Abstract
The Genome Consortium for Active Teaching (GCAT) facilitates the use of modern genomics methods in undergraduate education. Initially focused on microarray technology, but with an eye toward diversification, GCAT is a community working to improve the education of tomorrow's life science professionals. GCAT participants have access to affordable microarrays, microarray scanners, free software for data analysis, and faculty workshops. Microarrays provided by GCAT have been used by 141 faculty on 134 campuses, including 21 faculty that serve large numbers of underrepresented minority students. An estimated 9480 undergraduates a year will have access to microarrays by 2009 as a direct result of GCAT faculty workshops. Gains for students include significantly improved comprehension of topics in functional genomics and increased interest in research. Faculty reported improved access to new technology and gains in understanding thanks to their involvement with GCAT. GCAT's network of supportive colleagues encourages faculty to explore genomics through student research and to learn a new and complex method with their undergraduates. GCAT is meeting important goals of BIO2010 by making research methods accessible to undergraduates, training faculty in genomics and bioinformatics, integrating mathematics into the biology curriculum, and increasing participation by underrepresented minority students.
INTRODUCTION
Science and mathematics education plays a vital role in the preparation of tomorrow's scientists, teachers and parents, doctors and patients, and scientifically literate citizens. For years, many leaders in science and education have called for reform (Project Kaleidoscope [PKAL], 2001; National Research Council [NRC], 2003, 2005; Handelsman et al., 2004; Steen, 2005). To help guide the reform process, the NRC (2003) produced a report entitled BIO2010. The report concludes that although advances in technology have caused a dramatic transformation in biological research, undergraduate biology education has not kept pace. Among the remedies offered, four major recommendations are of critical importance: 1) integrate mathematics and physical science within cell and molecular biology courses; 2) redesign lab courses to be interdisciplinary and based on research projects, rather than canned labs with predictable outcomes; 3) provide faculty development in modern disciplines such as genomics and bioinformatics; and 4) increase the number of students from underrepresented minorities in the talent pool from which future scientists will emerge. These four recommendations present many challenges, but professional societies, institutions, departments, and forward-thinking faculty throughout the country are working to address them (Kumar, 2005; Campbell et al., 2006a; Kuldell, 2006; Pfund et al., 2006). One of these efforts is the Genome Consortium for Active Teaching (GCAT), the only laboratory-based model curriculum mentioned in BIO2010. This report documents the first 6 yr of GCAT activity and GCAT's progress toward accomplishing BIO2010 recommendations.
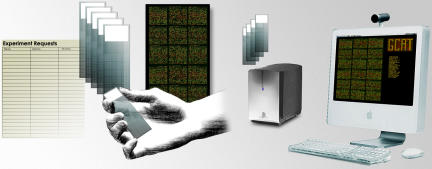
GCAT's mission is to bring modern genomics to undergraduate students, primarily through student research and research-based laboratory curricula. Our primary focus has been the use of DNA microarrays (sometimes referred to as chips) as a means to address the four BIO2010 recommendations outlined above (see Supplemental Material A for an overview of microarray methodology). The annual operational cycle of GCAT is illustrated in Figure 1. In the spring, GCAT solicits requests for DNA microarrays from participating faculty. Microarrays from 11 different species are currently available to GCAT members. GCAT contracts for the production of microarrays during the summer and distributes the microarrays in the fall. Faculty and students design and perform their experiments and ship their hybridized chips overnight for scanning on a GCAT community scanner purchased with support from the National Science Foundation (NSF), or on backup scanners available at other locations. GCAT then delivers tiff microarray data files to the student investigators by File Transfer Protocol (FTP). Students and faculty analyze their own data, and they have access to data produced by all other GCAT members. Many investigators use MAGIC Tool (Heyer et al., 2005), free software provided by GCAT (Heyer and Campbell, 2004a).
Figure 1.
Outline of GCAT microarray distribution plan. Faculty who teach undergraduates submit their requests for microarrays, which are produced by several academic labs. Students perform the experiments, then the chips are scanned and data are posted to an FTP server for students to analyze.
