Abstract
A universal mark of centromeric chromatin is its packaging by a variant of histone H3 known as centromeric H3 (CenH3). The mechanism by which CenH3s are incorporated specifically into centromere DNA or the specialized function they serve there is not known. In a genetic approach to identify factors involved in CenH3 deposition, we screened for dosage suppressors of a temperature-sensitive cse4 allele in Saccharomyces cerevisiae (Cse4 is the S. cerevisiae CenH3). Independent screens yielded ORF YDL139C, which we named SCM3. Dosage suppression by SCM3 was specific for alleles affecting the histone fold domain of Cse4. Copurification and two-hybrid studies showed that Scm3 and Cse4 interact in vivo, and chromatin immunoprecipitation revealed that Scm3, like Cse4, is found associated with centromere DNA. Scm3 contains two essential protein domains, a Leu-rich nuclear export signal and a heptad repeat domain that is widely conserved in fungi. A conditional scm3 allele was generated to allow us to deplete Scm3. Upon Scm3 depletion, cells undergo a Mad2-dependent G2/M arrest, and centromere localization of Cse4 is perturbed. We suggest that S. cerevisiae Scm3 defines a previously undescribed family of fungal kinetochore proteins important for CenH3 localization.
Keywords: centromeric H3, chromosome segregation, kinetochore, yeast, CENP-A
Centromeres are the chromosomal sites at which microtubules attach during mitotic and meiotic prometaphase. Mediating the attachment is the kinetochore, a complex structure composed of dozens of proteins, many of which are evolutionarily conserved across the plant, animal, and fungal kingdoms (1). A universal feature of centromere DNA is its packaging by nucleosomes containing a variant of histone H3 known as centromeric H3 (CenH3) (2). CenH3s [CENP-A in humans, Cid in Drosophila, HTR12 in Arabidopsis, and Cse4 in yeast (3)] are essential in all organisms tested, although the mechanism by which they are specifically deposited on the centromere DNA or the function they serve there are not known (4). In contrast to the conservation observed for many centromere proteins, centromere DNAs are diverse. They vary widely in size, and, with the exception of S. cerevisiae and its close relatives, no unique centromere identifier sequence has been defined (5). It is proposed that a unique chromatin structure conferred by CenH3 provides the epigenetic mark to signal kinetochore assembly (6).
The first centromeres cloned were the “point” centromeres of Saccharomyces cerevisiae. They are ≈125 bp in length and are characterized by conserved DNA elements (CDEs) unique to centromere (CEN) DNA in this organism (7). CDEI is the degenerate octanucleotide RTCACRTG, CDEII is 79–88 bp of highly AT-rich DNA, and CDEIII is a 24-bp sequence that binds CBF3, a four subunit, sequence-specific DNA binding protein that is essential for centromere function (8). Binding of CBF3 to CDEIII is thought to be the requisite first step in the ordered assembly of the S. cerevisiae kinetochore (9). The core CDEI-CDEII-CDEIII sequence is apparently assembled onto a single Cse4-containing nucleosome (10), although disagreement exists (11). Higher organisms and other yeasts have so-called “regional” centromeres which are larger, less well defined at the sequence level, and which often contain repetitive elements. For example, Schizosaccharomyces pombe centromeres are 40–100 kbp in length and consist of 4- to 7-kbp nonhomologous central cores flanked by repetitive sequences (12). Mammalian and higher plant centromeres consist of megabases of highly repetitive satellite DNAs (4, 13). The point centromeres of S. cerevisiae and close relatives seem to be a recent evolutionary invention, because the common ancestor to point and regional centromere yeasts seems to be a fungus with regional centromeres (1).
Here, we describe the discovery and characterization of Scm3, an essential S. cerevisiae centromere protein. Genetic and biochemical analysis shows that Scm3 is required for mitosis, possibly by serving as an assembly and/or targeting factor for Cse4. Scm3 contains an essential protein domain that is widely conserved in fungi, implying a function not limited to point centromeres. We suggest that S. cerevisiae Scm3 defines a previously undescribed family of proteins involved in CenH3 localization.
Results
YDL139C Is an Allele-Specific Suppressor of Temperature-Sensitive (Ts) cse4 Alleles.
Two independent screens for dosage suppressors of the S. cerevisiae Ts cse4-1 allele yielded, in addition to CSE4, only the gene YDL139C. The cse4-1 mutation causes an Ala to Thr substitution in helix 3 of the Cse4 histone fold domain (HFD). Suppressor screens of two other Ts cse4 HFD alleles, cse4-107 (Q219D) and cse4-162 (W178T), also yielded CSE4 and YDL139C, whereas YDL139C was not identified in any of several dosage suppressor screens performed using Ts cse4 alleles affecting the essential END domain located in the N terminus (14). Direct tests confirmed the allele-specificity [supporting information (SI) Fig. 6]. High-copy YDL139C plasmids also suppressed the cse4-1 chromosome missegregation phenotype (data not shown), so the suppressor gene was named SCM3 (suppressor of chromosome missegregation). SCM3 gene disruption was lethal, indicating that SCM3 is an essential gene. Introducing high-copy SCM3 plasmids into WT CSE4 strain backgrounds had no detectable phenotypic effect.
