Abstract
Loads on molecular motors regulate and coordinate their function. In a study that directly measures properties of internally strained myosin 2 heads bound to actin, we find that human nonmuscle myosins 2A and 2B show marked load-dependent changes in kinetics of ADP release but not in nucleotide binding. We show that the ADP release rate constant is increased 4-fold by the assisting load on one head and decreased 5-fold (for 2A) or 12-fold (for 2B) by the resisting load on the other. Thus these myosins, especially 2B, have marked mechanosensitivity of product release. By regulating the actin attachment of myosin heads, this provides a basis for energy-efficient tension maintenance without obstructing cellular contractility driven by other motors such as smooth muscle myosin. Whereas forward load accelerates the cycle of interaction with actin, resistive load increases duty ratio to favor tension maintenance by two-headed attachment.
Keywords: kinetics, load dependence, myosin, actomyosin, cytoskeleton
The mechanical performance of myosin 2 filaments interacting with actin is strongly determined by the duty ratio, which is the fraction of the mechanical cycle time that each myosin head is strongly attached to actin. If load changes the kinetics of the ATPase cycle that underlies the mechanical cycle, then the lifetimes and steady-state abundance of actin-bound and detached intermediates will change with profound effects on function.
Duty ratio varies widely in myosin 2. Skeletal muscle myosin 2 is the classical low duty ratio myosin, adapted for rapid contraction. It is able to sustain loads because it forms long, bipolar filaments containing ≈150 molecules in each half, so load can be sustained when only a small fraction of the heads are attached at any time. By contrast, nonmuscle myosin 2 (NM2), essential for cytokinesis, tension maintenance, and contractility of nonmuscle and smooth muscle cells (1–4), assembles into short filaments with only ≈10 molecules in each half (5, 6), so their duty ratios under load need to be higher to avoid slippage. The unloaded kinetic mechanisms of single-headed fragments of the NM2A and NM2B isoforms show that NM2A is the faster, lower duty ratio motor, whereas NM2B is slow, with a higher duty ratio suggesting a role in tension maintenance (7–9).
Load-dependent increase of NM2 duty ratio would make the distinction from skeletal muscle myosin more extreme. It could lead to both heads of the molecule being simultaneously attached to the same actin filament as occurs in the absence of ATP, a situation believed to be rare in active skeletal muscle. Kinetic and mechanical studies on smooth muscle myosin 2 have demonstrated load dependence of its lifetime of attachment (10, 11). Because ADP release is associated with axial translation in many myosins, load is expected to affect ADP release kinetics (12). Indeed, differential ADP affinities of the two heads of actin-bound smooth muscle heavy meromyosin have been indicated (13). However, there are no direct measurements of the kinetics of ADP release from mechanically loaded heads of any myosin 2 in solution. Moreover, the effects of internal strain in doubly actin-attached myosin 2 never have been directly determined. NM2 isoforms are best suited for the precise resolution of these processes because of their exceptionally high ADP affinity and slow ADP release (7–9). We have used thiophosphorylated NM2A and NM2B to define strain dependence in the active state of the molecule.
Results
Internal Strain in acto-Heavy Meromyosin (HMM) Can Be Exploited.
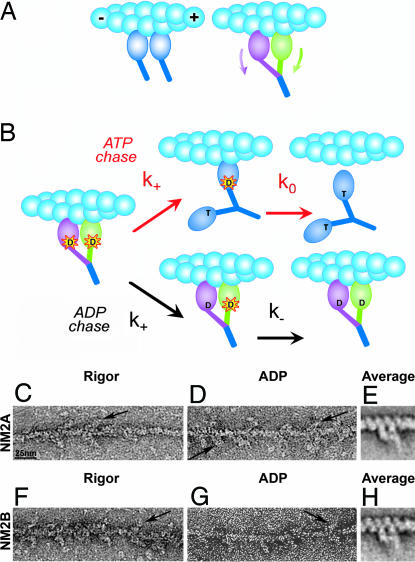
To address load-dependent ADP release, we have taken advantage of the fact that in contrast to an unloaded single-headed myosin fragment, the two heads of a double-headed HMM fragment undergo structural distortions when both heads bind to nearby sites on one actin filament (Fig. 1A). The “lead” (toward the plus end of actin) and “trail” heads will sense different loads that may result in different kinetic properties. This enables the measurement of load effects by comparing the kinetic properties of single- and double-headed constructs, according to the experimental scheme of Fig. 1B.
Fig. 1.
