Abstract
Natural killer (NK) cell lymphomas are a group of rare but highly aggressive malignancies. Clinically, they can be divided into nasal NK cell lymphomas, nonnasal NK cell lymphomas, and aggressive NK cell lymphoma/leukemia. To determine the patterns of genetic deletions in these tumors, we performed loss of heterozygosity (LOH) analysis on 15 cases (11 nasal and four nonnasal), and fluorescence in situ hybridization on three cases of aggressive lymphoma/leukemia. A panel of 41 microsatellite loci on chromosomes 6q, 11q, 13q, and 17p were investigated. LOH at chromosomes 6q and 13q was frequently detected in NK cell lymphomas, being found in 80 and 66.7% of cases, respectively. LOH at chromosomes 11q and 17p was less common, being found in 28.6 and 30.8% of cases, respectively. Most tumors showed multiple loci deletions at different chromosomal regions, but several patterns of LOH could be defined. LOH at chromosome 6q was found in 90.9% of nasal NK cell lymphomas, but only in 50% of nonnasal NK cell lymphomas. LOH at chromosome 13q was found in 63.6% of nasal NK cell lymphomas and 75% of nonnasal NK cell lymphomas. For nasal NK cell lymphomas, LOH at 13q was found in 33.3% of cases at presentation, but 100% of cases at relapse. Five tumors showed LOH in only one chromosomal region, involving 6q in three cases (two nasal and one nonnasal), and 13q in two cases (both nonnasal). For the three cases of aggressive NK cell lymphoma/leukemia studied by fluorescence in situ hybridization, DNA loss at 13q14 and 17p13 regions were demonstrated. 17p13 seemed to be more commonly involved in aggressive than nasal and nonnasal NK cell lymphomas. Our results suggested that consistent patterns of LOH could be defined in NK cell malignancies. These deleted loci may contain genes important in the initiation and progression of this lymphoma.
Natural killer (NK) cell lymphoma is a group of highly aggressive lymphoid malignancies that have been characterized in recent years. 1-6 Immunophenotypic and genotypic studies have confirmed that the tumor cells are of putative NK cell lineage. The tumor cells characteristically express CD2, cytoplasmic CD3ε (but not surface CD3 or the T-cell receptor), and CD56. The T cell receptor gene is in germline configuration, and there is an almost invariable association with monoclonal Epstein-Barr virus infection in the tumor cells. 7,8
Clinically, NK cell lymphomas can be classified into several categories depending on the initial sites of involvement. 3,4 In the majority of cases, the tumors initially involve the nasal and upper aerodigestive areas, presenting usually as nonhealing necrotic ulcers. These are referred to as nasal NK cell lymphoma. A minority involve primarily nonnasal areas, such as the liver, spleen, gastrointestinal tract, skin, testis, and muscle, and are referred to as nonnasal (nasal-type) NK cell lymphoma. Rarely, the lymphoma can be widely disseminated with a leukemic phase, in which case it is known as NK cell lymphoma/leukemia. These tumors are very rare diseases but show an interesting geographic predilection. They are reported mostly from Asia, Mexico, and South America, but are extremely rare in Western countries. 9
NK cell lymphomas have distinct clinicopathological features. However, very little is known of their cytogenetic and molecular changes. Therefore, there is a pressing need for data on genetic alterations in NK cell lymphomas, to understand the underlying pathogenetic mechanisms. Reported karyotypic data from our group and other groups 10,11 showed chromosomal deletions at 6q and 13q; and we have shown by comparative genomic hybridization lately that deletions of 11q and 17p also occurred at a high frequency. 12 However, small deletions and the exact localization of the deletions are below the limits of resolution of karyotyping and comparative genomic hybridization.
In this study of a large series of NK cell malignancies, we aimed at investigating the frequency of genetic deletions at chromosomes 6q, 11q, 13q, and 17p, as well as defining the specific loci of deletion. The findings were further correlated with the clinical subtypes of these malignancies.
Materials and Methods
Patients
All 18 cases were ethnic Chinese and were CD2-positive (+ve), surface CD3-negative/cytoplasmic CD3ε +ve, and CD56 +ve NK cell lymphoma. They included 11 cases of nasal NK cell lymphoma, four cases of nonnasal NK cell lymphoma, and three cases of NK cell lymphoma/leukemia. Among the 11 cases of nasal NK cell lymphomas, five cases were studied at relapse. Clinicopathological features of these cases are listed in Table 1 ▶ . Results of comparative genomic hybridization in seven cases (cases 1, 3, 8, 9, and 16 to 18) had been previously reported. 12
Table 1.