Essential Domains Within Scm3.
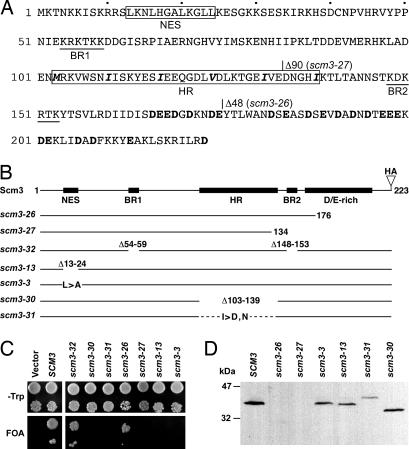
SCM3 encodes a 223-amino acid protein containing several sequence motifs of potential functional importance (Fig. 1A). Residues 13–24 resemble a Leu-rich nuclear export signal (NES), whereas short patches of basic residues, similar to nuclear localization sequences (NLSs), are found at positions 54–59 (BR1) and 148–153 (BR2) (15, 16). The C-terminal 58 aa are acid-rich (40% D + E). Finally, the central part of the protein resembles a coiled-coil domain in having repeating heptad units with hydrophobic residues occupying the fourth position and polar residues in the first position (17). Five such units repeat between residues 103–138.
Fig. 1.
Mutational analysis of Scm3. (A) Amino acid sequence of Scm3 showing potential functional motifs and site-directed mutations. The nuclear export signal (NES) and heptad repeat (HR) domain are boxed with the heptad-spaced hydrophobic residues shown in boldface italic. Potential nuclear localization sequences are underlined (BR1, BR2), and acidic residues in the C-terminal acidic (D/E-rich) region are shown in boldface. Deletion endpoints of the scm3-26 and scm3-27 alleles are indicated. (B) Schematic diagrams of HA-tagged WT SCM3 and site-directed scm3 mutants. (C) Plasmid shuffle complementation tests of mutants shown in B. Growth on 5-fluoroorotic acid (FOA) medium indicates the respective mutant allele provides Scm3 function. (D) Immunoblot of protein extracts from the indicated transformants detected with anti-HA antibody.
Site-directed mutations affecting each of these motifs were constructed (Fig. 1B) and the mutant proteins tested for function by using a plasmid shuffle assay (Fig. 1C). To monitor protein expression, all of the mutant alleles encoded a C-terminal triple-HA epitope tag. SCM3-HA fully complemented scm3::LEU2 and produced an Scm3-HA protein that migrated with an apparent molecular mass of 36 kDa, larger than the predicted 30 kDa (Fig. 1D). Removal of the C-terminal 48 aa (scm3-26), comprising most of the D/E-rich region, did not result in loss of complementing ability even though the truncated protein, undetectable by Western blot assay, was poorly expressed or unstable (Fig. 1 C and D). Deletion of 90 C-terminal residues (scm3-27), which removed all of the D/E-rich region and impinged on the heptad repeat region, yielded an inactive protein that, like Scm3-26, was undetectable. Deletion of potential NLSs BR1 and BR2 in mutant scm3-32 did not result in loss of complementing ability, but deletion of the putative NES (scm3-13) or the heptad repeat region (scm3-30) were lethal. The latter two mutant proteins were expressed at levels similar to WT Scm3 (Fig. 1D). We concluded that the NES homology domain and putative coiled-coil domain are essential for Scm3 function. The D/E-rich domain is also essential, although possibly only for protein stability.
Scm3 Heptad Repeat Domain Conserved in Fungi.
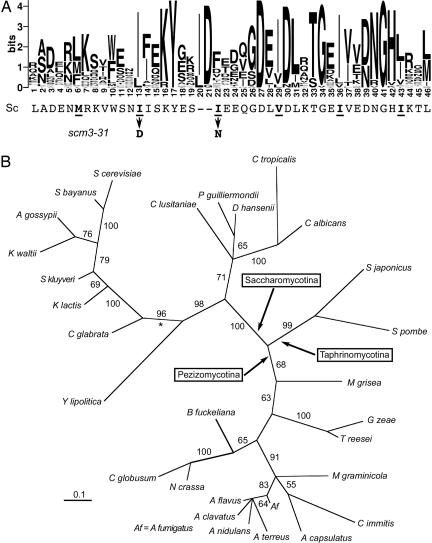
BLAST searches of public databases identified 45 potential Scm3 homologs, widely distributed in the Fungi. No homologs were found in metazoans, plants, or archae. Except for closely related sensu stricto and sensu latu yeasts, homology to S. cerevisiae Scm3 was limited to the central heptad repeat domain. An alignment of this region for 38 of the homologs is shown in Fig. 2A. The heptad repeat of hydrophobic residues located between positions 103 and 138 in the S. cerevisiae protein is conserved (positions 6–43 of the Logo diagram in Fig. 2A); however, alignment required insertion of one to three gaps, leading us to conclude that the structure of this domain is probably not an extended coiled coil. Nonetheless, the amino acid sequence is critical, because changing two of the Ile residues to polar amino acids (scm3-31) resulted in loss of Scm3 function (Fig. 1C). A phylogeny of the conserved protein domain is shown in Fig. 2B. The phylogeny is congruent with species phylogeny (18), suggesting orthology. All yeasts on the branch marked with the asterisk contain CDEI-CDEII-CDEIII point centromeres; thus, the ancestral Scm3 protein seems to have existed before the invention of point centromeres.