Experimental design and electron micrographs of doubly actin-bound NM2-HMM. (A) When two independent myosin heads (Left, blue) bind to nearby sites on one actin filament (cyan), the tips of their lever arms are separated. However, in doubly actin-attached HMM (Right, green and mauve heads), the lever arms must converge to a common stalk (blue) and thus the heads undergo different distortions (arrows). (B Left) Acto-HMM.dmADP ternary complex (dmADP, starred “D”). The trail head (mauve) will sense an assisting load (accelerated ADP release), whereas the resisting load imposed on the lead head (green) decelerates ADP release. Upper path (ATP chase in red arrows): Upon mixing acto-HMM.dmADP with excess ATP, the trail head will exchange dmADP to ATP (“T”) with an accelerated rate constant k+. ATP binding to this head causes rapid dissociation from actin, and the lead head will release ADP in an unstrained state (blue heads) with rate constant k0. Lower path (ADP chase in black arrows): Upon mixing with excess ADP, the first exchange will occur on the trail head (k+). The exchange of dmADP to ADP (“D”), however, does not cause dissociation from actin and, therefore, lead head ADP release will be decelerated (k−). (C–H) Negative staining electron micrographs. (C–E) NM2A-HMM on actin: in rigor (no nucleotide) (C), in 100 μM ADP (D), and average of 123 HMMs in ADP (E). (F–H) NM2B-HMM on actin: in rigor (F), in 100 μM ADP (G), and average of 89 HMMs in ADP (H). Arrows point out individual, doubly attached HMM molecules. HMM molecules were bound to single actin filaments by both heads, whether ADP was present. (E and H) show that motor domains are attached to adjacent actin subunits along a long pitch helix; the levers are weaker because of their variable shape. E and H are ×2 enlargements. (Scale bar: C, D, F, and G, 25 nm.)
Electron Microscopy Shows Two-Headed Attachment to Single Actin Filaments.
We verified the binding arrangement depicted in Fig. 1A of both NM2A and NM2B-HMM on actin and the lack of actin cross-linking artifacts by negative staining electron microscopy [Fig. 1 C–H and supporting information (SI) Fig. 5]. The complex of doubly actin-attached HMM with ADP readily forms in NM2A and NM2B because of their very high ADP affinity and the stability of the ternary complexes.
In the high-magnification images of Fig. 1 C–H, the wedge shape of each attached molecule appeared very similar to striated muscle myosin two-headed attachment (14). HMMs attached by one head with the other head free were only rarely observed. The density of HMM labeling of actin was undiminished by ADP, reflecting the high affinity of NM2-HMM.ADP for actin (7–9). We would not expect to detect any ADP-induced shape change in the HMM in raw images. Lever shape is insufficiently well defined in the averages to allow direct measurement of lever distortion in comparison with atomic structures of myosin 2 heads.
We found no bundling of actin filaments by NM2-HMM either in the absence of nucleotide or in ADP (SI Fig. 5). Our results contrast with the observation of frequent cross-linking of actin filaments by skeletal muscle HMM to form rafts (15). The different behavior of NM2 likely results from the markedly slower dissociation of nucleotide-free and ADP-bound NM2 heads from actin than in the case of the skeletal muscle isoform (7, 9, 16) and/or it may be related to the lower energy (less internal strain) of a HMM molecule bound to a single actin filament than to two different filaments at the protein concentrations used in our experiments.
Kinetics of ADP Release Show Marked Load Dependence.
N-methylanthraniloyl-2′-deoxy-ADP (dmADP) shows a large fluorescence increase on binding to NM2 while having very similar kinetics to unlabeled ADP (7–9). When NM2-bound dmADP was selectively excited via energy transfer from nearby tryptophans (using excitation at 280 nm and a 400 nm long-pass emission filter), signal enhancements as high as 27-fold (NM2-bound versus free dmADP) were attained and experiments could be performed at acceptable signal-to-noise ratios even at 30- to 40-fold excess of dmADP over NM2 heads (cf. Fig. 2).
Fig. 2.