Clinical Features and Summary of LOH Findings of 18 Cases of Natural Killer Cell Malignancies
| Case | Sex and age | Type of NK cell neoplasm | Tumor sample | LOH-6q | LOH-11q | LOH-13q | LOH-17p |
|---|---|---|---|---|---|---|---|
| 1 | M32 | Nasal | Nasopharynx (F) | − | − | − | − |
| 2 | M45 | Nasal | Nasopharynx (F) | + | − | − | + |
| 3 | F50 | Nasal | Nasopharynx (F) | + | − | − | − |
| 4 | M42 | Nasal | Nasal cavity (A) | + | + | − | + |
| 5 | M69 | Nasal | Nasopharynx (A) | + | + | + | + |
| 6 | M66 | Nasal | Nasal cavity (A) | + | − | + | − |
| 7 | M39 | Nasal | Intestine, relapse (A) | + | NI | + | NI |
| 8a | F48 | Nasal | Nasopharynx, relapse (F) | + | − | − | − |
| 8b | Marrow, relapse (F) | + | + | +FISH: loss | − | ||
| 9 | F45 | Nasal | Ovary, relapse (F) | + | + | + | + |
| 10 | M41 | Nasal | Testis, relapse (A) | + | − | + | − |
| 11 | M34 | Nasal | Appendix, relapse (A) | + | − | + | NI |
| 12 | F84 | Nonnasal | Small intestine (A) | − | − | + | − |
| 13 | F87 | Nonnasal | Skin (F) | + | − | − | − |
| 14 | M74 | Nonnasal | Skin, relapse (F) | − | − | + | − |
| 15 | M77 | Nonnasal | Buttock (F) | + | − | + | − |
| 16 | M37 | Aggressive NK cell lymphoma/leukemia | Marrow | Not done | Not done | FISH: loss | FISH: loss |
| 17 | M71 | Aggressive NK cell lymphoma/leukemia | Marrow | Not done | FISH: loss | FISH: loss | FISH: normal |
| 18 | M44 | Aggressive NK cell lymphoma/leukemia | Marrow | Not done | Not done | Not done | FISH: loss |
LOH analysis using fluorescent DNA technology was performed in cases 1 to 15. Normal tissue was not available in cases 16 to 18, and DNA loss was assessed by FISH using materials available.
F, fresh-frozen tissue; A, formalin-fixed paraffin-embedded archival tissue; +, LOH detected; −, LOH not detected; NI, noninformative or failed PCR. Results of FISH are as indicated in the corresponding boxes.
Loss of heterozygosity (LOH) analysis was performed in cases 1 to 15 in which paired normal/tumor samples were available. Among these cases, fresh-frozen tissues were available in eight paired cases. In seven of these cases, the uninvolved bone marrow/peripheral blood at presentation was used as the normal control for allele assignment. In one patient (case 8), the tumor involved both the nasopharynx and the marrow initially. Bone marrow obtained during complete remission after chemotherapy was used as normal control. In the remaining seven cases (cases 4 to 7 and 10 to 12), analysis was performed on formalin-fixed and paraffin-embedded archival samples. All samples were examined microscopically to confirm that they contained a significant proportion (>60%) of tumor cells. In cases 16 to 18, normal tissue was not available for LOH analysis because the tumor presented with a leukemic phase and multiorgan infiltration, and a remission was not achieved. Thus, fluorescence in situ hybridization (FISH) was performed to assess the status of DNA loss.
DNA Extraction
High molecular weight DNA was extracted from fresh tissue/blood samples with standard phenol-chloroform protocols. For archival samples, serial 5-μm sections were obtained. DNA was extracted with the QIAamp DNA mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. All DNA was quantified with the DynaQuant 200 fluorometer (Hoefer, Pharmacia Biotech, Stockholm, Sweden).
