Seareau M, Padoy E, Dutrillaux B: Near haploidy in breast cancer: a particular pathway of chromosome evolution. Cancer Genet Cytogenet 1998, 102:54-58 [DOI] [PubMed] [Google Scholar]
- 40.Mandahl N, Heim S, Arheden K, Rydholm A, Willen H, Mitelman F: Rings, dicentrics, and telomeric association in histiocytomas. Cancer Genet Cytogenet 1988, 30:23-33 [DOI] [PubMed] [Google Scholar]
- 41.Bullerdiek J, Bartnitzke S, Kahrs E, Schloot W: Further evidence for nonrandom chromosome changes in carcinoma cells: a report of 28 cases. Cancer Genet Cytogenet 1985, 16:33-43 [DOI] [PubMed] [Google Scholar]
- 42.Herrmann MA, Hay ID, Bartelt DH, Jr, Ritland SR, Dahl RJ, Grant CS, Jenkins RB: Cytogenetic and molecular genetic studies of follicular and papillary thyroid cancers. J Clin Invest 1991, 88:1596-1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Drouin V, Viguie F, Debesse B: Near-haploid karyotype in a squamous cell lung carcinoma. Genes Chromosom Cancer 1993, 7:209-212 [DOI] [PubMed] [Google Scholar]
- 44.Long PP, Hruban RH, Lo R, Yeo CJ, Morsberger LA, Griffin CA: Chromosome analysis of nine endocrine neoplasms of the pancreas. Cancer Genet Cytogenet 1994, 77:55-59 [DOI] [PubMed] [Google Scholar]
- 45.Noguera R, Navarro S, Carda C, Peydro-Olaya A, Llombart-Bosch A: Near-haploidy in a malignant sacrococcygeal teratoma. Cancer Genet Cytogenet 1999, 108:70-74 [DOI] [PubMed] [Google Scholar]
- 46.Orndal C, Mandahl N, Rydholm A, Willen H, Brosjo O, Mitelman F: Chromosome aberrations and cytogenetic intratumor heterogeneity in chondrosarcomas. J Cancer Res Clin Oncol 1993, 120:51-56 [DOI] [PubMed] [Google Scholar]
- 47.Pera F, Rainer B: Studies of multipolar mitoses in euploid tissue cultures. I. Somatic reduction to exactly haploid and triploid chromosome sets. Chromosoma 1973, 42:71-86 [DOI] [PubMed] [Google Scholar]
- 48.Oshimura M, Freeman AI, Sandberg AA: Chromosomes and causation of human cancer and leukemia. XXIII. Near-haploidy in acute leukemia. Cancer 1977, 40:1143-1148 [DOI] [PubMed] [Google Scholar]
- 49.Atkin NB, Baker MC: Favorable prognosis of solid tumors with very low chromosome numbers? Cancer Genet Cytogenet 1988, 34:121-123 [DOI] [PubMed] [Google Scholar]
- 50.Geirnaerdt MJA, Bloem JL, Eulderink F, Hogendoorn PCW, Taminiau AHM: Cartilaginous tumors: correlation of gadolineum-enhanced MR imaging and histopathologic findings. Radiology 1993, 186:813-817 [DOI] [PubMed] [Google Scholar]
- 51.Geirnaerdt MJA, Hogendoorn PCW, Bloem JL, Taminiau AHM, Van der Woude HJ: Cartilaginous tumors: fast contrast-enhanced MR imaging of cartilaginous tumors. Radiology 2000, 214:539-546 [DOI] [PubMed] [Google Scholar]
- 52.Bjornsson J, McLeod RA, Unni KK, Ilstrup DM, Pritchard DJ: Primary chondrosarcoma of long bones and limb girdles. Cancer 1998, 83:2105-2119 [PubMed] [Google Scholar]
- 53.Wuyts W, Van Hul W, Wauters J, Nemtsova M, Reyniers E, Van Hul E, De Boulle K, De Vries BBA, Hendrickx J, Herrygers I, Bossuyt P, Balemans W, Fransen E, Vits L, Coucke P, Nowak NJ, Shows TB, Mallet L, Van den Ouweland AMW, McGaughran J, Halley DJ, Willems PJ: Positional cloning of a gene involved in hereditary multiple exostoses. Hum Mol Genet 1996, 5:1547-1557 [DOI] [PubMed] [Google Scholar]
- 54.Raskind WH, Conrad EU, Chansky H, Matsushita M: Loss of heterozygosity in chondrosarcomas for markers linked to hereditary multiple exostoses loci on chromosomes 8 and 11. Am J Hum Genet 1995, 56:1132-1139 [PMC free article] [PubMed] [Google Scholar]
- 55.Hecht JT, Hogue D, Strong LC, Hansen MF, Blanton SH, Wagner M: Hereditary multiple exostosis and chondrosarcoma: linkage to chromosome 11 and loss of heterozygosity for EXT-linked markers on chromosomes 11 and 8. Am J Hum Genet 1995, 56:1125-1131 [PMC free article] [PubMed] [Google Scholar]
- 56.Bovee JVMG, Cleton-Jansen AM, Wuyts W, Caethoven G, Taminiau AHM, Bakker E, Van Hul W, Cornelisse CJ, Hogendoorn PCW: EXT-mutation analysis and loss of heterozygosity in sporadic and hereditary osteochondromas and secondary chondrosarcomas. Am J Hum Genet 1999, 65:689-698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Raskind WH, Conrad EU, Matsushita M: Frequent loss of heterozygosity for markers on chromosome arm 10q in chondrosarcomas. Genes Chromosom Cancer 1996, 16:138-143 [DOI] [PubMed] [Google Scholar]