Fig. 2.
Scm3 heptad repeat domain is evolutionarily conserved. (A) Alignment of 38 fungal protein sequences homologous to the heptad repeat domain of S. cerevisiae Scm3, shown in Logo format (42). The alignment requires insertion of one to three gaps at positions 20–22. The complete alignment in Clustal format is provided in SI Fig. 7. (B) Maximum likelihood phylogeny of the Scm3 homology domain of 29 representative ascomycetes derived by Bayesian inference (details in SI Materials and Methods). Numbers on the branches are the percentage of times that branch was present in the posterior distribution of trees. All members of the clade defined by the branch marked with the asterisk (∗) have point centromeres.
The Scm3 NES Motif Is a Functional NES.
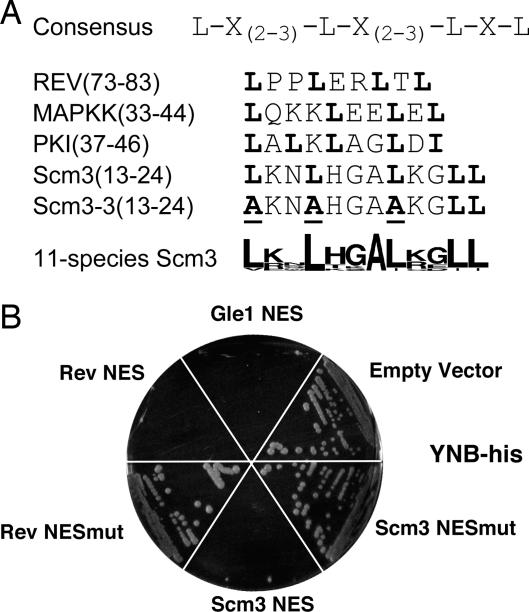
The putative NES at Scm3 residues 13–24 is homologous to other known NESs and conforms to the NES consensus (Fig. 3A) (15, 19, 20). The ability of this sequence to direct nuclear export in vivo was tested by using a genetic assay described by Shulga et al. (21). The assay strain contains an integrated HIS3 gene under control of a promoter containing LexA binding sites. The strain also expresses a chimeric transcriptional activator that reports nuclear location. Cells are His+ when the activator, comprising the LexA DNA binding domain fused to the pseudorabies virus transcription activation domain, is present in the nucleus; however, when a functional NES is fused to the activator, cells fail to grow on medium lacking His, because the fusion protein is efficiently exported from the nucleus. The putative Scm3 NES motif behaved like the authentic NESs of HIV Rev and the RNA export factor Gle1 in this assay (Fig. 3B). When three conserved Leu residues in the NES were mutated to Ala (Scm3mut), export failed. When introduced into full-length Scm3, the triple Ala substitution (scm3-3) had little effect on protein expression (Fig. 1D), but the mutant protein was unable to confer Scm3 function in the complementation assay (Fig. 1C). We conclude that the Scm3p NES motif directs nuclear export and that the ability to be exported from the nucleus is critical for Scm3 function.
Fig. 3.
Scm3p contains a functional NES. (A) The NES consensus sequence (15) and other known NESs are shown in comparison with the Scm3 NES and the mutated sequence of scm3-3. At the bottom is shown an alignment, in Logo format (42), of the NES region found in Scm3 orthologs of 11 close relatives of S. cerevisiae. (B) Results of the NES assay. Positive controls are the NESs of Gle1 and Rev. Negative controls are empty vector as well as a fusion containing a nonfunctional Rev sequence (Rev NESmut). Scm3mut is the fusion containing the mutated NES region of scm3-3.
Scm3 Interacts with Cse4 and CEN DNA.
Subcellular fractionation suggested that Scm3, like Cse4, is an integral chromatin protein. Originally present in the low-speed pellet, Scm3 and Cse4 were both released by limited micrococcal nuclease digestion (fraction 4) and pelleted in a 100,000 × g centrifugation (fraction 7; Fig. 4A). As a control, Cse1, a nuclear export protein known to be associated with the nuclear envelope, was monitored and found exclusively in the soluble fraction, i.e., fraction 2 (data not shown). The copurification of Scm3 and Cse4 prompted us to test for Scm3-Cse4 interaction. Yeast two-hybrid assays showed that Scm3 interacts with itself and with Cse4 (Table 1). Both interactions were abolished when baits carried the scm3-31 double missense mutation. Immunoblot analysis confirmed that the mutant DB-scm3-31 baits were stably expressed in the reporter cells (data not shown). Scm3-Cse4 interaction was mediated through the HFD of Cse4 and not the N terminus. The lethal cse4-286 H3 swap mutant, in which 9 Cse4 amino acids located in the C-terminal third of the HFD are exchanged with their histone H3 counterparts (22), failed to interact with Scm3, suggesting that the interaction occurs through the helix 2-helix 3 region of the HFD.