Polymerase Chain Reaction (PCR)
A total number of 41 fluorochrome (6-Fam, HEX, or NED)-labeled PCR primer pairs that amplified highly informative dinucleotide repeat microsatellite loci located on chromosomes 6q, 11q, 13q ,and 17p were obtained from the ABI Prism Linkage Mapping Set Version 2, which defined an approximately 10 cM resolution human index map (PE Biosystems, Foster City, CA). Cytogenetic location of the markers was obtained from the Genome Data Base (http://gdbwww.gdb.org/) and from the genetic location database (ftp://cedar.genetics.soton.ac.uk/publichtml). 13 The designation and cytogenetic location of the markers are shown in Figure 1 ▶ .
Figure 1.

Result of LOH on chromosomes 6q, 11q, 13q, and 17p in 14 cases of NK cell lymphoma at 41 polymorphic loci. Ideogram of chromosomes 6, 11, 13, and 17 are shown. Relative positions of the markers used in this study are shown to the right of each ideogram. Cytogenetic bands at the 400-band level are shown on the left. The case numbers are shown at the top row. The frequencies of LOH in informative cases are listed on the right column. Case 1 was not shown because no LOH could be detected in the loci tested. •, Informative with LOH; ○, informative without LOH; *, noninformative. Empty slots represent failed PCR.
PCR was initially performed using DNA from the patients’ normal tissues, and heterozygous loci informative for LOH analysis were then tested using DNA from the patients’ tumors. PCR was performed in a final volume of 15 μl containing 50 ng DNA, 0.2 μmol/L of each primer, 250 μmol/L of each of the four dNTP (Life Technologies, Inc., Gaithersburg, MD), 2.5 mmol/L MgCl2, 1 U AmpliTaq Gold, and 1× PCR Gold Buffer (PE Biosystems). After an initial heating at 95°C for 10 minutes, PCR cycles of denaturation, primer annealing, and extension were performed at 94°C for 15 seconds, 55°C for 15 seconds, and 72°C for 30 seconds for 22 cycles. This was followed by an additional 10 cycles at 90°C for 15 seconds, 55°C for 15 seconds, and 72°C for 30 seconds. All PCR reactions concluded with a final extension step at 72°C for 10 minutes. For amplification of archival paraffin samples, the denaturation, primer annealing, and extension time were extended to 20, 30, and 45 seconds, respectively, and 40 cycles to increase the amount of PCR product. PCR amplifications were performed in a Perkin Elmer 9700 thermal cycler (PE Biosystems).
LOH Analysis
PCR products were appropriately diluted and pooled. Pooled PCR product (1.5 μl) was mixed with a loading buffer containing deionized formamide, ethylenediaminetetraacetic acid-dextran, and ROX-labeled internal lane standard, denatured at 90°C for 10 minutes, snap-cooled on ice, and electrophoresed in a 5.3% denaturing Longranger sequencing gel (FMC Bioproducts, Rockland, ME) containing 6 mol/L urea in an automated DNA analyzer (ABI Prism 377, PE Biosystems) at a constant voltage of 3,000 V for 2.5 hours. Data were collected automatically with the ABI Prism 377 Collection version 2.5.1 and analyzed by the GeneScan software version 2.1.1 (PE Biosystems). Control DNA, CEPH 1347-02 (PE Biosystems), was amplified and analyzed to confirm that sufficient amplification and accurate sizing of alleles could be obtained in all microsatellite loci.
Calculation of LOH was based on the following formula: 14
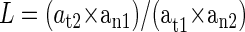 |
where a was the area, t was the tumor sample, n was the normal sample, 1 and 2 were the smaller and larger alleles, respectively. Values of L < 0.60 or ≥1.70 indicated that one of the alleles has decreased more than 40%, resulting in LOH. There was loss of the larger allele if L < 0.60 and the smaller allele when L ≥ 1.70. Cases in which either the PCR gave no signal or the allele peak height was <100 fluorescence units were designated as failed PCR.
FISH
FISH was performed on slides containing tumor cells fixed in Carnoy’s medium. Locus-specific probes for chromosome 6q were not commercially available. Probes specific for the loci 11q13, 13q14, 17p13.1, and Tel 17p (Vysis, Naperville, IL) were used according to the manufacturer’s protocols. For each slide, 200 cells were analyzed. The test loci were considered deleted when the percentage of cells with one hybridization signal was significantly (at least two times) more than the control from normal donors included in each experiment.