ADP release from NM2-HMM and acto-NM2-HMM monitored by dmADP fluorescence. The representative dmADP fluorescence traces shown were recorded on mixing HMM.dmADP or acto-HMM.dmADP complexes with a large excess of unlabeled “chaser” ATP or ADP in the stopped-flow apparatus. (Premix concentrations: 0.4 μM myosin heads, 10 μM dmADP, 0 or 2 μM actin, and 2 mM ATP or ADP.) Traces were offset along the ordinate axis for clarity. Note the identical logarithmic time scales of all panels. The traces shown contain all phases that were distinguishable from photobleaching on a time scale ranging up to 600 s. Slow fluorescence decrease components that are not discussed arose from photobleaching. (A) NM2A-HMM (no actin): ATP and ADP chase transients had similar kobs values of 3.3 s−1 and 4.1 s−1, respectively. (B) NM2B-HMM (no actin): kobs was 0.27 s−1 (ATP chase) and 0.33 s−1 (ADP chase). Note that all traces were single exponential (k−act, see Table 1) in the absence of actin. (C) Acto-NM2A-HMM: The ATP chase trace had a kobs of 2.9 s−1 (k+, k0), whereas the ADP chase transient was biphasic with rate constants of 11 s−1 (k+, 38% of total amplitude) and 0.86 s−1 (k−). (D) Acto-NM2B-HMM: The ATP chase transient showed a kobs of 0.22 s−1 (k+, k0), and the ADP chase trace was biphasic with rate constants of 0.59 s−1 (k+, 50% of total amplitude) and 0.021 s−1 (k−). (E) dmADP release from partially dmADP-saturated acto-NM2A-HMM (0.1 μM heads) at premix dmADP concentrations indicated, using 2 mM unlabeled ADP as a chaser. At 0.05 μM dmADP, a 0.18 s−1 transient (k−) with a small amplitude was detected. The transient at 1 μM dmADP had an 8-fold larger amplitude, but it was still single exponential with a kobs of 0.30 s−1 (k−). (F) dmADP release traces of acto-NM2B-HMM in the same conditions as in E. The transients were double exponential even at 0.05 μM dmADP, with a minor (7% of total amplitude) 1.0 s−1 phase (k+) and a major 0.007 s−1 phase (k−). At 1 μM dmADP, the fast phase (k+) became more expressed (36% of total amplitude, kobs = 0.34 s−1) relative to the slow (k−) phase (kobs = 0.019 s−1).
We recorded dissociation transients of dmADP from NM2A-HMM and NM2B-HMM in the absence and presence of actin and by using 2 mM ATP or ADP as a chase. When actin was not present, both NM2A-HMM and NM2B-HMM showed single-exponential dmADP release (with actin-free ADP release rate constant k-act), and the transients were the same when using ATP and ADP chase (Fig. 2 A and B). This shows that HMM heads behave uniformly in the absence of actin and load. With actin present, ATP chase (pathway defined by red arrows in Fig. 1B) traces of both NM2A-HMM and NM2B-HMM were still reasonably well fitted by single exponentials, implying that k+ and k0 are not very different from each other (Fig. 2 C and D and Table 1). Some differences between k+ and k0 can be inferred from the small systematic deviation of the traces from single exponentials, but clear resolution of two phases was not possible by using this method. Another way to measure k+ and k0 is to make a ternary complex of pyrene-labeled-actin, NM2-HMM, and ADP and use the large fluorescence increase of pyrene-labeled actin upon ATP-induced dissociation of the myosin head to indirectly monitor ADP release (7, 9). The improved signal to noise ratio in this method allowed clear resolution of k+ from k0 (Fig. 3) and showed that k+ is approximately 3–4 times faster than k0 (Table 1).
Table 1.
Rate constants of nucleotide binding and dissociation in NM2
| Isoform | Construct | ATP binding |
ADP binding |
ADP release |
|||
|---|---|---|---|---|---|---|---|
| kon (+ actin),* μM−1s−1 | kon (+ actin),† μM−1s−1 | k−act (− actin, unloaded), s−1‡ | k0, (+ actin, unloaded),* s−1 | k+ (+ actin, assisting load),*§ s−1 | k− (+ actin, resisting load),§ s−1 | ||
| NM2A | Double-headed (HMM)¶ | 0.23 ± 0.03 | 3.4 ± 0.1 | 3.6 ± 0.4 | 2.9 ± 1.0¶‖ | 11 ± 4¶‖ | 0.59 ± 0.09 |
| Single-headed** | 0.21 ± 0.04 | 2.7 ± 0.2 | 1.1 ± 0.3 | 2.7 ± 0.3 | |||
| NM2B | Double-headed (HMM)¶ | 0.25 ± 0.09 | 3.1 ± 0.1 | 0.31 ± 0.05 | 0.27 ± 0.06 | 1.1 ± 0.3 | 0.023 ± 0.003 |
| Single-headed†† | 0.40 ± 0.11 | 2.4 ± 0.1 | 0.48 ± 0.11 | 0.38 ± 0.09 | |||
Means ± SEM are shown for all values obtained from experiments using dmADP and pyrene-actin signals (n was between 3 and 9).
*Using pyrene-actin fluorescence (as in Fig. 3).
†Using dmADP fluorescence (SI Fig. 6).
‡Using dmADP fluorescence and ATP or ADP chase (Fig. 2 A and B).
§Using dmADP fluorescence and ADP chase (Fig. 2 C–F).
¶Present study.
‖Ref. 17.
**Ref. 7.
††Ref. 9.
Fig. 3.