- 58.El-Naggar AK, Dinh M, Tucker SL, Swanson D, Steck K, Vielh P: Numerical chromosomal changes in DNA hypodiploid solid tumors: restricted loss and gain of certain chromosomes. Cytometry 1999, 37:107-112 [DOI] [PubMed] [Google Scholar]
- 59.Kosaki K, Kosaki R, Craigen WJ, Matsuo N: Isoform-specific imprinting of the human PEG1/MEST gene. Am J Hum Genet 2000, 66:309-312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cahill DP, Lengauer C, Yu J, Riggins GJ, Willson JKV, Markowitz SD, Kinzler KW, Vogelstein B: Mutations of mitotic checkpoint genes in human cancers. Nature 1998, 392:300-303 [DOI] [PubMed] [Google Scholar]
- 61.Mertens F, Rydholm A, Kreicbergs A, Willen H, Jonsson K, Heim S, Mitelman F, Mandahl N: Loss of chromosome band 8q24 in sporadic osteocartilaginous exostoses. Genes Chromosom Cancer 1994, 9:8-12 [DOI] [PubMed] [Google Scholar]
- 62.Bridge JA, Nelson M, Orndal C, Bhatia P, Neff JR: Clonal karyotypic abnormalities of the hereditary multiple exostoses chromosomal loci 8q24.1 (EXT1) and 11p11–12 (EXT2) in patients with sporadic and hereditary osteochondromas. Cancer 1998, 82:1657-1663 [DOI] [PubMed] [Google Scholar]
- 63.Cooke HJ, Hindley J: Cloning of human satellite III DNA: different components are on different chromosomes. Nucleic Acids Res 1979, 6:3177-3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Waye JS, Willard HF: Chromosome specificity of satellite DNAs: short- and long-range organisation of a diverged dimeric subset of human alpha satellite from chromosome 3. Chromosoma 1989, 97:475-480 [DOI] [PubMed] [Google Scholar]
- 65.Jabs EW, Wolf SF, Migeon BR: Characterization of a cloned DNA sequence that is present at centromeres of all human autosomes and the X chromosome and shows polymorphic variation. Proc Natl Acad Sci USA 1984, 81:4884-4888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Waye JS, England SB, Willard HF: Genomic organisation of alpha satellite DNA on human chromosome 7: evidence for two distinct alphoid domains on a single chromosome. Mol Cell Biol 1987, 7:349-356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Donlon T, Wyman AR, Mulholland J, Barker D, Bruns G, Latt S, Botstein D: Alpha satellite-like sequences at the centromere of chromosome #8. Am J Hum Genet 1986, 39:A196 [Google Scholar]
- 68.Moyzis RK, Albright KL, Bartholdi MF, Cram LS, Deaven LL, Hildebrand CE, Joste NE, Longmire JL, Meyne J, Schwarzacher-Robinson T: Human chromosome-specific repetitive DNA sequences: novel markers for genetic analysis. Chromosoma 1987, 95:375-386 [DOI] [PubMed] [Google Scholar]
- 69.Waye JS, Creeper LA, Willard HF: Organization and evolution of alpha satellite DNA from human chromosome 11. Chromosoma 1987, 95:182-188 [DOI] [PubMed] [Google Scholar]
- 70.Looijenga LH, Smit VT, Wessels JW, Mollevanger P, Oosterhuis JW, Cornelisse CJ, Devilee P: Localization and polymorphism of a chromosome 12-specific alpha satellite DNA sequence. Cytogenet Cell Genet 1990, 53:216-218 [DOI] [PubMed] [Google Scholar]
- 71.Higgins MJ, Wang HS, Shtromas I, Haliotis T, Roder JC, Holden JJ, White BN: Organisation of a repetitive human 1.8 kb KpnI sequence localized in the heterochromatin of chromosome 15. Chromosoma 1985, 93:77-86 [DOI] [PubMed] [Google Scholar]
- 72.Waye JS, Willard HF: Molecular analysis of a deletion polymorphism in alpha satellite of human chromosome 17: evidence for homologous unequal crossing-over and subsequent fixation. Nucleic Acids Res 1986, 14:6915-6927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Devilee P, Cremer T, Slagboom P, Bakker E, Scholl HP, Hager HD, Stevenson AF, Cornelisse CJ, Pearson PL: Two subsets of human alphoid repetitive DNA show distinct preferential localization in the pericentric regions of chromosomes 13, 18 and 21. Cytogenet Cell Genet 1986, 41:193-201 [DOI] [PubMed] [Google Scholar]
- 74.Willard HF, Smith KD, Sutherland J: Isolation and characterization of a major tandem repeat family from the human X chromosome. Nucleic Acids Res 1983, 11:2017-2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cooke HJ, Gosden JR: Characterization of a human Y chromosome repeated sequence and related sequences in higher primates. Chromosoma 1982, 87:491-502 [DOI] [PubMed] [Google Scholar]