Results
Successful DNA amplification was obtained at each of the 41 loci tested in all but one case (case 15) of fresh tumor tissues (cases 1 to 3, 8, 9, and 13 to 15). Microscopic examination of case 15 showed severe tumor necrosis, and amplification was only successful at 27 loci. For the formalin-fixed archival samples (cases 4 to 7 and 10 to 12), DNA amplification was successful in all of the loci for case 6 that was stored for a few months. For the other cases stored from 1 year to >10 years, DNA amplification of the loci showed variable success. Thus, successful amplification for archival samples might be related to the age of the specimen and the size of the PCR product. 15,16 A summary of the LOH results are listed in Table 1 ▶ and Figure 1 ▶ , and representative electropherograms are shown in Figure 2 ▶ . The patterns of allelic loss based on clinical subtypes are listed in Table 2 ▶ .
Figure 2.
Electropherogram of normal/tumor pair in case 5 (Figure 2A) ▶ and case 9 (Figure 2B) ▶ . One locus from each chromosome is shown. The y axis represents the peak height in fluorescence units, and the LOH ratio is shown in parentheses. Upper tracing: amplification from normal tissue, the black arrows marking the heterozygous alleles. Lower tracing: amplification from tumor tissue, the red arrows marking the lost allele.
Table 2.
Pattern of Allelic Loss Based on Clinical Subtypes of Natural Killer Cell Malignancies
| 6q | 11q | 13q | 17p | |
|---|---|---|---|---|
| NK cell lymphoma | 12/15 (80) | 4/14 (28.6) | 10/15 (66.7) | 4/13 (30.8) |
| Nasal | 10/11 (90.9) | 4/10 (40) | 7/11 (63.6) | 4/9 (44.4) |
| At presentation | 5/6 (83.3) | 2/6 (33.3) | 2/6 (33.3) | 3/6 (50) |
| At relapse | 5/5 (100) | 2/4 (50) | 5/5 (100) | 1/3 (33.3) |
| Nonnasal | 2/4 (50) | 0/4 (0) | 3/4 (75) | 0/4 (0) |
| Aggressive NK cell lymphoma/leukemia | - | 1/1 (100) | 2/2 (100) | 2/3 (66.7) |
| Total | 12/15 (80) | 5/15 (33.3) | 12/17 (70.6) | 6/16 (37.5) |
Percentages are in parentheses.
LOH at Chromosome 6q
Analysis of informative loci showed LOH in 12 of 15 cases (80%), of which 10 occurred in 11 cases of nasal NK cell lymphomas (91%), and two occurred in four cases of nonnasal NK cell lymphomas (50%). LOH occurred in 57% of cases at 6q13-q14 (D6S434; four of seven cases), 31% at 6q21-q23 (D6S287, D6S262, D6S292; four of 13 cases), 50% at 6q24.3 (D6S441; seven of 14 cases), 67% at 6q25.3 (D6S1581; six of nine cases), and 45.5% at 6q27 (D6S281; five of 11 cases). LOH was not detected with marker D6S446, mapping to 6q27.
LOH at Chromosome 11q
LOH was detected in four of 14 (29%) cases. All four cases were nasal NK cell lymphomas. LOH was found in 10% of cases at 11q13.5-q14 (D11S937; one of 10 cases), and 31% at 11q21-q24 (D11S4175, D11S898, D11S908, D11S925, D11S1320; four of 13 cases).
LOH at Chromosome 13q
LOH was found in 10 of 15 cases (67%), of which seven occurred in 11 cases of nasal NK cell lymphomas (64%), and three occurred in four cases of nonnasal NK cell lymphomas (75%). LOH was found in 60% of cases at 13q12-q14 (D13S217, D13S171, D13S218, D13S263, D13S153; nine of 15 cases), 53% at 13q14 (D13S263, D13S153; eight of 15 cases), 14% at 13q21-q22 (D13S156; one of 7 cases), and 53% at 13q31-q34 (D13S170, D13S265, D13S159, D13S158, D13S173, D13S1265, D13S285; eight of 15 cases).
LOH at Chromosome 17p
LOH was found in four of 13 cases (31%). All four cases were nasal NK cell lymphomas. LOH was found in 36% at 17p13 (D17S849, D17S938, D17S1852; four of 11 cases) and 40% at 17p12 (D17S799; two of five). Case 9 had LOH at all of the three informative loci (D17S849, D17S938, D17S1852) mapping to region 17p13.1-p13.3.
