Abstract
The p53 homologue p63 encodes for different isotypes able to either transactivate p53 reporter genes (TAp63) or act as p53-dominant-negatives (ΔNp63). p63 is expressed in the basal cells of many epithelial organs and its germline inactivation in the mouse results in agenesis of organs such as skin appendages and the breast. Here, we show that prostate basal cells, but not secretory or neuroendocrine cells, express p63. In addition, prostate basal cells in culture predominantly express the ΔNp63α isotype. In contrast, p63 protein is not detected in human prostate adenocarcinomas. Finally, and most importantly, p63(−/−) mice do not develop the prostate. These results indicate that p63 is required for prostate development and support the hypothesis that basal cells represent and/or include prostate stem cells. Furthermore, our results show that p63 immunohistochemistry may be a valuable tool in the differential diagnosis of benign versus malignant prostatic lesions.
p63 is a recently cloned homologue of the p53 tumor suppressor gene. 1-4 In contrast to p53, the p63 gene encodes for at least six major isotypes. 4 Three isotypes (TAp63α, TAp63β, and TAp63γ) contain the transactivating (TA) domain and are able to transactivate p53 reporter genes and induce apoptosis. In contrast, the other three isotypes (ΔNp63α, ΔNp63β, and ΔNp63γ) are transcribed from an internal promoter localized within intron 3, lack the TA domain, and act as dominant-negatives to suppress transactivation by both p53 and TAp63 isotypes. p63 is selectively expressed in the basal cell compartment of a variety of epithelial tissues. 4,5 Further, p63-deficient mice show severe defects in the development of epithelial organs that express p63 protein, namely agenesis of squamous epithelia, mammary, salivary, and lachrymal glands. 6,7 Altogether, these results suggest that p63 may be essential for the maintenance of a stem cell population in various epithelial tissues.
Knowledge of the molecular events underlying prostate development is limited. Normal prostate epithelium consists of three different types of cells: secretory, basal, and neuroendocrine. A subset of cells that is morphologically and immunophenotypically intermediate between basal and secretory cells has also been identified within the normal prostate epithelium. 8-11 This observation has lead to the hypothesis that basal cells represent the precursors of secretory cells. In contrast to this hypothesis, cell kinetic studies in the rat prostate suggest that basal and secretory cells are independent lineages with self-renewal capacities. 12,13 Therefore, the existence of a prostate stem cell able to give rise to both basal and secretory cells remains highly controversial.
We have previously shown that both human and mouse prostate basal cells express p63 protein (see Yang et al 4 and data not shown). This result suggests that p63 may play a critical role in prostate development by maintaining a prostate stem cell population. However, because the mouse prostate starts to develop only during the last few days of gestation 14,15 and p63(−/−) mice die at birth, assessment of prostate development in these mice requires a very accurate morphological analysis and no data are currently available in the literature.
In the present study, we first confirmed that p63 represents a selective marker of basal cells within the prostatic epithelium by analyzing p63 expression in a series of normal prostates and in normal, basal PrEC prostate cells. Second, because it has been demonstrated that prostate cancers express markers of secretory cells and are usually negative for basal cell markers, 16,17 we analyzed p63 expression in a series of 130 prostatic carcinomas and in prostate cancer cell lines. Finally, to assess the role of p63 in prostate development we histologically analyzed the periurethral region in day 1, p63(−/−) male mice.
Our results show that p63 is a reliable prostate basal cell marker and that the ΔNp63α isotype is the most abundantly represented in normal prostate basal (PrEC) cells. Because p63 protein is consistently undetectable in prostate cancers, we propose that p63 expression may be used in the differential diagnosis between benign and malignant lesions of the prostate. Finally and most importantly, our results indicate that p63 expression is necessary for the normal development of the mouse prostate, suggesting that p63-positive basal cells may represent/include prostate stem cells.
Materials and Methods
Prostatectomy Specimens and Prostate Cell Lines
This study was performed after approval by the Institutional Review Board of the Dana Farber Cancer Institute and Brigham and Women’s Hospital.