ADP release and actomyosin dissociation in acto-NM2B-HMM monitored by pyrene-actin fluorescence. Pyrene-acto-NM2B-HMM (0.25 μM pyrene-actin, 0.2 μM myosin heads) was preincubated with various concentrations of ADP and then rapidly mixed with 500 μM ATP in the stopped-flow apparatus (premix concentrations stated). The fastest phase of the recorded pyrene-actin fluorescence increase transients resulted from ATP-induced dissociation of the ADP-free fraction of acto-HMM, whereas the two slower phases were assigned to rate-limiting ADP release rate constants k+ and k0. Note that with increasing ADP concentration in the premixture, the fast phase titrates away and also the fractional amplitude of k+ is increasing relative to that of k0. Fitting (black lines) yielded kobs values and percentage amplitudes as follows. At 0.5 μM ADP: 78 s−1 (ADP-free acto-HMM, amplitude: 45%), 1.5 s−1 (k+, 14%), 0.34 s−1 (k0, 41%); at 1 μM ADP: 81 s−1 (ADP-free acto-HMM, 16%), 1.3 s−1 (k+, 28%), 0.32 s−1 (k0, 56%); at 10 μM ADP: 95 s−1 (ADP-free acto-HMM, 7%), 1.2 s−1 (k+, 30%), 0.29 s−1 (k0, 63%). Traces were offset along the ordinate axis for clarity. The red line shows a fit to the transient at 10 μM ADP assuming monophasic ADP release, which shows clear systematic deviations from the experimental trace. Similar profiles to the ones shown were obtained in our earlier work on pyrene-acto-NM2A-HMM (17).
ADP chase (Fig. 1B, black arrows) traces of dmADP dissociation from acto NM2-HMM were clearly double exponential and allowed for measurement of k+ and k−. The faster rate constant (k+) was consistent with that measured above with pyrene fluorescence and the slower rate constant (k–) was markedly slower than k0 for both NM2A and NM2B (Fig. 2 C and D and Table 1). The effect of load on the slowing of ADP release from the lead head is particularly evident with NM2B where k− is 10 times lower than k0.
The fractional amplitudes of the two phases of the acto-HMM ADP chase traces of Fig. 2 C and D were close to the 0.5:0.5 expected, but consistently the slow phase had the larger amplitude (at a fast:slow amplitude ratio of approximately 0.45:0.55). In line with this finding, kinetic simulations of sequential release steps also showed an amplitude bias for the slow phase even if the actual fluorescence change is the same on both steps (data not shown). Generally, the ADP chase amplitudes were slightly larger than the ATP chase amplitudes. We attribute this to a modest contribution of the light scattering decrease occurring upon ATP-induced acto-HMM dissociation, which will slightly increase the final detected dmADP fluorescence level. All transients were the same within an actin concentration range of 2–10 μM and a dmADP concentration range of 10–100 μM, indicating that we achieved practically complete formation of the acto-HMM.dmADP ternary complex under the conditions applied.
Highly similar rate constants were obtained by using either the dmADP signal or the pyrene-actin signal where unlabeled ADP was used as the nucleotide. Thus, it appears that all kinetic properties of NM2A and NM2B related to dmADP are very similar to those related to unlabeled ADP and thus it is highly unlikely that dmADP would induce acto-HMM interactions different from those in unlabeled ADP (7–9, 17).
The kinetics of dmADP binding to acto-HMM was single exponential and very similar to that obtained earlier in single-headed constructs (Table 1 and SI Fig. 6) (7, 9). We interpret the monophasic dmADP binding data with a uniform, load-insensitive ADP binding behavior of NM2A-HMM and NM2B-HMM heads. It should be noted that we also examined an alternative interpretation of the dmADP binding and release transients. The single-exponential dmADP binding kinetics might imply that the traces represent dmADP binding to only the trail head of actin-bound HMM, and both dmADP binding and release from the lead head are extremely slow. In this case the kobs of the slow phase in the ADP chase transients of Fig. 2 C and D would not be determined by k−, but by the slow binding of a small amount of contaminating ATP (present in the ADP solution) to the trail head. The latter process would cause dissociation of the trail head from actin and, thus, relieve the intramolecular strain (as in the ATP chase experiments) and allow nucleotide exchange in the lead head. However, we find this scenario unlikely because (i) there is no sign of a slow dmADP binding component in the dmADP binding stopped-flow traces up to 60 s and thus dmADP binding to the lead head would have to be >2,500-fold slower than to the trail head, (ii) this large difference would have to be paralleled by a deceleration of dmADP release to almost precisely the same extent to produce the binding patterns of Fig. 2 E and F, and (iii), the ADP used for chase was of high purity (>99%).