FISH
Limited material was available for FISH analysis, so that it was performed on selected cases and regions only (11q13 in case 17; 13q14 in cases 8b, 16, and 17; and 17p in cases 16 to 18). 11q13 was deleted in case 17 (10% of cells with one signal versus 1.5% in control). 13q14 was deleted in cases 8b, 16, and 17 (16.5%, 49.5%, and 19.5% of cells with one signal versus 6.5% in control). 17p13.1 was deleted in case 16 (23% of cells with one signal versus 8.5% in control), but not in cases 17 and 18. However, FISH with a Tel 17p in case 18 showed a significant number of cells with one signal (42% versus 8.5% in control). In this case, DNA loss possibly involved a region distal to p13.1.
Discussion
So far there have been limited data on the genetic aberrations in NK cell malignancies. However, preliminary results from a small number of cases investigated by karyotyping and comparative genomic hybridization showed that deletions of chromosomes 6q, 11q, 13q, and 17p might be important. In this study, we aimed at investigating the frequencies of deletions of these chromosomal regions in a larger series of NK cell malignancies of various clinical subtypes, as well as defining the specific loci of deletion.
We have used PCR amplification of high-resolution polymorphic microsatellite markers with fluorochrome-labeled primers and automated DNA fragment analysis to map the loci of allelic loss. The four chromosomal regions were selected for several reasons. Although studied in small number of tumors, these regions have been consistently observed to be involved. Practically, biopsies of NK cell lymphomas are usually small, particularly from nasal areas. Often the amount of material left for investigations after completion of pathological diagnosis is scanty. A genome-wide search for allelic loss that requires large amounts of tumor materials is therefore not feasible.
In this study, LOH in chromosomes 6q, 13q, 17p, and 11q, occurring with overall frequencies of 80%, 70.6%, 37.5%, and 33.3%, respectively, showed heterogeneity in NK tumor subtypes (Table 2) ▶ . An examination of the patterns of aberrations showed that the heterogeneity might be related to the type as well as the stage of the tumor.
In nasal NK cell lymphomas, deletions of chromosome 6q were found in 10 of 11 cases (the only negative case, case 1, also did not show LOH at any other chromosomal loci tested, raising the possibility that there might be too many contaminating normal cells masking genetic loss in tumor cells). Deletions at D6S434, D6S441, and D6S1581 together identified all but one case (case 6) of NK cell lymphomas. D6S441 and D6S1581 map commonly to 6q24–6q25. Each was the only marker deleted in two cases (case 4, D6S1581; case 7, D6S441). D6S434 was also the only marker deleted in one other case (case 11). The results implied that these loci might be of primary pathogenetic significance. On the other hand, deletions of chromosome 13q were found only in 33.3% of cases at presentation, but 100% of cases at relapse. This implied that 13q loss might be a progression event in nasal NK cell lymphoma. Deletions at 17p and 11q were less frequent, occurring in 44.4% and 40% of cases, respectively.
In nonnasal NK cell lymphomas, deletions of chromosomes 13q and 6q were frequent events, occurring at 75 and 50% of cases, respectively. Interestingly, deletions of chromosome 11q and 17p were not found in any of these cases, implying that they might not be of primary pathogenetic significance in this group of tumors.
In aggressive NK cell lymphoma/leukemia, FISH analysis was performed in three cases. Deletion of 13q14 (covering the Rb gene) occurred in two of two cases, a frequency comparable to those of nasal and nonnasal NK cell lymphomas. However, deletion at 17p13.1 occurred in two of three cases, which seemed to be higher than that of 30% in nasal and nonnasal lymphomas. Whether deletion at 17p is more prevalent in NK cell lymphoma/leukemia or related to the aggressive nature of the lymphoma will have to be validated by future studies.
The heterogeneity of the location and frequencies of LOH at different chromosomal regions might also be of biological implications. In case 8, tumors at two locations were analyzed. This patient had a primary nasal NK cell lymphoma that relapsed locally and then metastasized. Analysis of the nasal lymphoma showed LOH at only one locus (D6S434), but the metastatic lymphoma in the marrow showed LOH at the same and other additional loci on 6q, which was consistent with clonal evolution. In case 9 where almost all loci on chromosome 13q were deleted, the LOH value ranged from 0.10 at D13S156 to 2.04 at D13S265, indicating that there might be subclones within the tumor with different percentage of cells showing deletions at any given locus.
