Abstract
Transglutaminase 1 (TGase 1) is a Ca2+-dependent enzyme which catalyzes ε-(γ-glutamyl)lysine cross-linking of substrate proteins such as involucrin and loricrin to generate the cornified envelope at the cell periphery of the stratum corneum. We have shown that disruption of the TGase 1 gene in mice results in neonatal lethality, absence of the cornified envelope, and impaired skin barrier function. Based on the importance of TGase 1 in epidermal morphogenesis, we have now assessed its role in wound healing. In neonatal mouse skin, TGase 1 mRNA as well as keratin 6α was induced in the epidermis at the wound edges as early as 2 hours after injury and that expression continued in the migrating epidermis until completion of re-epithelialization. The TGase 1 enzyme co-localized on the plasma membrane of migrating keratinocytes with involucrin, but not with loricrin, which suggests the premature assembly of the cornified envelope. Similar injuries to TGase 1 knockout mouse skins grafted on athymic nude mice showed substantial delays in wound healing concomitant with sustained K6α mRNA induction. From these results, we suggest that activation of the TGase 1gene is essential for facilitated repair of skin injury.
Transglutaminases (TGases) (EC 2.3.2.1.3) belong to a Ca2+-dependent enzyme family and catalyze the formation of ε-(γ-glutamyl)lysine cross-links between peptide-bound glutamine residues and the primary amine group of various amines. 1 Such cross-links are generally formed in or between proteins by reaction with the ε-amino group of lysine residues and they act to stabilize proteins via polymerization against chemicals, proteolytic enzymes, and physical disruption. TGases have been proposed to participate in a wide variety of biological events such as fertilization, development, differentiation, apoptosis, coagulation, and wound healing. 2
Cutaneous wound healing is a complex process consisting of three overlapping phases, inflammation, proliferation, and remodeling. 3 Skin injury elicits a temporary repair by formation of a clot that plugs the defect, followed by invasion of inflammatory cells and then fibroblasts and capillaries into the clot to form a contractile granulation tissue. Meanwhile, the cut epidermal edges migrate forward to cover the denuded wound surface. During those wound healing processes, TGases are activated in all layers of the skin corresponding to the epidermis, the dermis, and the panniculus carnosis. 4 Coagulation factor XIIIa (plasma TGase) is activated by thrombin and contributes to the formation of a fibrin clot at the site of injury. 5 Tissue TGase (TGase 2) is expressed in endothelial cells, macrophages, and skeletal muscle cells throughout the healing process. 6 TGase 2 might be induced in response to acute-phase injury cytokines, such as transforming growth factor-β, 7 interleukin-6, 8 and/or tumor necrosis factor-α, 9 because the TGase 2 gene has response elements to these cytokines. TGase 2 that binds to a cell surface complex composed of plasminogen, urokinase receptor, and mannose-6-phosphate receptor, converts latent transforming growth factor-β to its active form, providing a positive amplification loop for TGase 2 expression in wound sites. 10 TGase 2 and factor XIIIa may participate in the cross-linking of extracellular matrix molecules, such as fibrin/fibrinogen, 11,12 fibronectin, 13 collagen, 14 laminin/nidogen, 15 osteopontin, 16 and vitronectin. 17 Thus, TGases effectively construct a scaffold for migration of inflammatory and endothelial cells by stabilizing the extracellular matrix, 6 and the end product of the process is the replacement of the fibrin matrix with granulation tissue. Even after the wound has healed, those TGase isozymes possibly promote contraction throughout the wound by cross-linking extracellular matrix molecules. In addition to the important role of TGase 2 in dermal wound healing, the enzyme is also involved in the cross-linking of anchoring fibrils at the dermo-epidermal junction via ligation of collagen VII as a potential substrate. 18
In contrast to those TGase isoforms, little attention has been given to the role of TGase 1 (keratinocyte TGase) in wound healing since the report by Mansbridge and Knapp 19 that TGase immunoreactivity is detectable in the epidermis 15 hours after tape-stripping and 3 days after generation of a suction blister. TGase 1 is essential for the development and maturation of the stratum corneum, especially for production of the cornified envelope at the cell periphery of the stratum corneum, 20 by assembly of precursor proteins such as involucrin 21 and loricrin. 22 Furthermore, TGase 1 may be necessary to maintain skin barrier function by covalent cross-linking of ω-OH ceramides with involucrin. 23 The TGase 1 gene is one of the genes responsible for autosomal recessive lamellar ichthyosis and a variety of mutations in that gene have been identified in some families with the disorder. 24,25 The TGase 1 gene is expressed mainly in differentiating layers of normal stratified squamous epithelia and in inner root sheath cells, 26 although TGase 1 localizes not only in the stratified squamous epithelia but also concentrates at cadherin-based adherens junctions of simple epithelial cells. 27 In addition to these roles of TGase 1 in the skin, the unusual expression of the enzyme in the brain has been implicated in the pathogenesis of Alzheimer’s disease. 28
Recently, we have generated transgenic mice bearing a lacZ gene directed by a 2.5-kb 5′ upstream promoter of the human TGase 1 gene and we have analyzed β-galactosidase expression in these mice. 26 In a series of studies using those TGM1-lacZ transgenic mice, we have noticed that β-galactosidase staining is strongly induced in the epidermis close to the skin injury. Furthermore, based on the phenotype of our TGase 1 knockout mice, which have defective epidermal development and keratinization, lack the cornified envelope, and have impaired barrier function, 20 we have speculated on the importance of TGase 1 during skin regeneration. In this study, we characterize TGase 1 gene expression during wound healing of the skin and the results suggest the essential role of TGase 1 in that process.