Load-insensitive ADP binding and load-sensitive ADP release imply that the two heads of HMM have different ADP affinities. We verified the proposed model in experiments in which we could selectively populate one of the HMM heads with dmADP (Fig. 2 E and F). From the dmADP-on and -off rate constants of Table 1, it can be calculated that the lead and trail heads of acto-NM2A-HMM will have Kd values for dmADP of 0.17 and 3.2 μM, respectively, whereas the corresponding Kd values in NM2B-HMM will be 0.0074 μM and 0.35 μM, respectively. This implies that at 0.1 μM myosin head concentration, dmADP at a concentration of 0.05 μM should preferentially occupy the high-affinity lead head and, thus, only the k− phase would be visible in an ADP chasing experiment. This indeed is in line with our experimental findings shown in Fig. 2 E and F. In the case of acto-NM2A-HMM (Fig. 2E), the amplitude of the reaction at 0.05 μM dmADP was, as expected, severely reduced compared with that in records obtained at 1 μM dmADP, whereas in NM2B (Fig. 2F), the amplitude was already large even at 0.05 μM dmADP because of the higher affinity of the lead NM2B head. At 1 μM dmADP, as expected, the NM2A-HMM trace still showed a predominance of the k− phase (Kd trail = 3.2 μM, Fig. 2E), whereas the two heads were more evenly populated in NM2B-HMM (Kd trail = 0.35 μM, Fig. 2F).
We measured the ATP binding rate constant to apo acto-NM2A-HMM and acto-NM2B-HMM by mixing nucleotide-free pyrene-actoNM2A-HMM and pyrene-acto-NM2B-HMM with excess ATP in the stopped flow. The pseudo first-order traces were single exponential (data not shown) and the second-order ATP binding rate constants obtained were very similar to those obtained with single-headed constructs (Table 1) (7, 9). This indicates that there is no major difference between rate constants of ATP binding to unloaded and loaded (trail and possibly lead) heads of pyrene-acto-NM2-HMM.
Discussion
Structural Basis of Load-Dependent ADP Release.
Our data demonstrate that a marked load dependence of ADP release from acto-NM2 is a major determinant of its load-sensing mechanism, whereas nucleotide (ATP and ADP) binding is load insensitive. In this respect, myosin 2 is similar to myosin 5 and unlike myosin 6 and kinesin motors where load sensitivity of nucleotide binding is dominant (18–24). In doubly actin-attached myosin 2, the head distortions have both axial and transverse elements, which is similar to the situation during in vivo functioning (e.g., see tomograms in ref. 25). Therefore, the HMM structures used in our study are physiologically relevant.
Thermodynamic calculations based on our data indicate that upon double-headed actin binding, NM2-HMM heads undergo distortions in the range of several nanometers (Table 2). Load dependence of ADP release implies that the myosin lever produces a swing coupled to this process, which provides mechanosensitive gating (12). We could estimate the equilibrium constant between acto-NM2.ADP states with different lever orientations (SI Table 3). Indeed, our recent collaborative work on x-ray diffraction of acto-NM2 structures shows that both NM2A and NM2B exhibit an unprecedentedly large swing of the lever upon ADP release (4.6–4.8 nm displacement at the tip of the lever) (26).
Table 2.
Calculated displacements of the tips of the levers of doubly actin-bound NM2-HMM relative to those of unstrained single heads
| Method of calculation | NM2A |
NM2B |
||
|---|---|---|---|---|
| Strain energy* | Off-rate constant† | Strain energy* | Off-rate constant† | |
| Trail head, nm | 4.9 | 4.5 | 5.1 | 4.8 |
| Lead head, nm | 5.4 | 5.4 | 6.7 | 8.3 |
| Combined displacement, nm | 10.3 | 9.9 | 11.8 | 13.1 |
*Calculated from strain energies accounting for differences in the free energy change of ADP dissociation (according to ref. 12) as ΔΔG0 = RT ln(K/K′) = NAκΔL2/2, from which ΔL = ((2kBT/κ) ln(K/K′))1/2, where ΔΔG0 is the difference between the standard free energy changes of ADP dissociation from the actin-bound myosin head in a lower and a higher ADP affinity state, R is the universal gas constant, T is the absolute temperature, K and K′ are ADP dissociation constants from the actin-bound myosin head in a lower and higher ADP affinity state, respectively, NA is Avogadro's number, κ is the spring constant of the head [κ = 0.45 pN/nm of smooth muscle myosin S1 was used (11), which is the isoform closest to NM2 in both evolutionary and functional terms], and ΔL is the displacement of the tip of the lever between the two states.
†Calculated using ADP-off rate constants (according to and using data of ref. 11) as k′ = k exp(Fd0/kBT), from which F = kBT/d0 ln (k′/k) and ΔL = F/κ where k and k′ are ADP dissociation rate constants in unloaded and loaded conditions, respectively, F is the load exerted on the myosin head, and d0 is the distance parameter characteristic of the load dependence of the rate constant [d0 = 2.7 nm of smooth muscle myosin S1 was used (ref. 11)].