One hundred thirty prostate cancer specimens, were retrieved from the files of the departments of Pathology of the Brigham and Women’s Hospital, the Beth Israel Deaconess Medical Center, Boston, MA, and the University of Ancona, Italy, and used in immunohistochemistry experiments. The prostate cancer patients analyzed included 29 patients treated with total androgen ablation therapy for 3 months before surgery (Zoladex depot every 28 days plus Casodex 50 mg/day for 12 weeks) and 101 patients who did not receive any treatment before surgery.
Normal basaloid PrEC prostate cells (Clonetics, Walkersville, MD) and LNCaP, PC3, and DU145 prostate cancer cell lines (ATCC, Rockville, MD) were used in immunohistochemistry, immunoblotting, and real-time polymerase chain reaction (PCR) experiments. PC3 cells were also used for transfection experiments. Normal stromal PrSC prostate cells (Clonetics) were used in immunoblotting experiments.
Immunohistochemistry
Immunostaining was performed in all tissue specimens and paraffin-embedded cell lines using the 4A4 anti-p63 antibody, which recognizes all six p63 isotypes. 4 A subset of 58 samples was double-immunostained for p63 and HMWCK (34βE12; DAKO, Carpinteria, CA). Double-immunostaining for p63 and chromogranin A (Novocastra Laboratories Ltd., Newcastle on Tyne, UK) was performed in 10 samples.
For p63 immunostaining, 5-μm sections were deparaffinized, rehydrated, and subjected to microwaving in 10 mmol/L citrate buffer, pH 6.0 (BioGenex, San Ramon, CA) in a 750 W oven for 15 minutes. Slides were allowed to cool at room temperature for 30 minutes. The 4A4 antibody (1:50 dilution) was applied at room temperature for 2 hours in an automated stainer (Optimax Plus 2.0 bc; BioGenex, San Ramon, CA). Detection steps were performed by the instrument using the MultiLink-HRP kit (BioGenex). Peroxidase activity was localized using 3,3-diaminobenzidine or 3,3-diaminobenzidine-nickel chloride. Standardized development time periods allowed accurate comparison of all samples.
For double-immunostaining experiments, tissue sections were subsequently incubated with the second antibody (anti-HMWCK or anti-chromogranin A) at 1:50 dilution for 30 minutes. Detection was performed using alkaline-phosphatase-conjugated streptavidin and new fuchsin (BioGenex).
Substitution of the primary antibody with phosphate-buffered saline (PBS) served as negative-staining control.
Immunoblot Analysis
PrEC, PrSC, LNCaP, PC3, and DU145 cells were lysed in 200 μl of lysis buffer (50 mmol/L Tris, pH 7.5, 250 mol/L NaCl, 0.1% Triton X-100, 1 mmol/L ethylenediaminetetraacetic acid, 50 mmol/L NaF, 0.1 mmol/L Na3VO4) containing 1 mmol/L dithiothreitol, 1 mmol/L phenylmethyl sulfonyl fluoride, and protease inhibitor cocktail (Boehringer Mannheim, Indianapolis, IN). Immunoblotting was performed as previously described 18 using the 4A4 anti-p63 antibody at 1:250 dilution.
Cell Transfection
PC3 cells were maintained in F-12 nutrient mixture medium supplemented with 8% fetal bovine serum (Life Technologies, Inc., Gaithersburg, MD). A pCDNA3 vector encoding myc epitope-tagged ΔNp63α isotype 4 was transfected in PC3 cells using FuGENE 6 transfection reagent (Boehringer Mannheim) according to the manufacturer’s direction. Briefly, PC3 cells were incubated at 37°C in a 250-ml flask with a solution consisting of 20 μg of DNA, 30 μl of FuGENE 6 transfection reagent, and F-12 nutrient mixture medium supplemented with 8% fetal bovine serum. Cells were harvested 24 hours after transfection.
Real-Time Quantitative PCR (Taqman PCR)
RNA Extraction and Reverse Transcription (RT)
Total RNA was extracted from PrEC, LNCaP, PC3, and DU145 cells with the use of the RNeasy mini kit (Qiagen, Chatsworth, CA), according to manufacturer’s directions. For cDNA synthesis, 1 μg of total RNA was reverse-transcribed in a 20-μl reaction mixture containing 250 μmol/L of each dNTP, 20 U of RNase inhibitor, 50 U of MuLV reverse transcriptase, 2.5 μmol/L random hexamers, and 1× buffer (1.5 mmol/L MgCl2) (all reagents were purchased from PE Applied Biosystems, Foster City, CA). The reaction mix was incubated at 42°C for 45 minutes and then denatured at 99°C for 5 minutes. For each sample, a control reaction not containing the reverse transcriptase enzyme was also performed.