Materials and Methods
Materials
Rat monoclonal TGase 1 antibody was kindly supplied by Dr. Hiiragi, Department of Cell Biology, Kyoto University Graduate School of Medicine, Kyoto, Japan. 27 Rabbit polyclonal mouse loricrin and mouse involucrin antibodies were from Covance Research Products Inc. (Richmond, CA); Cy3-conjugated anti-rat IgG and fluorescein isothiocyanate-conjugated anti-mouse IgG were from Jackson ImmunoResearch Laboratories, Inc. (West Grove, PA); fluorescein isothiocyanate-conjugated and Texas Red-conjugated anti-rabbit IgGs were from Vector Laboratories, Inc. (Burlingame, CA).
Experimental Wounding, Harvesting, and Sectioning
All studies involving animals were reviewed by the Animal Use and Care Committee of the Kyoto Prefectural University of Medicine. BDF1 neonatal mice were anesthetized and a 10-mm linear full-thickness incision was made on the dorsal skin. Wounds were harvested from three mice at 1, 2, 3, 4, 8, 12, 16, 20, 24 hours and 2, 3, 4, 5, 6 days after wounding. For skin grafting, nine normal controls and eight TGase 1−/− neonatal mice were killed by decapitation and their dorsal skins were excised and transplanted onto athymic nude mice. Two weeks after the skin grafting, a round wound 3 mm in diameter was made at the center of each graft with a biopsy punch. Wounds made on the grafted skins were harvested for histological examinations at 48 hours and 5, 10, and 11 days after wounding.
Wound tissues were fixed in 10% formaldehyde in phosphate-buffered saline (PBS), then embedded in paraffin. Those tissues were sectioned at 4-μm thickness and were used for hematoxylin and eosin staining and for in situ hybridization. For immunofluorescence, tissues were embedded in OCT compound (Miles Inc., Elkhart, IN) and were frozen at −80°C, then sectioned at 5-μm thickness at −20°C.
In Situ Hybridization
pMTG1–1.1 containing a 1.1-kb mouse TGase 1 cDNA 26 was linearized by SalI digestion and antisense TGase 1 cRNA was synthesized with T7 RNA polymerase, using a digoxigenin RNA Labeling Kit (Roche Diagnostics Corp., Basel, Switzerland). Mouse keratin 6α (K6α) and keratin 6β (K6β) cRNA probes were prepared as described previously. 29
In situ hybridization was performed using a method described elsewhere. 26 Tissue sections were deparaffinized in xylene and dehydrated through a graded ethanol series. After proteinase K digestion (18 μg/ml), the sections were postfixed with 4% (w/v) paraformaldehyde in PBS for 10 minutes and were then treated with 0.1 mol/L triethanolamine-HCl (pH 8.0) for 1 minute. After acetylation for 10 minutes, the sections were dehydrated, air-dried, and then incubated at 50°C in a hybridization buffer composed of 50% formamide, 10 mmol/L Tris-HCl (pH 7.5), 1 mg/ml yeast tRNA (Sigma Chemical Co., St. Louis, MO), 1× Denhardt’s solution (Sigma Chemical Co.), 10% polyethylene glycol 6,000, 600 mmol/L NaCl, 0.25% sodium dodecyl sulfate, 1 mmol/L ethylenediaminetetraacetic acid (pH 8.0), and 0.2 μg/ml of probe. After hybridization, the sections were washed at 45°C for 1 hour in 50% formamide and 2× standard saline citrate, and were then digested with 20 μg/ml RNase (Sigma Chemical Co.) in 10 mmol/L Tris-HCl (pH 8.0) and 500 mmol/L NaCl at 37°C for 30 minutes. Hybridized digoxigenin-labeled probes were visualized with a Nucleic Acid Detection Kit (Roche Diagnostics Corp.).
Immunofluorescence and Confocal Fluorescence Imaging
Cryosections mounted on glass slides were air-dried and rinsed with PBS containing 1% bovine serum albumin for 15 minutes, and were then incubated for 1 hour with rat TGase 1 (1:2) and rabbit involucrin antibodies (1:600) or with rabbit loricrin antibodies (1:500). Sections were then rinsed with PBS containing 1% bovine serum albumin and 0.15% Triton X-100, followed by incubation for 1 hour with Cy3-conjugated anti-rat IgG and fluorescein isothiocyanate-conjugated anti-rabbit IgG or Texas Red-conjugated anti-rabbit IgG. A confocal laser-scanning microscope, Olympus Fluoview (Olympus Optical Co., Ltd., Tokyo, Japan) was used for immunofluorescence imaging.