When examined as a group, some interesting findings are observed. There were five cases with LOH at only one chromosomal region (6q in three cases; 13q in two cases). As illustrated in case 8, the lymphoma at the nasal site showed LOH only at one locus in 6q, whereas the metastatic marrow lesion showed LOH at multiple loci at 6q, 11q, and 13q, implying that these additional deletions were secondary changes. As such, LOH at 13q was found as the sole abnormality in only nonnasal NK cell lymphomas (cases 12 and 14). More cases of nonnasal NK cell lymphomas therefore will have to be investigated to define if LOH at 13q may also be an important primary aberration.
LOH at these chromosomal regions suggested that potential tumor suppressor genes might be involved. Chromosome 6q is frequently deleted in lymphoid malignancies. Several commonly deleted regions involving 6q21, 6q23, and 6q25-q27, 17 6q23-q24, 18 6q15-q21, 19 6q21 20 have been reported. Alteration of the c-myb proto-oncogene (mapping to 6q22-q23) expression has been described in cases with deletion in chromosome 6q, 21 and more recently a candidate tumor suppressor gene, hZAC, has also been mapped to 6q24-q25. 22 Moreover, breaks at 6q21-q25 were associated with a decreased probability of achieving remission in other types of lymphomas. 23 Thus, in addition to a pathogenetic role, LOH at 6q might also be associated with the highly malignant nature of NK cell lymphomas. At chromosome 13q, in addition to the retinoblastoma (Rb) gene, 24 other tumor suppressor genes have also been proposed to be present. 25,26 At chromosome 11q, the commonly deleted region was 11q21-q24. A candidate tumor suppressor gene, ATM, has been mapped to 11q22.3-q23.1. 27 Moreover, the NCAM gene, coding for the CD56 NK cell marker, is also localized at 11q23. 28,29 In chromosome 17p, the commonly deleted region was 17p13. The tumor suppressor gene TP53 is localized to this region. 30 Further studies are needed to delineate if any of these genes are involved in the pathogenesis of NK cell malignancies.
In summary, using high-resolution microsatellite markers, we have defined consistent patterns of genetic deletions in NK cell lymphomas. Our findings suggest specific deletion patterns to be associated with different subtypes of the lymphoma, and the occurrence of distinct molecular pathways of tumor progression. The high frequency of LOH at chromosome 6q may serve as a molecular marker of NK cell lymphoma, particularly the nasal subtype. Refinement of allelic loss at 6q might lead to the identification of novel tumor suppressor genes involved in the tumorigenesis of NK cell lymphoma. Finally, it will be interesting to examine NK cell lymphomas from different geographic localities to define if similar patterns of genetic lesions are involved.
Acknowledgments
The authors thank Ms. Chris Tam for technical assistance.
Footnotes
Address reprint requests to Dr. Y. L. Kwong, University Department of Medicine, Professorial Block, Queen Mary Hospital, Pokfulam Road, Hong Kong, China. E-mail: ylkwong@hkucc.hku.hk.
Supported by the Kadoorie Charitable Foundation.