GCAT members are free to pursue their own research or research-style teaching without any limitations by GCAT. The only requirements for participation are 1) only undergraduates may use the microarrays; 2) faculty and students must participate in assessment; and 3) all data and protocols are open access for the GCAT community. Faculty training in microarray laboratory protocols and data analysis methods is provided by workshops. NSF has funded three workshops to date, and it has recently awarded GCAT a new grant to fund three more workshops during summers 2007, 2008, and 2009. New and veteran GCAT faculty alike appreciate the collective expertise and support of the GCAT community, as evidenced by the high level of activity on the GCAT-Listserv (GCAT-L) e-mail distribution list.
Based on assessment data from students and faculty, GCAT is having a significant impact. Faculty report very strong support for GCAT, and students report learning gains and attitudinal changes as a result of their GCAT experiences. Faculty self-reported substantial gains in their scholarship and teaching activities as well as overall satisfaction with GCAT. This report documents these successes and identifies new ways in which GCAT can reach a wider audience.
MATERIALS AND METHODS
DNA Microarray Resources
Currently, DNA microarrays are purchased with funds from Howard Hughes Medical Institute (HHMI) awarded to Grinnell, Pomona, Swarthmore, and Davidson. Chips were produced at numerous academic labs. The cost to participating faculty is $50 for the first microarray and $20 for each additional microarray per species to cover the costs of shipping and scanning. For the first 3 yr, before the advent of HHMI funding, academic labs donated the chips to GCAT free of charge; this service provided crucial support necessary for launching GCAT (from Patrick Brown at Stanford University [Stanford, CA] in year 1 and Leroy Hood at the Institute for Systems Biology [Seattle, WA] in years 2 and 3).
MAGIC Tool software was developed using funds from Davidson College, HHMI, and NSF. The software is written in Java, so it works on all operating systems (i.e., Macintosh OS X, Windows, and Linux), is freely available for downloading (Heyer and Campbell, 2004a), and is open source. Computers must have at least 512 MB of RAM to run MAGIC Tool, but we recommend 1–2 GB of RAM for optimal performance. GCAT offered NSF-funded workshops during summers 2003, 2004, and 2005. In 2004, we offered one complete workshop (data analysis and wet lab components) as well as some sessions for data analysis only (Campbell et al., 2006a). The workshop participants produced and analyzed two-color microarray data from the yeast diauxic shift from anaerobic to aerobic metabolism. In that way, the participants were able to obtain data directly comparable with a ground-breaking published study (DeRisi et al., 1997). Professors using variations on this experiment in their classes can augment their student data by adding the public domain data before analysis (Heyer and Campbell, 2004b). The two concurrent workshops in 2005 were 5 d long, including training in both data production and data analysis (GCAT, 2005). NSF funding has now been obtained for additional workshops in 2007, 2008, and 2009 to cover both aspects of microarray work. All workshop participants receive materials to take home, including a CD containing MAGIC Tool software, and all raw and analyzed data produced at the workshop; the MAGIC Tool user's guide; a data analysis exercise for further practice; guided activities in comparing and clustering gene expression profiles; a reading quiz on the DeRisi paper; laboratory protocols with annotations; guidelines for faculty timing of weekly labs; and notes on reagents and suppliers. The hands-on, collaborative nature of the workshop ensures that participants have experienced the microarray process from beginning to end and have had a chance to learn from mistakes in a supportive environment. Continued support for faculty participants is provided after the workshops via e-mail (GCAT, 2003) and by a helpdesk (staffed by NSF-funded undergraduates), through 2009. In addition, some of us have led other less comprehensive workshops at a variety of locations.