Fig. 4.
Scm3 copurifies with Cse4 and cross-links to CEN3. (A) Subcellular fractionation and immunoblot analysis of HA-tagged Scm3 and Cse4. (B) Immunoblot of proteins copurifying with Cse4 from extracts of cells coexpressing Protein A-tagged Cse4 (Cse4SpA) and HA-tagged Scm3 or Scm3 mutant proteins. Purification was performed with IgG-agarose beads and eluted proteins were analyzed by immunoblotting with anti-HA antibody. Cse4SpA protein was detected by its binding to the enzyme-conjugated secondary antibody. T, total cell extract; P, affinity eluate. (C) Formaldehyde cross-linked, solubilized chromatin was immunoprecipitated with anti-HA monoclonal antibody. Total (T) and coprecipitated (P) DNA was used for multiplex PCR with the primer sets shown above the gel image.
Table 1.
Results of yeast two-hybrid tests
| BD fusion | AD fusion | Growth on -His |
|---|---|---|
| Scm3 | Empty | No |
| Empty | Scm3 | No |
| Scm3 | Scm3 | Yes |
| Scm3–31 | Scm3 | No |
| Scm3 | Cse4 | Yes |
| Scm3–31 | Cse4 | No |
| Scm3 | Cse4–286 | No |
| Scm3 | Cse4 HFD | Yes |
| Empty | Cse4 HFD | No |
| Cse4 N term | Scm3 | No |
BD, binding domain; AD, activation domain.
The two-hybrid results were confirmed by affinity purification. A strain was constructed that expressed both HA-tagged Scm3 and a functional Cse4-Protein A fusion (Cse4SpA) under control of the GAL1-10 promoter (14). Cse4SpA and associated proteins were purified from cell extracts by using IgG-conjugated agarose beads and the protein components analyzed by immunoblot analysis. Scm3-HA and Cse4SpA copurified (Fig. 4B). Copurification was abolished by deletion of the Scm3 heptad repeat domain (scm3-30HA) but not by mutations affecting the NES (scm3-3HA, scm3-13HA), although relatively less of the NES mutant proteins copurified. Whereas this difference could mean that the NES contributes to but is not essential for the interaction, it is more likely that the yield of NES mutant complexes is reduced by the presence of WT Scm3 in the strains. Because the NES mutations are lethal, the mutant alleles must be tested in a WT SCM3 background.
Chromatin immunoprecipitation (ChIP) was used to determine whether Scm3 is associated with centromeres. Yeast cells expressing Scm3-HA were treated with formaldehyde and the sheared chromatin fractionated by using anti-HA conjugated Sepharose beads. DNA in the Scm3HA and control fractions was analyzed by multiplex PCR by using primer sets designed to amplify CEN and non-CEN sequences on chromosome III. Three DNA bands are resolved by agarose gel electrophoresis, the middle band (275-bp) diagnostic for CEN3 (Fig. 4C). Scm3-HA as well as Cse4-HA immunoprecipitates were enriched for CEN3 sequences, indicative of CEN DNA specificity. CEN3 DNA was not enriched in control immunoprecipitates prepared from cells expressing Scm3 with a different epitope tag (Scm3-Myc). Thus, Scm3p, like Cse4p, selectively associates with CEN DNA.
Scm3 Depletion Results in G2/M Arrest and Aberrant Cse4 Localization.
A conditional scm3 allele was generated by using the S. cerevisiae degron system of Moqtaderi et al. (23). In this genetic background, addition of CuSO4 to the culture medium results in rapid degradation of preexisting Scm3 and repression of scm3 transcription. Strains carrying the scm3deg degron allele rapidly arrest their cell cycle upon CuSO4 addition, accumulating as distinctive large-budded cells indicative of G2/M arrest. The terminally arrested cells, operationally dubbed “dumbbells”, are distinguishable from large-budded G2/M cells in untreated cultures by their larger size and bi-lobed appearance. The arrest depends on MAD2, suggesting that the defect in Scm3-depleted cells is monitored by the spindle assembly checkpoint (SI Fig. 8).