Results
Time Course of TGase 1 Gene Expression Induced by Skin Injury
A linear 10-mm full-thickness incision was made with a scalpel on the dorsal skin of BDF1 neonatal mice and the expression of the TGase 1 gene in the epidermis was compared with that of the K6α and K6β genes. Histological changes at the wound edges were stained with hematoxylin and eosin and are shown in Figure 1 ▶ (left panels A, D, G, J, M, and P). The morphological changes noted throughout the time course were basically similar to those in adult mice reported by Croft and Tarin. 30 Before wounding, TGase 1 mRNA was evident in the upper spinous and granular layers of the epidermis (Figure 1B) ▶ , but K6α mRNA was undetectable there (Figure 1C) ▶ . One hour after wounding, no obvious induction of TGase 1, K6α, or K6β mRNA was observed. However, 2 hours after injury, TGase 1 mRNA was focally increased in a few layers of suprabasal keratinocytes near the wound edge (Figure 1E) ▶ . At that time, K6α mRNA was induced in a few suprabasal spinous layers (Figure 1F) ▶ . The TGase 1 mRNA signals were accentuated in keratinocytes near the wound edge from 4 to 8 hours after wounding (Figure 1H) ▶ and at later times TGase 1 mRNA was distributed over the epidermis around the wound. This event occurred earlier than did massive leukocyte infiltration at the injured sites, which was found from 8 hours after the injury. The strongest induction of TGase 1 mRNA was observed in migrating keratinocytes 20 hours after incision (Figure 1K) ▶ , at which time the epidermis was thickening and re-epithelialization was becoming obvious under the scab. At 48 hours, when intercellular spaces between keratinocytes were widened (Figure 1M) ▶ , intense signals were still evident there, but the expression of TGase 1 mRNA was decreasing in the epidermis away from the migrating edge (Figure 1N) ▶ . Ninety-six hours after wounding, TGase 1 mRNA expression was back to normal in the thickened re-epithelializing epidermis (Figure 1Q) ▶ . At 144 hours, the re-epithelialized epidermis was arranged in regular layers and the cutaneous surface became flattened (data not shown). The induction of K6α mRNA was found at 2 hours (Figure 1I) ▶ and the distribution of K6α mRNA seemed to be optimal at 20 hours after wounding (Figure 1L) ▶ . At 48 hours, K6α mRNA expression was detected in the initial wound site and in the regenerating epidermis (Figure 1O) ▶ and was still evident 96 hours after the injury (Figure 1R) ▶ , by which time the induction of TGase 1 mRNA had normalized. The expression of K6β mRNA began 2 hours after injury as did K6α, and strong signals were observed 48 hours after the injury (data not shown). K6α mRNA often extended down to the basal layer, whereas K6β mRNA was primarily restricted to the suprabasal layers of the epidermis.
Figure 1.

Induction of TGase 1 and K6α mRNAs during wound healing in neonatal mouse skin. A 10-mm, full-thickness skin wound was made in BDF1 dorsal skin. The skin was harvested just before (A–C), 2 hours (D–F), 4 hours (G–I), 20 hours (J–L), 48 hours (M–O), and 96 hours (P–R) after the injury. The tissues were stained with H&E (A, D, G, J, M, P), or were subjected to in situ hybridization of TGase 1 (B, E, H, K, N, Q) and K6α mRNA (C, F, I, L, O, R). The induction of TGase 1 and K6α mRNAs begins in the epidermis at the wound edge as early as 2 hours after the injury (E and F). The induction of TGase 1 mRNA becomes more pronounced (H) and is evident even in migrating keratinocytes (K). The induction of TGase 1 mRNA is almost normalized until 96 hours after the injury (Q), at which time K6α mRNA expression is still evident (R). Scale bar, 50 μm.
Coordinated Induction of Involucrin with TGase 1 Enzyme in Injured Epithelia
The expression of TGase 1 enzyme and its substrate proteins, involucrin and loricrin, during wound healing were assessed in five control and four injured neonatal mice by confocal immunofluorescence imaging. In control mice, TGase 1 enzyme was evident in a few layers under the stratum corneum and in the inner root sheath of hair follicles (Figure 2A) ▶ . Involucrin and loricrin were also located in the subcorneal layers, but loricrin was not evident in hair follicles (Figure 2, B and C) ▶ . The expression of those substrate proteins was virtually unchanged 8 hours after the injury (data not shown). At 48 hours, TGase 1 enzyme was abundantly expressed suprabasally in the thickened epidermis near the wound edge and in migrating keratinocytes (Figure 2D) ▶ . The fluorescence of TGase 1 enzyme was clearly localized at the cell periphery, suggesting membrane translocation of the enzyme (Figure 2G) ▶ . At that time, the expression of involucrin was detectable in the upper layers of the thickened epidermis and in migrating keratinocytes (Figure 2, E and H) ▶ . Involucrin was also observed in differentiated layers of the epidermis (Figure 2H) ▶ , and almost completely co-localized with TGase 1 enzyme at the cell periphery (Figure 2I) ▶ . Some fluorescein isothiocyanate-staining of cell debris and scabs at the wound sites and the stratum corneum was nonspecific. In contrast, loricrin expression was not evident at the wound edge and migrating epidermis (Figure 2F) ▶ . Even in the thickened epidermis, where the abundant expression of TGase 1 and involucrin was observed, loricrin expression was limited to a few layers under the stratum corneum. The expression patterns of TGase 1 enzyme, involucrin, and loricrin were unchanged on the opposite side of the wound edge examined in those samples (data not shown).