Real-Time PCR
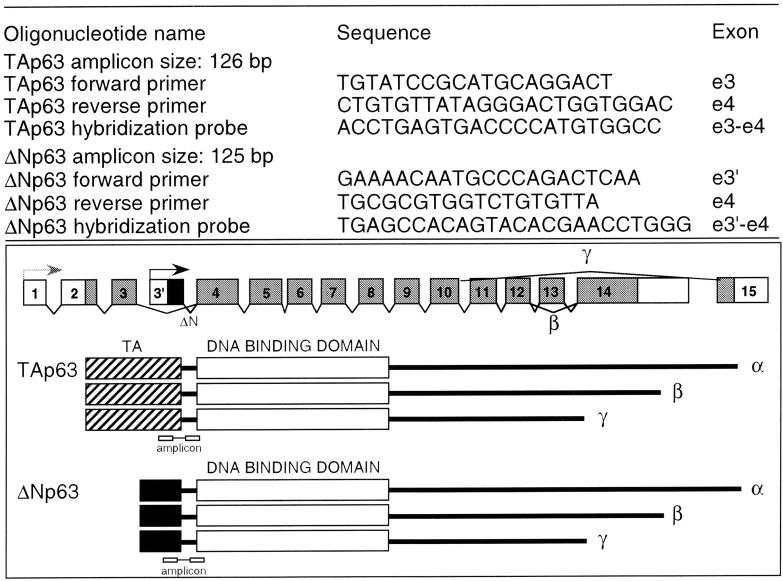
Specific primers and probe sets for TAp63 and ΔNp63 isotypes (Figure 1) ▶ were designed from sequences in the GenBank database using the Primer Express 1.0 Software (PE Applied Biosystems). The primers and hybridization probes spanned an intron to exclude annealing to genomic DNA. The housekeeping glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene was used as endogenous control to standardize the amount of RNA in each reaction (Taqman GAPDH control reagents). All primers and probes were synthesized by PE Applied Biosystems. PCR was performed on the cDNA samples using an ABI PRISM 7700 Sequence Detector (PE Applied Biosystems). The Taqman PCR Core Reagent Kit (PE Applied Biosystems) was used according to the manufacturer’s protocol with the following modifications: dUTP was replaced by dTTP and incubation with AmpErase was omitted. For each sample tested, PCR reaction was performed in a 50 μl volume containing 2 μl of cDNA reaction (equivalent to 100 ng of template RNA) and 2.5 U of AmpliTaq Gold. Oligonucleotide primers and fluorogenic probes were added to a final concentration of 100 nmol/L each. The amplification step consisted of 60 cycles of 94°C for 45 seconds, 54°C for 45 seconds, and 64°C for 1 minute.
Figure 1.
Sequences, amplicon sizes, and exon localizations of primers and probes used for Taqman® PCR experiment.
In each experiment, additional reactions with seven serial twofold dilutions of PrEC cDNA as template were performed with each set of primers and probes (TAp63, ΔNp63, and GAPDH) on the same 96-well plate to generate standard curves, which related the threshold cycle (CT) to the log input amount of template. All samples were amplified in triplicates. The relative amount of each p63 transcript in each cell line was determined by using the standard curve method and by normalizing for GAPDH mRNA expression levels, as described in detail in the ABI PRISM Sequence Detection System User Bulletin No. 2 (PE Applied Biosystems) and elsewhere. 19 The amplification efficiencies for ΔNp63 and TAp63 transcripts were also calculated. Because they were approximately similar (TA/ΔN relative efficiency curve had a slope value of 0.91), ΔNp63 and TAp63 transcripts levels could be compared with reasonable accuracy.