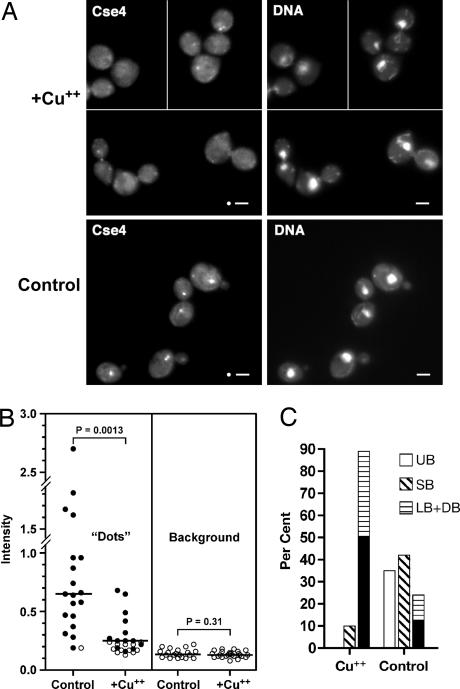
To assess the effect of Scm3 depletion on Cse4 localization, a GFP-tagged Cse4 allele was introduced into the scm3deg background. Cells were synchronized with α-factor and released from the G1 arrest after depleting Scm3. Ninety minutes after release, 90% of Scm3-depleted cells had failed to complete mitosis, with about half showing the terminal dumbbell phenotype; in contrast, control cells had doubled, few dumbbell figures were observed, and the bud morphology profile resembled that of an asynchronous population (Fig. 5C). Cse4 localization in the control cells exhibited the distinctive “dot” pattern typical of kinetochore proteins (Fig. 5A Lower), i.e., the 16 centromeres cluster as a single dot in G1 cells and as two dots after CEN DNA replication and spindle pole body separation in early S phase (24). Scoring four microscopic fields, 19 of 20 control cells were found to have one or two dots associated with the DAPI-stained mass. In Scm3-depleted cultures, about half of the cells (12 of 25 cells scored in four fields) lacked distinct Cse4 dots (P < 10−4 by χ2), and where Cse4 dots were observed, they seemed to be smaller and less intense (Fig. 5A Upper). This judgment was confirmed by quantitating the fluorescence intensity of the kinetochore dots. As seen in Fig. 5B, the median intensity of kinetochore dots in Scm3-depleted cells was significantly less than that of the controls, with the majority of distinguishable dots (solid circles) having intensities less than twice background. By comparison, only two of 19 control kinetochore dots failed to exceed the twice-background threshold (P < 10−4 by χ2). Background fluorescence did not differ between control and Scm3-depleted cells. The cell cycle arrest and Cse4 mislocalization phenotypes observed upon Scm3 depletion are not restricted to cells previously arrested in G1. Qualitatively similar results were obtained when Scm3 was depleted from cells in asynchronous culture or cells previously arrested in G2 (SI Table 2).
Fig. 5.
Scm3 depletion leads to G2/M arrest and aberrant Cse4 localization. (A) Strain R421 (scm3deg CSE4GFP) was arrested with α-factor for 2 h after which half of the culture was transferred to a separate flask and CuSO4 added to a final concentration of 0.5 mM to induce the degron and deplete Scm3. After an additional 60 min, cells from the control and Scm3-depleted cultures were rapidly filtered, washed, and resuspended in medium lacking α-factor with (Scm3-depleted) or without (control) CuSO4. Ninety minutes after release from the α-factor arrest, cells were processed for fluorescence microscopy. Images of the Scm3-depleted cells (Upper) are from a contiguous microscopic field moved digitally to minimize empty space. (Scale bars: 2 μm.) Circles, mask used for quantitating fluorescence. (B) Fluorescence intensity of Cse4-GFP dots. Filled circles, measurements of distinguishable kinetochore dots; open circles, measurements from cells lacking a distinguishable kinetochore dot. The latter were excluded from the statistical analysis. Lines show median (“Dots”) or mean (Background) values; significance of differences was assessed by Mann–Whitney or unpaired t tests, respectively. (C) Bud morphologies of samples from A. UB, unbudded; SB, small-budded; LB, large-budded; DB, dumbbell. Black shading in the LB + DB bars indicates the proportion of DB.
Discussion
Taken together, our results suggest that S. cerevisiae Scm3 plays some role in the targeting of Cse4 to CEN chromatin or its maintenance there. SCM3 is a dosage suppressor of Ts cse4 mutations, and affinity purification and two-hybrid assays demonstrate that Scm3 and Cse4 interact in vivo. The Cse4-Scm3 interaction occurs via the Cse4 HFD, and SCM3 dosage suppression is specific for cse4 HFD alleles; it is the Cse4 HFD where CEN targeting information resides (25). ChIP assays show that Scm3 and Cse4 are colocalized at CEN DNA. Finally, Scm3-depleted cells are unable to complete mitosis, and Cse4 localization is aberrant. Many cells in the arrested population lack the distinctive Cse4 dots indicative of normal kinetochore localization, and where dots are observed at the DNA margin, their Cse4 content is significantly less than that of WT cells. We interpret this finding to mean that Cse4 is missing from many but not all of the 32 G2 chromatids. Because a single nonfunctional kinetochore is able to activate the mitotic checkpoint (26), loss of Cse4 from only a few centromeres would suffice to effect the near complete cell cycle arrest we observe.
Assembly of Cse4 into CEN DNA nucleosomes requires several steps that might require a specialized assembly factor. By analogy to canonical nucleosome assembly, the first step would be association of Cse4 with H4 to form (Cse4-H4)2 tetramers. To accomplish this step, Cse4 must compete with an excess of H3 to bind H4, then Cse4-H4 dimers must selectively self-associate to avoid formation of mixed tetramers (i.e., Cse4-H4/H3-H4), which are not observed (27, 28). Next, (Cse4-H4)2 tetramers must target one of sixteen 125-bp CENs, which together comprise only 0.01% of the genomic DNA. Scm3 could confer Cse4-Cse4 specificity by binding to and stabilizing (Cse4-H4)2 tetramers, or it could provide Cse4-CEN DNA specificity by binding Cse4 (either as a monomer or Cse4–H4 complex) and delivering it to the site of CEN chromatin assembly, perhaps through protein-protein interaction with another centromere protein. An attractive partner would be CBF3, the inner kinetochore protein that binds CDEIII and on which the incorporation of all other kinetochore components depends (9). The observed protein-protein interaction between Scm3 and Cse4 is consistent with either scenario.