Figure 2.
Expression of TGase 1, involucrin, and loricrin in injured neonatal mouse skin. Intact skins (A–C) or injured skins 48 hours after wounding (D–F) were stained with TGase 1 (A and D), involucrin (B and E), or loricrin (C and F) antibodies and were analyzed by confocal fluorescence imaging as described under Materials and Methods. G and H: A higher magnification of D and E showing the migrating edge of keratinocytes expressing TGase 1 and involucrin, respectively. I: A composite image of G over H, showing co-localization of TGase 1 and involucrin at the cell periphery. The arrows show the initial wound sites and the arrowheads indicate the migrating edge of the epithelium. Scale bar, 100 μm (A–F); 25 μm (G–I).
Retardation of Wound Healing with Sustained K6α Gene Expression in the TGase 1−/− Grafted Skins
To elucidate the contribution of TGase 1 in wound healing, we examined the effect of TGase 1 deficiency on epidermal regeneration. Because of the neonatal lethality of TGase 1−/− mice, 20 we grafted their skins onto athymic nude mice and examined histological changes and K6α mRNA expression during wound healing of those TGase 1−/− skin grafts. Two weeks after transplantation, skin grafts from normal neonates resembled normal adult mouse skin (Figure 3A) ▶ , where the expression of K6α mRNA was barely detectable in the epidermis (Figure 3B) ▶ . In contrast, grafted skins from TGase 1−/− neonates showed thickened epidermis with massive stratum corneum and immature hair follicles (Figure 3G) ▶ . K6α mRNA expression was evident in the epidermis (Figure 3H) ▶ , suggesting hyperproliferation of the epidermis. An injury was then made in the center of each graft, and those tissues were harvested up to 11 days after wounding. At 48 hours after the injury, the wounds were covered with bloody crusts but no apparent difference was observed between wounds in control and in TGase 1−/− skin. In the control grafts, epithelial spurs were elongated from the wound edges and had invaded under the scab, and finally closed 5 days after the injury (Figure 3C) ▶ . At that time, K6α mRNA remained around the wound site, but its expression was decreasing in the re-epithelialized epidermis (Figure 3D) ▶ . On the other hand, at that same time, wounded TGase 1−/− skin grafts still had large clots in the center, under which epithelial migration was apparently interrupted (Figure 3I) ▶ . A strong induction of K6α mRNA was evident in the migrating epidermis (Figure 3J) ▶ . At 11 days after wounding, the control grafts had matured with hair follicles (Figure 3E) ▶ and the K6α mRNA expression had subsided. However, in TGase 1−/− skin grafts, epithelial spurs had just fused in each center of the wound (Figure 3K) ▶ and a diffuse expression pattern of K6α mRNA remained. Thus, the regeneration of the epidermis was markedly delayed and the expression of K6α was uncontrolled in TGase 1−/− grafted skins.
Figure 3.
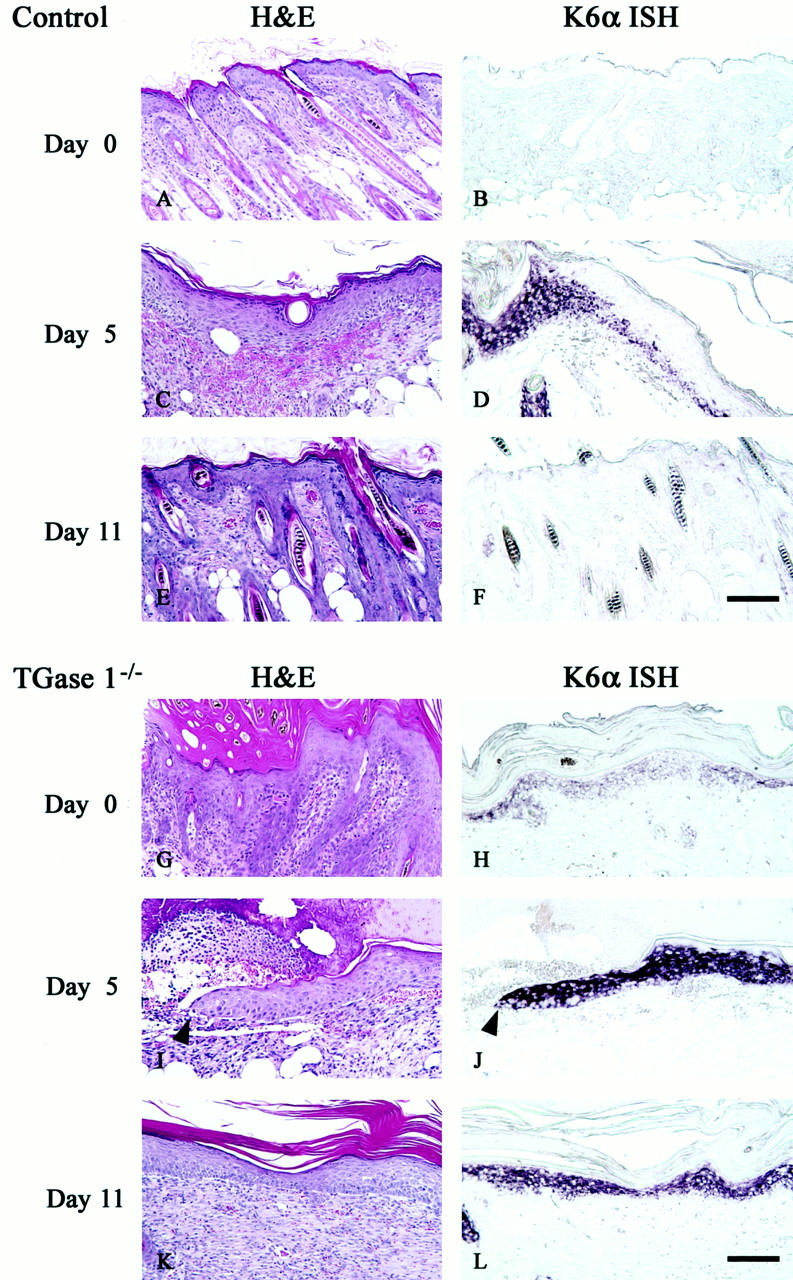
Wound healing and induction of K6α mRNA of control and TGase 1−/− skins grafted on nude mice. Skins from control (A–F) and TGase 1−/− neonates (G–L) were grafted onto nude mice. Two weeks after transplantation (A, B, G, H), an injury was made with a 3-mm diameter punch in the center of each graft. Wounded grafts were harvested at 5 (C, D, I, J) and 11 days (E, F, K, L) after the injury. The tissues were stained with H&E (A, C, E, G, I, K), or were subjected to in situ hybridization of K6α mRNA (B, D, F, H, J, L). The control skin grafts mimic adult mouse skin (A) and K6α mRNA is hardly evident (B). The wounds are already closed at day 5 (C), at which time K6α mRNA remains around the initial wound site and is seen only weakly in the regenerated epidermis (D). The control skin grafts are completely remodeled (E) and the K6α mRNA expression subsides (F) 11 days after wounding. On the contrary, the TGase 1−/−-grafted skins show acanthosis with markedly thickened stratum corneum and irregular hair follicles (G), where K6α mRNA is evident (H). At day 5, the wounded TGase 1−/− grafts are covered with a large clot under which epithelial migration is apparently interrupted (I) and K6α mRNA is strongly induced in the migrating epidermis (J). Arrowheads indicate the tip of the migrating epithelium. The re-epithelialization of the TGase 1−/−-grafted skins has been completed at 11 days after wounding (K), but the expression of K6α mRNA is still sustained (L). Scale bar, 100 μm.