Prostate Analysis of p63(−/−) and Wild-Type Newborn Mice
p63(−/−) mice were generated as previously described. 6 The periurethral region of newborn p63(−/−) male mice and of newborn wild-type B57Bl/6 male mice (controls) was analyzed. Specifically, the caudal portion of each animal was sectioned along a coronal plane, fixed in 10% buffered formalin, and embedded in paraffin. Histological examination was performed on paraffin sections stained with hematoxylin and eosin. Coronal sections of the entire length of the urethra were examined. In addition, the periurethral region of both newborn p63(−/−) and wild-type male mice was immunostained for p63 as described in the Immunohistochemistry section.
Results
p63 Protein Is Selectively Expressed in the Nuclei of Basal Cells of Normal Prostate Glands
Prostatectomy Specimens
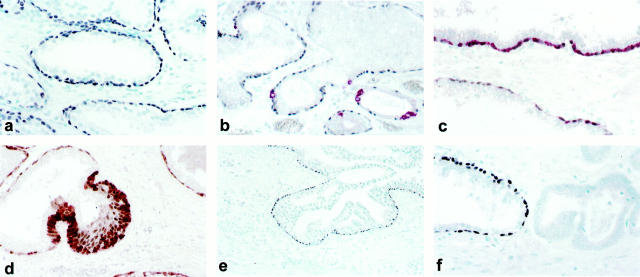
We first determined the expression of p63 protein in normal prostate tissue. All prostatectomy specimens showed universal p63 immunostaining of basal cell nuclei, whereas secretory cells were consistently negative (Figure 2a) ▶ .
Figure 2.
a: Normal prostate gland showing selective p63 nuclear expression in basal cells. b: Double-immunostaining for p63 (black nuclear staining) and chromogranin A (red cytoplasmic staining): neuroendocrine cells do not express p63. c: Double-immunostaining for p63 (black nuclear staining) and HMWCK (red cytoplasmic staining): the majority of basal cells co-express p63 and HMWCK. A subset of basal cells expresses p63 only. d: Prostate gland with basal cell hyperplasia double-immunostained for p63 (brown nuclear staining) and HMWCK (red cytoplasmic staining): a subset of p63-positive and HMWCK-negative basal cells is identified. e: Dysplastic cells from high-grade prostatic intraepithelial neoplasia glands are negative for p63 expression whereas a rim of residual p63-positive basal cells can be identified. f: Prostate cancer cells do not express p63 (left) whereas an adjacent normal prostate gland (right) shows p63 expression in basal cells.
To assess p63 expression in neuroendocrine cells and to test for p63 and HMWCK co-expression in basal cells, double-immunostaining for p63/chromogranin A and p63/HMWCK, respectively, was performed. Neuroendocrine cells did not express p63 protein (Figure 2b) ▶ . Double-immunostaining for p63 and HMWCK showed co-localization of the two antigens in the majority of basal cells. However, a minority (∼1 to 5%) of p63-positive/HMWCK-negative basal cells was identified (Figure 2, c and d) ▶ .
Normal Prostate Cells (PrEC and PrSC)
PrEC and PrSC cells are commercially available, normal prostate basal and stromal cells, respectively. We confirmed by immunohistochemistry the basal cell phenotype of PrEC cells (positivity for HMWCK and negativity for androgen receptor and prostate-specific antigen) (Signoretti and Loda, unpublished data).
By immunohistochemistry, 80% of PrEC cells expressed p63 protein in the nuclei (Figure 3a) ▶ . Immunoblot analysis of PrEC cell lysate with anti-p63 antibody showed a major band at ∼80 kd and a fainter band at ∼60 kd (Figure 3e) ▶ . In contrast, PrSC cells were negative for p63 expression by immunoblotting (not shown). Immunoblotting of PC3 cells transfected with a mammalian expression vector encoding myc epitope-tagged ΔNp63α isotype generated a band that migrated slightly slower than the 80 kd band obtained with PrEC cells (not shown). These results are consistent with our previous data obtained in primary human foreskin keratinocytes. 4 Taken together our results suggest that ΔNp63α is the predominant p63 isotype in PrEC cells.