Could Scm3 be a Cse4-specific histone chaperone? Histone chaperones are acidic proteins that associate with core histones and facilitate nucleosome assembly—NAP-1, CAF-1, and HIRA are well-known examples (29). In Drosophila, Cid (the Drosophila Cse4 ortholog) is found in a soluble complex with histone H4 and the chaperone RbAp48, and it is proposed that the Cid/H4/RbAp48 complex is responsible for deposition of the CenH3 in vivo (30). But RbAp48 is not centromere-specific. It is also a component of H3.1/CAF-1 and H3.3/HIRA complexes responsible for replication-dependent and replication-independent nucleosome assembly pathways, respectively (31). Likewise the fission yeast RbAp48 homolog, Mis16, is required for deposition of Cnp1 (the S. pombe CenH3), but Mis16 localization is not restricted to centromere DNA (32). It is proposed that Mis16 centromere specificity is conferred by Mis18 (32). The S. cerevisiae RbAp48 homolog is Msi1/Cac3—a subunit of yeast CAF-1 (33)—not Scm3; however, Scm3 could serve a Mis18-like function, lending CEN specificity to a Cse4-chaperone complex. In this model, interaction between Scm3 and Cse4 is indirect; both are components of the same Cse4 chaperone complex. Alternatively, Scm3 could be the Cse4-binding subunit in a CEN-specific, CAF-1-like assembly complex. The presence in Scm3 of the essential D/E-rich domain is consistent with a histone chaperone function.
The Leu-rich NES homology domain in Scm3 directs nuclear export in vivo. The NES is conserved in the Scm3 proteins of other point centromere yeasts but is not found in the orthologs of more diverged hemiascomycetes or other fungi. Interestingly, NAP-1 contains a Leu-rich NES and is known to shuttle between nucleus and cytoplasm (34). Mutations that abolish NAP-1 shuttling disrupt transcription, presumably because of defects in histone deposition (34). Possibly, Scm3 acts as a carrier to shuttle Cse4 into and out of the nucleus. The Scm3 NLS was not identified. Whereas Scm3 contains two short stretches of basic amino acids that are potential NLSs, they are essential neither alone nor in combination; thus, some other region in the Scm3 protein can supply NLS function. Scm3 must gain access to the nucleus, because fractionation studies indicate that Scm3 is predominantly chromatin-bound, and ChIP shows it to be associated with CEN DNA.
Scm3 contains a conserved protein domain found in proteins of 44 other fungi including budding and fission yeasts, filamentous fungi, and basidiomycetes. In the Pfam database (35), the Scm3 homology domain defines protein family B_19394, although only nine members are annotated. In the S. cerevisiae protein, the domain consists of heptad repeats of hydrophobic amino acids predicted by COILS (36) to have some coiled-coil character. Coiled coils often provide the interface for specific protein-protein interaction (17), and mutation of hydrophobic residues in the heptad repeat (scm3-31) result in loss of Scm3-Scm3 and Scm3-Cse4 interactions detected by two-hybrid analysis. The scm3-31 mutation is lethal, suggesting that one or both interactions are essential for Scm3 function. In other homologs, amino acid sequence is conserved but spacing is not, leading us to doubt the original proposition of a coiled-coiled structure. The phylogeny of the Scm3 homology domain is consistent with species phylogeny, suggesting orthology; however, outside this 50 aa domain, little homology is observed except between orthologs of close relatives. The proteins range in size from ≈200 aa in the Saccharomyces yeasts to >1,000 aa in N. crassa and C. cinerea.
Orthologs of several S. cerevisiae kinetochore proteins are found in insects, plants, and metazoans including humans, whereas others are restricted to yeasts having point centromeres (1). Conversely, some kinetochore proteins are conserved between fungi and metazoans but are limited to organisms having regional centromeres and are not found in S. cerevisiae and other point centromere yeasts. Finally, other proteins are found associated with both point and regional CENs but only in fungi. SCM3 joins the fungi-restricted group. Our biochemical and genetic characterization of Scm3 function in S. cerevisiae suggests that Scm3 plays a role in the targeting of Cse4 to CEN chromatin or its maintenance there. Future work will be needed to determine whether the fungal orthologs are kinetochore proteins in their respective organisms and whether they execute an analogous function in localizing CenH3.