Discussion
In the present study, we provide evidence that the TGase 1 gene is activated at very early stages of wound healing. The early induction of TGase 1 gene expression might result from cytokines released from keratinocytes in response to cell damage or mechanical stimuli (the free edge effect), although such triggering factors are still unidentified. TGase 1 gene induction at the wound site occurs earlier than the massive infiltration of leukocytes. However, cytokines or lymphokines released by those infiltrating cells, such as tumor necrosis factor-α, 31,32 interferon-γ, 33,34 interleukin-1, 34 and/or activin A, 35,36 might participate in the maintenance of TGase 1 gene expression during wound healing, because those cytokines are able to activate the TGase 1 gene.
The regulation of TGase 1 and K6 genes differ from each other in normal interfollicular epidermis because the TGase 1 gene is primarily expressed at late stages of keratinization, whereas K6 gene activation is not evident except for limited epithelia. 26,37 The machinery for K6α and β gene activation may not be identical to each other because the induction of those mRNAs do not necessarily co-localize in the wounded epidermis. However, the following evidence suggests that regulatory systems common to TGase 1 and those K6 genes are activated in the wound healing process: 1) up-regulation of the TGase 1 gene occurs as early as the K6 genes in injured neonatal mouse skin; 2) the time course of TGase 1 gene expression overlaps that of the K6 genes during wound healing; and 4) the 5′ upstream promoter of those genes are responsive to treatment with the phorbol ester 12-O-tetradecanoylphorbol-13-acetate in the epidermis of TGM1-LacZ and K6-LacZ transgenic mice. 26,37,38 The Jun transcription factor might mediate signals for induction of those genes, 39,40 but further study of the complex mechanisms involved in gene regulation is necessary to explore systems orchestrating epidermal regeneration.
The expression of K16, a type I partner for the K6 protein, leads to retraction of tonofilaments in keratinocytes and allows cell movement for re-epithelialization during wound healing. 41 There is no doubt that the abundant generation of TGase 1 enzyme is required to prepare for the rapid remodeling of the stratum corneum, but there are no obvious reasons for its expression in the leading edges of migrating keratinocytes. The expression of TGase 1 enzyme occurs with involucrin expression in migrating keratinocytes, and eventually they are co-localized on the plasma membrane to form premature cornified envelopes. In contrast, loricrin is not expressed in the leading edges and its expression is restricted to the granular layers behind the migrating epidermis. Those premature cornified envelopes might provide mechanical strength to migrating epidermal cells dissecting the wound between the collagenous dermis and the fibrin eschar. During the process, leading-edge keratinocytes release tissue plasminogen activator, 42 urokinase-type PA, 43 and various matrix metalloproteinases 44 that degrade the extracellular matrix for the dissection. Stabilized membrane structures reinforced by protein cross-linking might protect keratinocytes themselves from proteolytic damage. Similar premature expression of the TGase 1 gene 45,46 and involucrin 47 have been found in hyperplastic psoriatic epidermis, where it is evident that thinner premature cornified envelopes are generated. 48 Analysis of the physical and biochemical nature of the prematurely cross-linked cornified envelope will be necessary to elucidate the role of TGase 1 in wound healing as well as in psoriasis.