Our results show that piconewton range forces arising in double-headed actin-bound NM2 structures cause an ≈4-fold acceleration of ADP release from heads loaded in the forward direction (assisting load) in both NM2A and NM2B, whereas the deceleration caused by backward (resisting) load is significantly different in the case of the two isoforms (5-fold in NM2A, but as high as 12-fold in NM2B; Table 1). The internal consistency of the kinetic data are indicated by the calculated displacements of the tips of the levers of the heads of doubly actin-bound NM2-HMM relative to those of unstrained actin-bound single heads. The data indicate an ≈5-nm displacement of each tip (trail and lead) from an unstrained state, both when calculated from ADP binding equilibrium constants and from changes in the ADP release rate constants (Table 2). Although the molecular mechanical parameters of NM2 (including head stiffness) remain to be determined and thus we used smooth muscle myosin mechanical data in these calculations, the results are in excellent agreement with earlier data of Chakrabarty et al. (27) on chicken skeletal muscle myosin 2 and also our atomic models of scallop striated and chicken skeletal myosin 2 head structures docked onto actin (the tip-to-tip distance of adjacent independent single myosin heads was between 8.8 and 9.4 nm in all cases) (ref. 27; K.T., S. A. Burgess, J. Trinick, H. D. White, and P.J.K., unpublished results).
The larger change in the ADP release rate constant from NM2B heads under resisting load yields an apparently larger distortion in this case (≈7–8 nm; Table 2). We verified by electron microscopic analysis that the binding arrangement of HMM to actin is very similar in NM2A and NM2B (Fig. 1 C–H) and thus it is likely that, rather than there being structural differences between the two isoforms, the mechanical properties of NM2B are such that it will have especially high load sensitivity in this direction to enable highly efficient deceleration of ADP release by resisting loads (see also SI Table 3).
Physiological Role of the Load-Dependent Mechanism.
Marked slowing of ADP release by load will result in remarkably long lifetimes of actin-attachment of NM2 heads during functioning. In addition to the load effect, rapid ADP binding to acto-NM2 (10–15-fold faster than ATP binding; Table 1) enables actin attachment lifetimes to increase still further in response to an increase in cellular ADP levels from 10 μM to 100 μM even in the presence of physiological ATP concentrations (SI Fig. 7). Thus rates of ATP usage <0.01 ATP per head per second may be achieved during tension maintenance. Loaded heads spend practically all of their cycle time in a load-bearing, strongly actin-bound state and, in consequence, we can predict a frequent occurrence of doubly actin-attached myosin molecules similar to the acto-HMM.ADP complexes produced in our experiments (Fig. 1 C–H) under physiological conditions. This leads to the novel conclusion that under resisting load, the myosin molecules in the short NM2 filaments only rarely would detach from actin completely, with lead and trail heads undergoing several slow ATPase cycles without the molecule moving on actin, or with stepwise forward or backward movement, depending on the magnitude of the load (Fig. 4).
Fig. 4.
Structural cycles of NM2 on actin. At left an actin filament tethered at its right end interacts with NM2 under low load. Sequential release of phosphate and ADP from a strongly attached head is associated with a two-stage translocation of the myosin tail along the actin, and one head at a time interacts with actin (orange, single head attached states; white, detached state). Under resisting load (indicated by arrowhead on end of NM2 tail), the attached head structure is distorted, slowing release of ADP and allowing progression to the doubly attached ADP-ADP state (gray shading; blue, doubly attached states), which we can observe by EM in the absence of ATP (Fig. 1 E and H). The lead and trail heads of this NM2 molecule then can continue to cycle while the molecule stays attached to actin under load. The lead head of the complex is predicted to release ADP especially slowly and, thus, lead head cycles will be much less frequent (thin arrow) than trail head cycles (thick arrow). Acceleration of trail head ADP release by the lead head, reported in this study, is expected to be reversed by the load. The blue arrow linking the lead and trail head cycles indicates that these two cycles can interconvert by axial movement of the detached head. This allows both cycles to respond to changes in load. It also provides a route by which processive forward or backward stepping of an attached NM2 molecule could occur. T, ATP; DP, ADP+phosphate; D, ADP.
NM2 (and especially NM2B) has been implicated for smooth muscle function, namely in the prolonged force maintenance phase (tonic contraction) (1, 2). Load sensitivity of NM2 kinetics reported here thus may vitally contribute to its efficient functioning both under assisting and resisting loads. Under assisting loads, acceleration of ADP release will result in shorter actin attachment lifetimes and prevent NM2 from hindering contraction driven by the faster smooth muscle myosin. Under resisting loads, extremely long actin attachment lifetimes of NM2 (especially NM2B) will reach into the minutes timescale and thus enable tension maintenance and structural anchoring at a very low energetic cost.
Methods
Proteins.
NM2A-HMM and NM2B-HMM were produced as described in refs. 28 and 29. We confirmed by SDS/PAGE that HMM preparations contained <3% of potentially single-headed proteolytic products. HMM was thiophosphorylated as in refs. 17 and 30. HMM showed phosphorylation-dependent actin-activated MgATPase activity as in our earlier reports (17, 28). Actin was prepared and pyrene-labeled (with a labeling ratio ≈0.7) as in refs. 31 and 32. HMM concentrations are expressed in terms of myosin heads.