Materials and Methods
Strain CDL151 (MATa ade2-101 his3-11,15 leu2-3 lys2-801 trp1-901 ura3-52 scm3::LEU2) carrying the episomal SCM3-URA3 plasmid pMU3 was used for scm3 complementation tests. CDL151 was transformed with TRP1 plasmids carrying scm3 alleles to be tested. Transformants were then plated onto 5-flouroorotic acid (FOA) medium to select for loss of pMU3. Noncomplementing mutant alleles produce no FOAr progeny, because loss of pMU3 is lethal. Strain PJ69-4A was used for two-hybrid assays as described (37). The pGAD and pGBD plasmids were cotransformed, and interaction was assessed on His dropout medium containing 1 mM 3-aminotriazole. The Scm3 degron allele in strain R421 was obtained as described by Moqtaderi et al. (23); the CSE4GFP allele was integrated at the CSE4 locus. Details of plasmid construction and yeast genetic procedures can be found in SI Materials and Methods.
The genetic assay for NES function was described by Shulga et al. (21). A DNA fragment encoding the Scm3 NES and flanking amino acids (nucleotides 4–156 of the SCM3 ORF) was obtained by PCR and cloned between the EcoRI and BamHI sites of the reporter plasmid (pSW715). An analogous construct was made for the scm3-3 triple Ala substitution mutant. These plasmids, along with pSW715 (no insert), pSW716 (Gle1 NES insert), pSW722 (Rev NES), and pSW723 (Rev nonfunctional NES) were transformed into test strain L40. Transformants were streaked on Trp dropout and Trp/His double dropout media and scored for growth after 3 days.
Yeast transformations were carried out by the lithium acetate procedure (38). Media were as described (39). For α-factor arrest, cells were incubated at a cell density of 1 × 106 cells per ml in medium containing 3 × 10−6 M α-factor. Coprecipitation of protein A-tagged Cse4, immunoblotting, and ChIP were carried out as described (14). Subcellular fractionation was by the method of Liang and Stillman (40). Bud morphology was scored as follows: unbudded, no bud; small-budded, bud < 50% size of mother; large-budded, bud > 50% size of mother; dumbbell, mother and bud approximately equal in size, both rounded rather than oval in shape.
Fluorescence microscopy was performed on a Nikon Eclipse E800 microscope with ×100 oil immersion objective (1.40 N.A.). Samples were prepared as described (41). Images were captured on a Santa Barbara Instrument Group ST-8 CCD camera by using identical exposure times for control and Scm3-depleted samples. Fluorescence intensity of Cse4-GFP kinetochore dots was quantitated by using NIH Image 1.63 (http://rsb.info.nih.gov/nih-image) after converting the raw image files to 8-bit TIFFs, keeping brightness and contrast settings equal for control and experimental images, and then inverting the images. Average pixel intensities in a 5.9-pixel (0.5 μm) diameter circular mask were measured and expressed [as optical density (O.D.)]. Budded cells were counted as a single cell with the measurement taken in the bud containing the DNA. All cells were scored. In cases where a kinetochore dot was not observed, a reading was taken at the nuclear periphery where the fluorescence appeared most intense. Where two dots were observed (G2/M cells), both were measured, the values were summed, and 1× background was subtracted. Background readings were taken over cytoplasmic regions where the fluorescence appeared most dim. Statistical analysis was performed by using Prism 4 software (GraphPad, San Diego, CA).
Supplementary Material
Acknowledgments
We thank Christina Defalco, Julianna Layzer, Jeffery Marblestone, and Yinhuai Chen for contributions in the initial stages of the project, Cindy Gingrich for help in characterizing the degron strains, and Sue Biggins for encouragement and helpful comments on the manuscript. This work was supported by National Institutes of Health Grants GM54766 (to M.F.-H.) and GM61120 (to R.E.B).
Abbreviations
- CenH3
centromeric H3
- CEN
centromere
- CDE
conserved DNA element
- HFD
histone fold domain
- NES
nuclear export signal
- NLS
nuclear localization sequence
- Ts
temperature-sensitive.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0703178104/DC1.