We also demonstrate the substantial delay in epidermal migration with sustained expression of the K6α gene during wound repair of TGase 1−/− grafted skins. The evidence suggests that TGase 1 is essential to early wound healing of the injured skin. Neonatal TGase 1−/− mice show severely impaired skin barrier function, 20 but it is unlikely that the aberrant skin barrier function directly affects epidermal regeneration, because skin barrier function has been restored to normal levels in grafted TGase 1−/− skins, possibly by the massive thick covering of scales even though the cornified envelope is lacking (unpublished data). Hiiragi and colleagues 27 have shown that TGase 1 is concentrated in adherence junctions in simple epithelial cells. The minor expression of TGase 1 in simple epithelia, including endothelial cells other than keratinocytes, might play a role in wound healing. However, TGase 1 expression is not evident in any components of neonatal mouse skin other than the epidermis and inner root sheaths.
In conclusion, TGase 1 is important not only for development and maturation of the epidermis, but also for the progression of cutaneous regeneration. The TGase 1 gene is an early responder gene to acute cutaneous injury, and is possibly involved in major changes in keratinocyte cytoarchitecture for migration into the wound site.
Footnotes
Address reprint requests to Kiyofumi Yamanishi, Department of Dermatology, Kyoto Prefectural University of Medicine, Kamigyo-ku, Kyoto 602-8566, Japan. E-mail: kyamanis@derm.kpu-m.ac.jp.
Supported in part by a grant-in-aid from the Ministry of Education, Science, Sports, and Culture of Japan and by the Nakatomi Foundation.
References
- 1.Folk JE: Transglutaminases. Annu Rev Biochem 1980, 49:517-531 [DOI] [PubMed] [Google Scholar]
- 2.Greenberg CS, Birckbichler PJ, Rice RH: Transglutaminases: multifunctional cross-linking enzymes that stabilize tissues. FASEB J 1991, 5:3071-3077 [DOI] [PubMed] [Google Scholar]
- 3.Singer AJ, Clark RA: Cutaneous wound healing. N Engl J Med 1999, 341:738-746 [DOI] [PubMed] [Google Scholar]
- 4.Bowness JM, Tarr AH, Wong T: Increased transglutaminase activity during skin wound healing in rats. Biochim Biophys Acta 1988, 967:234-240 [DOI] [PubMed] [Google Scholar]
- 5.Muszbek L, Adany R, Mikkola H: Novel aspects of blood coagulation factor XIII. I. Structure, distribution, activation, and function. Crit Rev Clin Lab Sci 1996, 33:357-421 [DOI] [PubMed] [Google Scholar]
- 6.Haroon ZA, Hettasch JM, Lai TS, Dewhirst MW, Greenberg CS: Tissue transglutaminase is expressed, active, and directly involved in rat dermal wound healing and angiogenesis. FASEB J 1999, 13:1787-1795 [DOI] [PubMed] [Google Scholar]
- 7.George MD, Vollberg TM, Floyd EE, Stein JP, Jetten AM: Regulation of transglutaminase type II by transforming growth factor-β 1 in normal and transformed human epidermal keratinocytes. J Biol Chem 1990, 265:11098-11104 [PubMed] [Google Scholar]
- 8.Suto N, Ikura K, Sasaki R: Expression induced by interleukin-6 of tissue-type transglutaminase in human hepatoblastoma HepG2 cells. J Biol Chem 1993, 268:7469-7473 [PubMed] [Google Scholar]
- 9.Kuncio GS, Tsyganskaya M, Zhu J, Liu SL, Nagy L, Thomazy V, Davies PJ, Zern MA: TNF-α modulates expression of the tissue transglutaminase gene in liver cells. Am J Physiol 1998, 274:G240-G245 [DOI] [PubMed] [Google Scholar]
- 10.Nunes I, Gleizes PE, Metz CN, Rifkin DB: Latent transforming growth factor-β binding protein domains involved in activation and transglutaminase-dependent cross-linking of latent transforming growth factor-β. J Cell Biol 1997, 136:1151-1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greenberg CS, Achyuthan KE, Borowitz MJ, Shuman MA: The transglutaminase in vascular cells and tissues could provide an alternate pathway for fibrin stabilization. Blood 1987, 70:702-709 [PubMed] [Google Scholar]
- 12.Achyuthan KE, Rowland TC, Birckbichler PJ, Lee KN, Bishop PD, Achyuthan AM: Hierarchies in the binding of human factor XIII, factor XIIIa, and endothelial cell transglutaminase to human plasma fibrinogen, fibrin, and fibronectin. Mol Cell Biochem 1996, 162:43-49 [DOI] [PubMed] [Google Scholar]