Figure 3.
a: p63 protein is expressed in the nuclei of ∼80% of normal basal prostate PrEC cells by immunohistochemistry. LNCaP (b), PC3 (c), and DU145 (d) cells do not express p63 protein. e: Immunoblotting of PrEC cell lysate (lane 1) with anti-p63 antibody shows a major band at ∼80 kd and a fainter band at ∼60 kd. LNCaP (lane 2), PC3 (lane 3) and DU145 (lane 4) cells are negative for p63 protein expression by immunoblotting.
p63 Protein Is Not Expressed in Prostatic Intraepithelial Neoplasia and Invasive Prostate Cancer
Prostatectomy Specimens
We next examined p63 protein expression in 130 human prostate cancers. HMWCK expression was concomitantly analyzed in a subset of 58 cases by double-immunostaining.
High-grade prostatic intraepithelial neoplasia was detected in 48 of 130 prostatectomy specimens. In all prostatic intraepithelial neoplasia cases, dysplastic cells were negative for both p63 and HMWCK, but a rim of residual p63-positive basal cells could be identified (Figure 2e) ▶ .
One hundred twenty-six of 130 (97%) invasive prostate cancers were negative for p63 (Figure 2f) ▶ , whereas in four cases, <1% of cells were positive for both p63 and HMWCK (not shown). In contrast, in three cases a minority of cells (<5%) was positive for HMWCK but negative for p63 (not shown).
Prostate Cancer Cell Lines (LNCaP, PC3, DU145)
By both immunoblot and immunohistochemical analysis LNCaP, PC3, and DU145 cells were negative for p63 protein expression (Figure 3, b–e) ▶ .
ΔNp63 Is the Most Abundant p63 Isotype in PrEC Cells
Immunoblot analysis showed that ΔNp63α likely represents the most abundant p63 protein in PrEC cells whereas prostate tumor cell lines are negative for p63 expression. To investigate the relative abundance of p63 isotypes at the RNA level, ΔNp63 and TAp63 transcripts levels were compared in PrEC, LNCaP, PC3, and DU145 cells by real-time relative quantitative PCR (TaqMan-PCR). As compared to PrEC cells and after normalization for GAPDH mRNA levels, ΔNp63 mRNA levels were at least 2,000 times lower in PC3 cells and were undetectable in LNCaP and DU145 cells. TAp63 mRNA levels were similar in PrEC and PC3 cells (ratio PC3/PrEC = 1.3) and were significantly lower in LNCaP and DU145 cells with 14-fold and sevenfold decreases, respectively. Reactions in which the reverse transcriptase enzyme was omitted did not yield significant amplification (data not shown). These results indicate that ΔNp63 transcripts are virtually absent in prostate cancer cell lines, whereas TAp63 mRNA levels are detectable in varying amounts in all three cancer cell lines tested.
In PrEC cells, the threshold cycle (CT) for ΔNp63 amplification was significantly lower as compared to the CT for TAp63 (23 versus 30 when 100 ng of RNA were used as template) (not shown), indicating that ΔNp63 transcripts are expressed at significantly higher levels than TAp63 transcripts in these cells.
p63 Is Essential for Normal Prostate Development in the Mouse
Histological examination of the entire length of the urethra in newborn p63(−/−) male mice revealed absence of the prostate. Specifically, no ducts or epithelial budding structures could be identified either in the ventral or in the dorsolateral region of the periurethral mesenchyma (Figure 4b) ▶ . Conversely, in control mice, epithelial structures were present in both the ventral and dorsolateral portions of the periurethral region (Figure 4a) ▶ . The epithelial structures consisted of both solid buds and small glands. By immunohistochemistry, both urothelial and prostatic basal cells of wild-type mice expressed p63 protein (Figure 4a ▶ , inset). In contrast, p63 expression could not be detected in either epithelial or stromal cells of p63(−/−) mice (Figure 4b ▶ , inset).
Figure 4.