References
- 1.Meraldi P, McAinsh AD, Rheinbay E, Sorger PK. Genome Biol. 2006;7:R23. doi: 10.1186/gb-2006-7-3-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choo KH. Dev Cell. 2001;1:165–177. doi: 10.1016/s1534-5807(01)00028-4. [DOI] [PubMed] [Google Scholar]
- 3.Malik HS, Henikoff S. Nat Struct Biol. 2003;10:882–891. doi: 10.1038/nsb996. [DOI] [PubMed] [Google Scholar]
- 4.Sullivan KF. Curr Opin Genet Dev. 2001;11:182–188. doi: 10.1016/s0959-437x(00)00177-5. [DOI] [PubMed] [Google Scholar]
- 5.Henikoff S, Dalal Y. Curr Opin Genet Dev. 2005;15:177–184. doi: 10.1016/j.gde.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 6.Sullivan BA, Blower MD, Karpen GH. Nat Rev Genet. 2001;2:584–596. doi: 10.1038/35084512. [DOI] [PubMed] [Google Scholar]
- 7.Hegemann JH, Fleig UN. Bioessays. 1993;15:451–460. doi: 10.1002/bies.950150704. [DOI] [PubMed] [Google Scholar]
- 8.Sorger PK, Doheny KF, Hieter P, Kopski KM, Huffaker TC, Hyman AA. Proc Natl Acad Sci USA. 1995;92:12026–12030. doi: 10.1073/pnas.92.26.12026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Wulf P, McAinsh AD, Sorger PK. Genes Dev. 2003;17:2902–2921. doi: 10.1101/gad.1144403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meluh PB, Yang P, Glowczewski L, Koshland D, Smith MM. Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- 11.Espelin CW, Simons KT, Harrison SC, Sorger PK. Mol Biol Cell. 2003;14:4557–4568. doi: 10.1091/mbc.E02-08-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steiner NC, Hahnenberger KM, Clarke L. Mol Cell Biol. 1993;13:4578–4587. doi: 10.1128/mcb.13.8.4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hosouchi T, Kumekawa N, Tsuruoka H, Kotani H. DNA Res. 2002;9:117–121. doi: 10.1093/dnares/9.4.117. [DOI] [PubMed] [Google Scholar]
- 14.Chen Y, Baker RE, Keith KC, Harris K, Stoler S, Fitzgerald-Hayes M. Mol Cell Biol. 2000;20:7037–7048. doi: 10.1128/mcb.20.18.7037-7048.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bogerd HP, Fridell RA, Benson RE, Hua J, Cullen BR. Mol Cell Biol. 1996;16:4207–4214. doi: 10.1128/mcb.16.8.4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jans DA, Xiao CY, Lam MH. BioEssays. 2000;22:532–544. doi: 10.1002/(SICI)1521-1878(200006)22:6<532::AID-BIES6>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 17.Lupas A. Trends Biochem Sci. 1996;21:375–382. [PubMed] [Google Scholar]
- 18.James TY, Kauff F, Schoch CL, Matheny PB, Hofstetter V, Cox CJ, Celio G, Gueidan C, Fraker E, Miadlikowska J, et al. Nature. 2006;443:818–822. doi: 10.1038/nature05110. [DOI] [PubMed] [Google Scholar]
- 19.Fukuda M, Gotoh I, Gotoh Y, Nishida E. J Biol Chem. 1996;271:20024–20028. doi: 10.1074/jbc.271.33.20024. [DOI] [PubMed] [Google Scholar]
- 20.Wen W, Meinkoth JL, Tsien RY, Taylor SS. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 21.Shulga N, James P, Craig EA, Goldfarb DS. J Biol Chem. 1999;274:16501–16507. doi: 10.1074/jbc.274.23.16501. [DOI] [PubMed] [Google Scholar]
- 22.Keith KC, Baker RE, Chen Y, Harris K, Stoler S, Fitzgerald-Hayes M. Mol Cell Biol. 1999;19:6130–6139. doi: 10.1128/mcb.19.9.6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moqtaderi Z, Bai Y, Poon D, Weil PA, Struhl K. Nature. 1996;383:188–191. doi: 10.1038/383188a0. [DOI] [PubMed] [Google Scholar]
- 24.He X, Rines DR, Espelin CW, Sorger PK. Cell. 2001;106:195–206. doi: 10.1016/s0092-8674(01)00438-x. [DOI] [PubMed] [Google Scholar]
- 25.Morey L, Barnes K, Chen Y, Fitzgerald-Hayes M, Baker RE. Eukaryot Cell. 2004;3:1533–1543. doi: 10.1128/EC.3.6.1533-1543.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wells WA, Murray AW. J Cell Biol. 1996;133:75–84. doi: 10.1083/jcb.133.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Westermann S, Cheeseman IM, Anderson S, Yates JR, 3rd, Drubin DG, Barnes G. J Cell Biol. 2003;163:215–222. doi: 10.1083/jcb.200305100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shelby RD, Vafa O, Sullivan KF. J Cell Biol. 1997;136:501–513. doi: 10.1083/jcb.136.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loyola A, Almouzni G. Biochim Biophys Acta. 2004;1677:3–11. doi: 10.1016/j.bbaexp.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 30.Furuyama T, Dalal Y, Henikoff S. Proc Natl Acad Sci USA. 2006;103:6172–6177. doi: 10.1073/pnas.0601686103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 32.Hayashi T, Fujita Y, Iwasaki O, Adachi Y, Takahashi K, Yanagida M. Cell. 2004;118:715–729. doi: 10.1016/j.cell.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 33.Game JC, Kaufman PD. Genetics. 1999;151:485–497. doi: 10.1093/genetics/151.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mosammaparast N, Ewart CS, Pemberton LF. EMBO J. 2002;21:6527–6538. doi: 10.1093/emboj/cdf647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, et al. Nucleic Acids Res. 2006;34:D247–51. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lupas A, Van Dyke M, Stock J. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 37.James P, Halladay J, Craig EA. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gietz RD, Woods RA. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- 39.Baker RE, Masison DC. Mol Cell Biol. 1990;10:2458–2467. doi: 10.1128/mcb.10.6.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang C, Stillman B. Genes Dev. 1997;11:3375–3386. doi: 10.1101/gad.11.24.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Biggins S, Severin FF, Bhalla N, Sassoon I, Hyman AA, Murray AW. Genes Dev. 1999;13:532–544. doi: 10.1101/gad.13.5.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Crooks GE, Hon G, Chandonia JM, Brenner SE. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.