- 13.Barsigian C, Stern AM, Martinez J: Tissue (type II) transglutaminase covalently incorporates itself, fibrinogen, or fibronectin into high molecular weight complexes on the extracellular surface of isolated hepatocytes: use of 2-[(2-oxopropyl)thio] imidazolium derivatives as cellular transglutaminase inactivators. J Biol Chem 1991, 266:22501-22509 [PubMed] [Google Scholar]
- 14.Mosher DF, Schad PE: Cross-linking of fibronectin to collagen by blood coagulation Factor XIIIa. J Clin Invest 1979, 64:781-787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aeschlimann D, Paulsson M: Cross-linking of laminin-nidogen complexes by tissue transglutaminase: a novel mechanism for basement membrane stabilization. J Biol Chem 1991, 266:15308-15317 [PubMed] [Google Scholar]
- 16.Prince CW, Dickie D, Krumdieck CL: Osteopontin, a substrate for transglutaminase and factor XIII activity. Biochem Biophys Res Commun 1991, 177:1205-1210 [DOI] [PubMed] [Google Scholar]
- 17.Sane DC, Moser TL, Pippen AM, Parker CJ, Achyuthan KE, Greenberg CS: Vitronectin is a substrate for transglutaminases. Biochem Biophys Res Commun 1988, 157:115-120 [DOI] [PubMed] [Google Scholar]
- 18.Raghunath M, Hopfner B, Aeschlimann D, Lüthi U, Meuli M, Altermatt S, Gobet R, Bruckner-Tuderman L, Steinmann B: Cross-linking of the dermo-epidermal junction of skin regenerating from keratinocyte autografts: anchoring fibrils are a target for tissue transglutaminase. J Clin Invest 1996, 98:1174-1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mansbridge JN, Knapp AM: Changes in keratinocyte maturation during wound healing. J Invest Dermatol 1987, 89:253-263 [DOI] [PubMed] [Google Scholar]
- 20.Matsuki M, Yamashita F, Ishida-Yamamoto A, Yamada K, Kinoshita C, Fushiki S, Ueda E, Morishima Y, Tabata K, Yasuno H, Hashida M, Iizuka H, Ikawa M, Okabe M, Kondoh G, Kinoshita T, Takeda J, Yamanishi K: Defective stratum corneum and early neonatal death in mice lacking the gene for transglutaminase 1 (keratinocyte transglutaminase). Proc Natl Acad Sci USA 1998, 95:1044-1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eckert RL, Green H: Structure and evolution of the human involucrin gene. Cell 1986, 46:583-589 [DOI] [PubMed] [Google Scholar]
- 22.Mehrel T, Hohl D, Rothnagel JA, Longley MA, Bundman D, Cheng C, Lichti U, Bisher ME, Steven AC, Steinert PM, Yuspa SH, Roop DR: Identification of a major keratinocyte cell envelope protein, loricrin. Cell 1990, 61:1103-1112 [DOI] [PubMed] [Google Scholar]
- 23.Nemes Z, Marekov LN, Fesus L, Steinert PM: A novel function for transglutaminase 1: attachment of long-chain ω-hydroxyceramides to involucrin by ester bond formation. Proc Natl Acad Sci USA 1999, 96:8402-8407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huber M, Rettler I, Bernasconi K, Frenk E, Lavrijsen SPM, Ponec M, Bon A, Lautenschlager S, Schorderet DF, Hohl D: Mutations of keratinocyte transglutaminase in lamellar ichthyosis. Science 1995, 267:525-528 [DOI] [PubMed] [Google Scholar]
- 25.Russell LJ, DiGiovanna JJ, Rogers GR, Steinert PM, Hashem N, Compton JG, Bale SJ: Mutations in the gene for transglutaminase 1 in autosomal recessive lamellar ichthyosis. Nat Genet 1995, 9:279-283 [DOI] [PubMed] [Google Scholar]
- 26.Yamada K, Matsuki M, Morishima Y, Ueda E, Tabata K, Yasuno H, Suzuki M, Yamanishi K: Activation of the human transglutaminase 1 promoter in transgenic mice: terminal differentiation-specific expression of the TGM1-lacZ transgene in keratinized stratified squamous epithelia. Hum Mol Genet 1997, 6:2223-2231 [DOI] [PubMed] [Google Scholar]
- 27.Hiiragi T, Sasaki H, Nagafuchi A, Sabe H, Shen SC, Matsuki M, Yamanishi K, Tsukita S: Transglutaminase type 1 and its cross-linking activity are concentrated at adherens junctions in simple epithelial cells. J Biol Chem 1999, 274:34148-34154 [DOI] [PubMed] [Google Scholar]
- 28.Kim SY, Grant P, Lee JH, Pant HC, Steinert PM: Differential expression of multiple transglutaminases in human brain: increased expression and cross-linking by transglutaminases 1 and 2 in Alzheimer’s disease. J Biol Chem 1999, 274:30715-30721 [DOI] [PubMed] [Google Scholar]
- 29.Takahashi K, Yan B, Yamanishi K, Imamura S, Coulombe PA: The two functional keratin 6 genes of mouse are differentially regulated and evolved independently from their human orthologs. Genomics 1998, 53:170-183 [DOI] [PubMed] [Google Scholar]
- 30.Croft CB, Tarin D: Ultrastructural studies of wound healing in mouse skin I. Epithelial behaviour. J Anat 1970, 106:63-77 [PMC free article] [PubMed] [Google Scholar]