Negative Staining Electron Microscopy.
In electron microscopic experiments, the ratio of HMM heads to actin subunits of 1:5 was designed to match that in kinetic experiments; thus the actin filaments were only partially decorated with HMM. Thawed proteins were diluted in 100 mM KCl/0.1 mM EGTA/5 mM MgCl2,/10 mM Mops, pH 7.0 at 20°C. For the ADP experiment, concentrated HMMs (1.71 μM NM2A-HMM; 1.04 μM NM2B-HMM) and actin (53 μM) were treated with 0.05 unit/ml hexokinase/1 mM glucose/100 μM ADP for 40 min at 20°C. For rigor, concentrated HMMs (3.4 μM NM2A-HMM; 2.1 μM NM2B-HMM) and actin (106 μM) were treated with 0.14 unit/ml apyrase to remove ATP and ADP (HMM for 1 min at 20°C and actin for 15 min on ice). Proteins were diluted to 0.4 μM HMM and 2 μM actin, then equal volumes were combined and negatively stained with 1% uranyl acetate as described in ref. 33. Specimens were prepared 30 s and 30 min after mixing HMM and actin. Micrographs were recorded on film at a nominal magnification of ×40,000 by using a JEOL 1200EX microscope at 80 kV and digitized at a resolution referred to the specimen of 0.5 nm per pixel. Image averaging used the SPIDER software suite.
Kinetic Measurements.
Stopped-flow experiments were done at 25°C in 10 mM Mops (pH 7.0)/2 mM MgCl2/0.15 mM EGTA/100 mM KCl in a KinTek SF-2001 apparatus as described in refs. 7 and 9. Acto-HMM had <10% bound ADP contamination before dmADP addition. HPLC-purified ADP (>99% pure) was used. In experiments starting from nucleotide-free acto-HMM, preincubation with 0.01 unit/ml apyrase for 30 min at 25°C was applied to remove ATP and ADP contamination. The stopped-flow mixing volume ratio was 1:1. Premix concentrations are stated. To ensure that none of the analyzed phases results from photobleaching of dmADP, we recorded photobleaching controls for all experiments. The extent of photobleaching was smaller than 10% in amplitude compared with any reported dmADP dissociation phases.
Supplementary Material
Acknowledgments
We thank Dr. Howard White (Eastern Virginia Medical School, Norfolk, VA) for the gift of dmADP. M.K. is supported by National Institutes of Health Research Grant D43 TW006230 (1 R01 TW007241–01) funded by the Fogarty International Center; the National Heart, Lung, and Blood Institute; a European Molecular Biology Organization–Howard Hughes Medical Institute Startup Grant; and the Bolyai Fellowship of the Hungarian Academy of Sciences. K.T. is supported by the Wellcome Trust.
Abbreviations
- dmADP
N-methylanthraniloyl-2′-deoxy-ADP
- HMM
heavy meromyosin
- NM2
nonmuscle myosin 2.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0701181104/DC1.
References
- 1.Rhee AY, Ogut O, Brozovich FV. Pflügers Arch. 2006;452:766–774. doi: 10.1007/s00424-006-0091-4. [DOI] [PubMed] [Google Scholar]
- 2.Morano I, Chai GX, Baltas LG, Lamounier-Zepter V, Lutsch G, Kott M, Haase H, Bader M. Nat Cell Biol. 2000;2:371–375. doi: 10.1038/35014065. [DOI] [PubMed] [Google Scholar]
- 3.Conti MA, Even-Ram S, Liu C, Yamada KM, Adelstein RS. J Biol Chem. 2004;279:41263–41266. doi: 10.1074/jbc.C400352200. [DOI] [PubMed] [Google Scholar]
- 4.Wylie SR, Chantler PD. Nat Cell Biol. 2001;3:88–92. doi: 10.1038/35050613. [DOI] [PubMed] [Google Scholar]
- 5.Niederman R, Pollard TD. J Cell Biol. 1975;67:72–92. doi: 10.1083/jcb.67.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verkhovsky AB, Svitkina TM, Borisy GG. J Cell Biol. 1995;131:989–1002. doi: 10.1083/jcb.131.4.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kovacs M, Wang F, Hu A, Zhang Y, Sellers JR. J Biol Chem. 2003;278:38132–38140. doi: 10.1074/jbc.M305453200. [DOI] [PubMed] [Google Scholar]