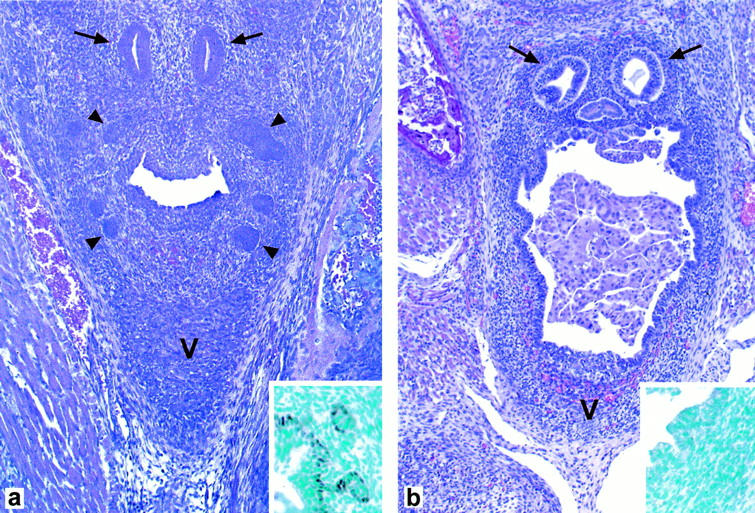
a: A coronal section of the periurethral region of a wild-type mouse shows the presence of prostatic buds (arrowheads) in both the ventral (V) and dorsolateral periurethral region. Arrows indicate ejaculatory ducts. By immunohistochemistry, basal urothelial and prostatic cells express p63 (inset). b: A coronal section of the periurethral region of a p63(−/−) mouse shows complete absence of prostatic ducts or budding structures in both the ventral (V) and dorsolateral region of the periurethral mesenchyma. p63 expression could not be detected in either epithelial or stromal cells by immunohistochemistry (inset). Ejaculatory ducts (arrows) are present in the posterior periurethral region. Detached urothelial cells are present within the urethral lumen.
Discussion
As previously demonstrated in the hematopoietic system, cancers arise from cells arrested at various stages of differentiation. 20 As a result, the identification of stem cells and subsequent stages of differentiation in normal tissues is an essential step in understanding the mechanisms involved in neoplastic transformation.
In normal prostate epithelium, the lineage relationships between secretory, basal, and neuroendocrine cells, and the existence of a common precursor, are still matters of debate. The vast majority of prostate cancers express markers of secretory cells such as androgen receptor and prostate-specific antigen, and are negative for basal cell markers. 16,17 Consequently, it has been generally accepted that prostate cancer arises from malignant transformation of secretory cells. However, prostate carcinomas may also express genes characteristic of basal cells. 17,21-25 Thus, the cell from which prostate cancer arises is still unknown.
The p63 gene is expressed exclusively in the basal cells of several epithelial organs and has been suggested to play a major role in the maintenance of the stem cell compartment in these organs. 4,6,7 Here, in a large series of prostate specimens, we confirm that the p63 protein is selectively expressed in the nuclei of basal cells of normal prostate glands. In addition, we show that homozygous inactivation of this gene in the mouse results in prostate agenesis.
Because p63 is selectively expressed in adult prostate basal cells and it is undetectable in adult prostate stromal cells both in vivo and in vitro, the defect in prostate development in p63(−/−) mice is likely ascribed to the epithelial component. However, p63 may be essential for either maintaining a prostate epithelial stem cell population or sustaining a basal cell population, which does not represent a stem cell compartment per se but is essential for prostate development (eg, signaling to the surrounding mesenchyma).
The p63(−/−) mouse is the first engineered animal model demonstrating prostate agenesis. Further manipulation of this animal model will certainly provide a unique tool to study prostate development, differentiation, and neoplastic transformation.
In prostate basal cells grown in vitro, the predominantly expressed p63 isotype is the ΔNp63α that lacks the transactivating domain. This isotype has been shown to act as a dominant-negative on the transactivation by both TAp63 and p53. 4 We have previously shown that keratinocytes differentiation in vitro is associated with down-regulation of the ΔNp63 transcripts. 5 Because secretory cells do not express p63, we speculate that down-regulation of the ΔNp63α isotype may be required for differentiation of basal cells into secretory cells. These findings are interesting inasmuch as p53-mediated transcriptional activity increases in a variety of differentiating cells including muscle cells and keratinocytes, despite unchanged 26 or decreased p53 protein levels. 27 In addition, transfection of mutant p53 27 or dominant-negative p53 26 results in a lack of p53-dependent transactivation and differentiation, suggesting that transcription of p53-regulated genes is necessary for differentiation. However, because terminal differentiation occurs in p53(−/−) cells, p53 homologues, including TAp63, may have overlapping functions to those of p53 in differentiation. Altogether, these data suggest that relief of the dominant-negative function of ΔNp63 in basal cells may be necessary for activating the p53-dependent gene transcription machinery required for the differentiation of these cells into secretory cells.