- 31.Bikle DD, Pillai S, Gee E, Hincenbergs M: Tumor necrosis factor-alpha regulation of 1,25-dihydroxyvitamin D production by human keratinocytes. Endocrinology 1991, 129:33-38 [DOI] [PubMed] [Google Scholar]
- 32.Hubner G, Brauchle M, Smola H, Madlener M, Fassler R, Werner S: Differential regulation of pro-inflammatory cytokines during wound healing in normal and glucocorticoid-treated mice. Cytokine 1996, 8:548-556 [DOI] [PubMed] [Google Scholar]
- 33.Saunders NA, Jetten AM: Control of growth regulatory and differentiation-specific genes in human epidermal keratinocytes by interferon γ: antagonism by retinoic acid and transforming growth factor β1. J Biol Chem 1994, 269:2016-2022 [PubMed] [Google Scholar]
- 34.Wei L, Debets R, Hegmans JJ, Benner R, Prens EP: IL-1β and IFN-γ induce the regenerative epidermal phenotype of psoriasis in the transwell skin organ culture system. IFN-γ up-regulates the expression of keratin 17 and keratinocyte transglutaminase via endogenous IL-1 production. J Pathol 1999, 187:358-364 [DOI] [PubMed] [Google Scholar]
- 35.Hübner G, Hu Q, Smola H, Werner S: Strong induction of activin expression after injury suggests an important role of activin in wound repair. Dev Biol 1996, 173:490-498 [DOI] [PubMed] [Google Scholar]
- 36.Seishima M, Nojiri M, Esaki C, Yoneda K, Eto Y, Kitajima Y: Activin A induces terminal differentiation of cultured human keratinocytes. J Invest Dermatol 1999, 112:432-436 [DOI] [PubMed] [Google Scholar]
- 37.Takahashi K, Coulombe PA: Defining a region of the human keratin 6a gene that confers inducible expression in stratified epithelia of transgenic mice. J Biol Chem 1997, 272:11979-11985 [DOI] [PubMed] [Google Scholar]
- 38.Heyden A, Lutzow-Holm C, Clausen OP, Brandtzaeg P, Huitfeldt HS: Expression of keratins K6 and K16 in regenerating mouse epidermis is less restricted by cell replication than the expression of K1 and K10. Epithelial Cell Biol 1994, 3:96-101 [PubMed] [Google Scholar]
- 39.Jiang CK, Magnaldo T, Ohtsuki M, Freedberg IM, Bernerd F, Blumenberg M: Epidermal growth factor and transforming growth factor α specifically induce the activation- and hyperproliferation-associated keratins 6 and 16. Proc Natl Acad Sci USA 1993, 90:6786-6790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamada K, Yamanishi K, Kakizuka A, Kibe Y, Doi H, Yasuno H: Transcriptional regulation of human transglutaminase 1 gene by signaling systems of protein kinase C, RAR/RXR and Jun/Fos in keratinocytes. Biochem Mol Biol Int 1994, 34:827-836 [PubMed] [Google Scholar]
- 41.Takahashi K, Folmer J, Coulombe PA: Increased expression of keratin 16 causes anomalies in cytoarchitecture and keratinization in transgenic mouse skin. J Cell Biol 1994, 127:505-520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grøndahl-Hansen J, Lund LR, Ralfkiær E, Ottevanger V, Danø K: Urokinase- and tissue-type plasminogen activators in keratinocytes during wound reepithelialization in vivo. J Invest Dermatol 1988, 90:790-795 [DOI] [PubMed] [Google Scholar]
- 43.Morioka S, Lazarus GS, Baird JL, Jensen PJ: Migrating keratinocytes express urokinase-type plasminogen activator. J Invest Dermatol 1987, 88:418-423 [DOI] [PubMed] [Google Scholar]
- 44.Vaalamo M, Weckroth M, Puolakkainen P, Kere J, Saarinen P, Lauharanta J, Saarialho-Kere UK: Patterns of matrix metalloproteinase and TIMP-1 expression in chronic and normally healing human cutaneous wounds. Br J Dermatol 1996, 135:52-59 [PubMed] [Google Scholar]
- 45.Michel S, Bernerd F, Jetten AM, Floyd EE, Shroot B, Reichert U: Expression of keratinocyte transglutaminase mRNA revealed by in situ hybridization. J Invest Dermatol 1992, 98:364-368 [DOI] [PubMed] [Google Scholar]
- 46.Nonomura K, Yamanishi K, Hosokawa Y, Doi H, Hirano J, Fukushima S, Yasuno H: Localization of transglutaminase 1 mRNA in normal and psoriatic epidermis by non-radioactive in situ hybridization. Br J Dermatol 1993, 128:23-28 [DOI] [PubMed] [Google Scholar]
- 47.Bernard BA, Reano A, Darmon YM, Thivolet J: Precocious appearance of involucrin and epidermal transglutaminase during differentiation of psoriatic skin. Br J Dermatol 1986, 114:279-283 [DOI] [PubMed] [Google Scholar]
- 48.Ishida-Yamamoto A, Iizuka H: Differences in involucrin immunolabeling within cornified cell envelopes in normal and psoriatic epidermis. J Invest Dermatol 1995, 104:391-395 [DOI] [PubMed] [Google Scholar]