References
- 1.Wong KF, Chan JKC, Ng CS, Lee KC, Tsang WYW, Cheung MMC: CD56 (NKH1)-positive hematolymphoid malignancies: an aggressive neoplasm featuring frequent cutaneous/mucosal involvement, cytoplasmic azurophilic granules and angiocentricity. Hum Pathol 1992, 23:798-804 [DOI] [PubMed] [Google Scholar]
- 2.Kern WF, Spier CM, Hanneman EH, Miller TP, Matzner M, Grogan TM: Neural cell adhesion molecule-positive peripheral T-cell lymphoma: a rare variant with a propensity for unusual sites of involvement. Blood 1992, 79:2432-2437 [PubMed] [Google Scholar]
- 3.Chan JKC, Sin VC, Wong KF, Ng CS, Tsang WYW, Chan CH, Cheung MMC, Lau WH: Non-nasal lymphoma expressing the natural killer cell marker CD56: a clinicopathologic study of 49 cases of an uncommon aggressive neoplasm. Blood 1997, 89:4501-4513 [PubMed] [Google Scholar]
- 4.Kwong YL, Chan ACL, Liang R, Chiang AKS, Chim CS, Chan TK: CD56+ NK lymphomas: clinicopathologic features and prognosis. Br J Haematol 1997, 97:821-829 [DOI] [PubMed] [Google Scholar]
- 5.Cheung MMC, Chan JKC, Lau WH, Foo W, Chan PT, Ng CS, Ngan RKC: Primary non-Hodgkin’s lymphoma of the nose and nasopharynx: clinical features, tumor immunophenotype, and treatment outcome in 113 patients. J Clin Oncol 1998, 16:70-77 [DOI] [PubMed] [Google Scholar]
- 6.Jaffe ES, Harris NL, Diebold J, Muller-Hermelink H: World Health Organization classification of neoplastic diseases of the hematopoietic and lymphoid tissues. A progress report. Am J Clin Pathol 1999, 111(Suppl 1):S8-S12 [PubMed] [Google Scholar]
- 7.Tsang WYW, Chan JKC, Yip TTC, Ng CS, Wong KF, Poon YF, Ma VWS: In situ localization of Epstein-Barr virus encoded RNA in non-nasal/nasopharyngeal CD56-positive and CD56-negative T-cell lymphomas. Hum Pathol 1994, 25:758-765 [DOI] [PubMed] [Google Scholar]
- 8.Chiang AKS, Tao Q, Srivastava G, Ho FCS: Nasal NK- and T-cell lymphomas share the same type of Epstein-Barr virus latency as nasopharyngeal carcinoma and Hodgkin’s disease. Int J Cancer 1996, 68:285-290 [DOI] [PubMed] [Google Scholar]
- 9.Chan JKC: Natural killer cell neoplasms. ASCP Rev Pathol (Anat Pathol) 1998, 3:77-145 [PubMed] [Google Scholar]
- 10.Wong KF, Chan JKC, Kwong YL: Identification of del(6)(q21q25) as a recurring chromosomal abnormality of putative NK cell lymphoma/leukaemia. Br J Haematol 1997, 98:922-926 [DOI] [PubMed] [Google Scholar]
- 11.Tien H, Su I, Tang J, Liu M, Lee F, Chen Y, Chuang S: Clonal chromosomal abnormalities as direct evidence for clonality in nasal T/natural killer cell lymphoma. Br J Haematol 1997, 97:621-625 [DOI] [PubMed] [Google Scholar]
- 12.Siu LLP, Wong KF, Chan JKC, Kwong YL: Comparative genomic hybridization analysis of natural killer cell lymphoma/leukemia—recognition of consistent patterns of genetic alteration. Am J Pathol 1999, 155:1419-1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collins A, Frezal J, Teague J, Morton NE: A metric map of humans: 23,500 loci in 850 bands. Proc Natl Acad Sci USA 1996, 93:14771-14775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Canzian F, Salovaara R, Hemminki A, Kristo P, Chadwick RB, Aaltonen LA, de la Chapelle A: Semiautomated assessment of loss of heterozygosity and replication error in tumors. Cancer Res 1996, 56:3331-3337 [PubMed] [Google Scholar]
- 15.Greer CE, Peterson SL, Kiviat NB, Manos MM: PCR amplification from paraffin-embedded tissues—effects of fixative and fixation time. Am J Clin Pathol 1991, 95:117-124 [DOI] [PubMed] [Google Scholar]
- 16.Diss TC, Pan L, Peng H, Wotherspoon AC, Isaacson PG: Sources of DNA for detecting B cell monoclonality using PCR. J Clin Pathol 1994, 47:493-496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Offit K, Parsa NZ, Gaidano G, Filippa DA, Louie D, Pan D, Jhanwar SC, Dalla-Favera R, Chaganti RSK: 6q deletions define distinct clinico-pathologic subsets of non-Hodgkin’s lymphoma. Blood 1993, 82:2157-2162 [PubMed] [Google Scholar]