We have demonstrated that p63 is not expressed in prostate carcinomas. This finding supports the hypothesis that prostatic carcinomas have a secretory phenotype. Because we have shown that p63 is expressed in virtually all basal cells, including a cell subset negative for HMWCK, p63 immunohistochemistry may be a valuable tool for the differential diagnosis of benign versus malignant prostatic lesions.
In conclusion, the basal cell marker p63 is essential for prostate development in the mouse suggesting that prostate basal cells may represent and/or include prostate stem cells. In addition, because of the universal expression of p63 by basal cells, p63 immunohistochemistry may be a useful adjunct to morphological analysis in the prostate surgical pathology setting.
Footnotes
Address reprint requests to Massimo Loda, Department of Adult Oncology, Dana Farber Cancer Institute, Dana 740B, 44 Binney St., Boston, MA 02215. E-mail: massimo_loda@dfci.harvard.edu.
Supported by grants from CaPCURE, National Cancer Institute (CA, 81755–03), and Department of Defense (DAMD 17 98-8574).
References
- 1.Osada M, Ohba M, Kawahara C, Ishioka C, Kanamaru R, Katoh I, Ikawa Y, Nimura Y, Nakagawara A, Obinata M, Ikawa S: Cloning and functional analysis of human p51, which structurally and functionally resembles p53. Nat Med 1998, 4:839-843 [DOI] [PubMed] [Google Scholar]
- 2.Senoo M, Seki N, Ohira M, Sugano S, Watanabe M, Inuzuka S, Okamoto T, Tachibana M, Tanaka T, Shinkai Y, Kato H: A second p53-related protein, p73L, with high homology to p73. Biochem Biophys Res Commun 1998, 248:603-607 [DOI] [PubMed] [Google Scholar]
- 3.Trink B, Okami K, Wu L, Sriuranpong V, Jen J, Sidransky D: A new human p53 homologue. Nat Med 1998, 4:747-748 [DOI] [PubMed] [Google Scholar]
- 4.Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V, Andrews NC, Caput D, McKeon F: p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell 1998, 2:305-316 [DOI] [PubMed] [Google Scholar]
- 5.Parsa R, Yang A, McKeon F, Green H: Association of p63 with proliferative potential in normal and neoplastic human keratinocytes. J Invest Dermatol 1999, 113:1099-1105 [DOI] [PubMed] [Google Scholar]
- 6.Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, McKeon F: p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 1999, 398:714-718 [DOI] [PubMed] [Google Scholar]
- 7.Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A: p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 1999, 398:708-713 [DOI] [PubMed] [Google Scholar]
- 8.Verhagen AP, Aalders TW, Ramaekers FC, Debruyne FM, Schalken JA: Differential expression of keratins in the basal and luminal compartments of rat prostatic epithelium during degeneration and regeneration. Prostate 1988, 13:25-38 [DOI] [PubMed] [Google Scholar]
- 9.Jones EG, Harper ME: Studies on the proliferation, secretory activities, and epidermal growth factor receptor expression in benign prostatic hyperplasia explant cultures. Prostate 1992, 20:133-149 [DOI] [PubMed] [Google Scholar]
- 10.Peehl DM, Leung GK, Wong ST: Keratin expression: a measure of phenotypic modulation of human prostatic epithelial cells by growth inhibitory factors. Cell Tissue Res 1994, 277:11-18 [DOI] [PubMed] [Google Scholar]
- 11.Robinson EJ, Neal DE, Collins AT: Basal cells are progenitors of luminal cells in primary cultures of differentiating human prostatic epithelium. Prostate 1998, 37:149-160 [DOI] [PubMed] [Google Scholar]
- 12.English HF, Santen RJ, Isaacs JT: Response of glandular versus basal rat ventral prostatic epithelial cells to androgen withdrawal and replacement. Prostate 1987, 11:229-242 [DOI] [PubMed] [Google Scholar]