- 8.Rosenfeld SS, Xing J, Chen LQ, Sweeney HL. J Biol Chem. 2003;278:27449–27455. doi: 10.1074/jbc.M302555200. [DOI] [PubMed] [Google Scholar]
- 9.Wang F, Kovacs M, Hu A, Limouze J, Harvey EV, Sellers JR. J Biol Chem. 2003;278:27439–27448. doi: 10.1074/jbc.M302510200. [DOI] [PubMed] [Google Scholar]
- 10.Cremo CR, Geeves MA. Biochemistry. 1998;37:1969–1978. doi: 10.1021/bi9722406. [DOI] [PubMed] [Google Scholar]
- 11.Veigel C, Molloy JE, Schmitz S, Kendrick-Jones J. Nat Cell Biol. 2003;5:980–986. doi: 10.1038/ncb1060. [DOI] [PubMed] [Google Scholar]
- 12.Nyitrai M, Geeves MA. Philos Trans R Soc London B. 2004;359:1867–1877. doi: 10.1098/rstb.2004.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berger CE, Fagnant PM, Heizmann S, Trybus KM, Geeves MA. J Biol Chem. 2001;276:23240–23245. doi: 10.1074/jbc.M100524200. [DOI] [PubMed] [Google Scholar]
- 14.Craig R, Szent-Gyorgyi AG, Beese L, Flicker P, Vibert P, Cohen C. J Mol Biol. 1980;140:35–55. doi: 10.1016/0022-2836(80)90355-1. [DOI] [PubMed] [Google Scholar]
- 15.Trinick J, Offer G. J Mol Biol. 1979;133:549–556. doi: 10.1016/0022-2836(79)90407-8. [DOI] [PubMed] [Google Scholar]
- 16.Taylor EW. J Biol Chem. 1991;266:294–302. [PubMed] [Google Scholar]
- 17.Kovacs M, Toth J, Nyitray L, Sellers JR. Biochemistry. 2004;43:4219–4226. doi: 10.1021/bi036007l. [DOI] [PubMed] [Google Scholar]
- 18.Altman D, Sweeney HL, Spudich JA. Cell. 2004;116:737–749. doi: 10.1016/s0092-8674(04)00211-9. [DOI] [PubMed] [Google Scholar]
- 19.Conibear PB, Geeves MA. Biophys J. 1998;75:926–937. doi: 10.1016/S0006-3495(98)77581-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robblee JP, Olivares AO, De La Cruz EM. J Biol Chem. 2004;279:38608–38617. doi: 10.1074/jbc.M403504200. [DOI] [PubMed] [Google Scholar]
- 21.Rosenfeld SS, Sweeney HL. J Biol Chem. 2004;279:40100–40111. doi: 10.1074/jbc.M402583200. [DOI] [PubMed] [Google Scholar]
- 22.Veigel C, Schmitz S, Wang F, Sellers JR. Nat Cell Biol. 2005;7:861–869. doi: 10.1038/ncb1287. [DOI] [PubMed] [Google Scholar]
- 23.Rosenfeld SS, Fordyce PM, Jefferson GM, King PH, Block SM. J Biol Chem. 2003;278:18550–18556. doi: 10.1074/jbc.M300849200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valentine MT, Gilbert SP. Curr Opin Cell Biol. 2007;19:75–81. doi: 10.1016/j.ceb.2006.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen LF, Winkler H, Reedy MK, Reedy MC, Taylor KA. J Struct Biol. 2002;138:92–104. doi: 10.1016/s1047-8477(02)00013-8. [DOI] [PubMed] [Google Scholar]
- 26.Iwamoto H, Oiwa K, Kovacs M, Sellers JR, Suzuki T, Wakayama J, Tamura T, Yagi N, Fujisawa T. J Mol Biol. 2007;369:249–264. doi: 10.1016/j.jmb.2007.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chakrabarty T, Xiao M, Cooke R, Selvin PR. Proc Natl Acad Sci USA. 2002;99:6011–6016. doi: 10.1073/pnas.082024299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim KY, Kovacs M, Kawamoto S, Sellers JR, Adelstein RS. J Biol Chem. 2005;280:22769–22775. doi: 10.1074/jbc.M503488200. [DOI] [PubMed] [Google Scholar]
- 29.Wang F, Harvey EV, Conti MA, Wei D, Harvey EV, Conti MA, Wei D, Sellers JR. Biochemistry. 2000;39:5555–5560. doi: 10.1021/bi000133x. [DOI] [PubMed] [Google Scholar]
- 30.Facemyer KC, Cremo CR. Bioconjug Chem. 1992;3:408–413. doi: 10.1021/bc00017a009. [DOI] [PubMed] [Google Scholar]
- 31.Cooper JA, Walker SB, Pollard TD. J Muscle Res Cell Motil. 1983;4:253–262. doi: 10.1007/BF00712034. [DOI] [PubMed] [Google Scholar]
- 32.Spudich JA, Watt S. J Biol Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- 33.Burgess SA, Walker ML, Thirumurugan K, Trinick J, Knight PJ. J Struct Biol. 2004;147:247–258. doi: 10.1016/j.jsb.2004.04.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.