- 18.Zhang Y, Weber-Matthiesen K, Siebert R, Matthiesen P, Schlegelberger B: Frequent deletions of 6q23–24 in B-cell non-Hodgkin’s lymphomas detected by fluorescence in situ hybridization. Genes Chromosom Cancer 1997, 18:310-313 [DOI] [PubMed] [Google Scholar]
- 19.Hatta Y, Yamada Y, Tomonaga M, Miyoshi I, Said JW, Koeffler HP: Detailed deletion mapping of the long arm of chromosome 6 in adult T-cell leukemia. Blood 1999, 93:613-616 [PubMed] [Google Scholar]
- 20.Zhang Y, Matthiesen P, Harder S, Siebert R, Castoldi G, Calasanz MJ, Wong KF, Rosenwald A, Ott G, Atkin NB, Schlegelberger B: A 3-cM commonly deleted region in 6q21 in leukemias and lymphomas delineated by fluorescence in situ hybridization. Genes Chromosom Cancer 2000, 27:52-58 [DOI] [PubMed] [Google Scholar]
- 21.Barletta C, Pelicci PG, Kenyon LC, Smith SD, Dalla-Favera R: Relationship between the c-myb locus and the 6q-chromosomal aberration in leukemias and lymphomas. Science 1987, 235:1064-1067 [DOI] [PubMed] [Google Scholar]
- 22.Varrault A, Ciani E, Apiou F, Bilanges B, Hoffmann A, Pantaloni C, Bockaert J, Spengler D, Journot L: hZAC encodes a zinc finger protein with antiproliferative properties and maps to a chromosomal region frequently lost in cancer. Proc Natl Acad Sci USA 1998, 95:8835-8840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Offit K, Wong G, Filippa DA, Tao Y, Chaganti RSK: Cytogenetic analysis of 434 consecutively ascertained specimens of non-Hodgkin’s lymphoma: clinical correlations. Blood 1991, 77:1508-1515 [PubMed] [Google Scholar]
- 24.Friend SH, Bernards R, Rogelj S, Weinber RA, Rapaport JM, Albert DM, Dryja TP: A human DNA segment with properties of the gene that predisposes to retinoblastoma and osteosarcoma. Nature 1986, 323:643-646 [DOI] [PubMed] [Google Scholar]
- 25.Kalachikov S, Migliazza A, Cayanis E, Fracchiolla NS, Bonaldo MF, Lawton L, Jelenc P, Ye X, Qu X, Chien M, Hauptschein R, Gaidano G, Vitolo U, Saglio G, Resegotti L, Brodjansky V, Yankovsky N, Zhang P, Soares MB, Russo J, Edelman IS, Efstratiadis A, Dalla-Favera R, Fischer SG: Cloning and gene mapping of the chromosome 13q14 region deleted in chronic lymphocytic leukemia. Genomics 1997, 42:369-377 [DOI] [PubMed] [Google Scholar]
- 26.Stilgenbauer S, Nickolenko J, Wilhelm J, Wolf S, Weitz S, Dohner K, Boehm T, Dohner H, Lichter P: Expressed sequences as candidates for a novel tumor suppressor gene at band 13q14 in B-cell chronic lymphocytic leukemia and mantle cell lymphoma. Oncogene 1998, 16:1891-1897 [DOI] [PubMed] [Google Scholar]
- 27.Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, Tagle DA, Smith S, Uziel T, Sfez S, Ashkenazi M, Pecker I, Frydman M, Harnik R, Sankhavaram RP, Simmons A, Clines GA, Sartiel A, Gatti RA, Chessa L, Sanal O, Lavin MF, Jaspers NGJ, Taylor AMR, Arlett CF, Miki T, Weissman SM, Lovett M, Collins FS, Shiloh Y: A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science 1995, 268:1749-1753 [DOI] [PubMed] [Google Scholar]
- 28.Nguyen C, Mattei MG, Mattei JF, Santoni MJ, Goridis C, Jordan BR: Localization of the human NCAM gene to band q23 of chromosome 11: the third gene coding for a cell interaction molecule mapped to the distal portion of the long arm of chromosome 11. J Cell Biol 1986, 102:711-715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lanier LL, Testi R, Bindl J, Phillips JH: Identity of Leu-19 (CD56) leukocyte differentiation antigen and neural cell adhesion molecule. J Exp Med 1989, 169:2233-2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Isobe M, Emanuel BS, Givol D, Oren M, Croce CM: Localization of gene for human p53 tumour antigen to band 17p13. Nature 1986, 320:84-85 [DOI] [PubMed] [Google Scholar]