- 13.Evans GS, Chandler JA: Cell proliferation studies in the rat prostate: II. The effects of castration and androgen-induced regeneration upon basal and secretory cell proliferation. Prostate 1987, 11:339-351 [DOI] [PubMed] [Google Scholar]
- 14.Cunha GR, Donjacour AA, Cooke PS, Mee S, Bigsby RM, Higgins SJ, Sugimura Y: The endocrinology and developmental biology of the prostate. Endocr Rev 1987, 8:338-362 [DOI] [PubMed] [Google Scholar]
- 15.Bhatia-Gaur R, Donjacour AA, Sciavolino PJ, Kim M, Desai N, Young P, Norton CR, Gridley T, Cardiff RD, Cunha GR, Abate-Shen C, Shen MM: Roles for Nkx3.1 in prostate development and cancer. Genes Dev 1999, 13:966-977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagle RB, Ahmann FR, McDaniel KM, Paquin ML, Clark VA, Celniker A: Cytokeratin characterization of human prostatic carcinoma and its derived cell lines. Cancer Res 1987, 47:281-286 [PubMed] [Google Scholar]
- 17.Liu AY, True LD, LaTray L, Nelson PS, Ellis WJ, Vessella RL, Lange PH, Hood L, van den Engh G: Cell-cell interaction in prostate gene regulation and cytodifferentiation. Proc Natl Acad Sci USA 1997, 94:10705-10710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pagano M, Tam SW, Theodoras AM, Beer-Romero P, Del Sal G, Chau V, Yew PR, Draetta GF, Rolfe M: Role of the ubiquitin-proteasome pathway in regulating abundance of the cyclin-dependent kinase inhibitor p27. Science 1995, 269:682-685 [DOI] [PubMed] [Google Scholar]
- 19.Fink L, Seeger W, Ermert L, Hanze J, Stahl U, Grimminger F, Kummer W, Bohle RM: Real-time quantitative RT-PCR after laser-assisted cell picking. Nat Med 1998, 4:1329-1333 [DOI] [PubMed] [Google Scholar]
- 20.Sawyers CL, Denny CT, Witte ON: Leukemia and the disruption of normal hematopoiesis. Cell 1991, 64:337-350 [DOI] [PubMed] [Google Scholar]
- 21.McDonnell TJ, Troncoso P, Brisbay SM, Logothetis C, Chung LW, Hsieh JT, Tu SM, Campbell ML: Expression of the protooncogene bcl-2 in the prostate and its association with emergence of androgen-independent prostate cancer. Cancer Res 1992, 52:6940-6944 [PubMed] [Google Scholar]
- 22.Colombel M, Symmans F, Gil S, O’Toole KM, Chopin D, Benson M, Olsson CA, Korsmeyer S, Buttyan R: Detection of the apoptosis-suppressing oncoprotein bc1–2 in hormone-refractory human prostate cancers. Am J Pathol 1993, 143:390-400 [PMC free article] [PubMed] [Google Scholar]
- 23.Pisters LL, Troncoso P, Zhau HE, Li W, von Eschenbach AC, Chung LW: c-met proto-oncogene expression in benign and malignant human prostate tissues. J Urol 1995, 154:293-298 [PubMed] [Google Scholar]
- 24.Reiter RE, Gu Z, Watabe T, Thomas G, Szigeti K, Davis E, Wahl M, Nisitani S, Yamashiro J, Le Beau MM, Loda M, Witte ON: Prostate stem cell antigen: a cell surface marker overexpressed in prostate cancer. Proc Natl Acad Sci USA 1998, 95:1735-1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gu Z, Thomas G, Yamashiro J, Shintaku IP, Dorey F, Raitano A, Witte ON, Said JW, Loda M, Reiter RE: Prostate stem cell antigen (PSCA) expression increases with high Gleason score, advanced stage and bone metastasis in prostate cancer. Oncogene 2000, 19:1288-1296 [DOI] [PubMed] [Google Scholar]
- 26.Tamir Y, Bengal E: p53 protein is activated during muscle differentiation and participates with MyoD in the transcription of muscle creatine kinase gene. Oncogene 1998, 17:347-356 [DOI] [PubMed] [Google Scholar]
- 27.Weinberg WC, Azzoli CG, Chapman K, Levine AJ, Yuspa SH: p53-mediated transcriptional activity increases in differentiating epidermal keratinocytes in association with decreased p53 protein. Oncogene 1995, 10:2271-2279 [PubMed] [Google Scholar]