Surveys and Statistics
All student and faculty surveys were conducted online (Tonidandel, 2004). A pre- and postterm design was used for the student assessment, whereas faculty completed an assessment only at the end of the term. The preterm assessment (see Supplemental Material B) was completed by students at the beginning of a semester. The assessment asks students to respond with basic demographic information and to complete an 11-item test of prior knowledge (see Supplemental Material D). When the semester was over, students' knowledge was again assessed along with their attitudes about using the GCAT materials (see Supplemental Material C). The online survey for faculty asked whether they used the GCAT materials in their classes, and it assessed their attitudes regarding the GCAT experience in their particular course. A mixed factorial analysis of variance was used to evaluate statistical significance.
Of 52 professors identified by students as supervising the use of GCAT materials at their home institutions, 43 professors completed the faculty survey at the end of the program. Three faculty members responded to the survey twice because they used GCAT materials in both semesters of the 2005–2006 school year. In July 2004, GCAT offered a series of NSF-sponsored hands-on workshops for faculty interested in curricular innovations to include gene expression analysis via microarrays. Thirty-seven participants attended one of two 1.5-d dry lab workshops that introduced the microarray method and covered data analysis by using public domain data. Participants learned to work with the open-source MAGIC Tool spot-finding and analysis software, along with other free packages, to analyze public domain data sets in short projects. Twenty-three additional participants continued with a 2.5-d hybridization workshop, which involved hands-on preparation of fluorescently labeled probes for yeast expression microarrays, their hybridization, data acquisition, and data analysis by using the methods presented in the earlier workshop. In June 2005, the GCAT team conducted a follow-up evaluation to assess the degree to which the participants met their goals. An e-mail invitation was sent to 39 faculty members in late May 2005, requesting that they respond to a 10-item online survey assessing participants' retrospective evaluation of the workshops and their use of the microarray tools during the 2004–2005 academic year. Twenty-five people (64%) returned surveys by June 10; of those, 10 participated in the wet and dry labs, 14 only in the dry labs, and 1 was unable to attend but gathered and implemented the workshop materials.
RESULTS
Origin and Growth of GCAT
The concept of GCAT was inspired by a 1999 presentation given by Dr. Pat Brown of Stanford University. Two of us (A.M.C. and M.L.L.) realized that this technology embodied the power of genome-wide strategies and could be affordable for undergraduate institutions if we pooled our resources. Brown agreed to provide us with 144 yeast DNA microarrays. With this promise, we used the annual meetings of PKAL and the Council on Undergraduate Research (CUR) to recruit faculty who would be willing to take a collective leap of faith and learn how to conduct microarray experiments together but on different campuses. None of us had ever performed such an experiment, but the procedure was conceptually accessible and seemed relatively straightforward. Twenty-three faculty agreed to participate in the inaugural year (2000–2001) of GCAT.
During GCAT's first year, we realized that two potential limitations might prevent widespread adoption of microarray strategies: the cost of microarrays themselves and access to a microarray scanner. A collaborative grant from NSF allowed us to purchase a scanner in fall 2001 with additional funding from Missouri Western State University, Pomona College, and Davidson College. User fees of $20 per microarray were collected to cover the expense of its service contract. After the first year of demonstrated success using the microarrays with undergraduates, other academic labs donated additional microarrays. Dr. Hood, for example, donated 400 yeast DNA microarrays over 2 yr. We used Michael Eisen's ScanAlyze (Eisen, 2006) and commercial GeneSpring software (Agilent Technology, Santa Clara, CA) to analyze the data, because they were offered to GCAT members free of charge for educational purposes.
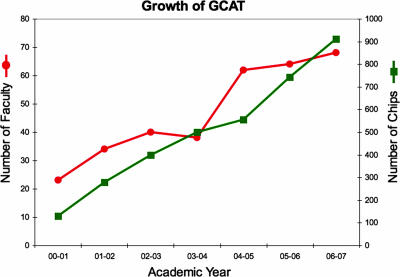
Because GCAT relied on donated microarrrays, we were hesitant to advertise in any formal manner. However, a number of our institutions were invited to participate in the HHMI competition for undergraduate institutions in 2003. Working with guidance from Stephen Barkanic of HHMI, the proposed budgets in applications from 24 GCAT member institutions included funds to support direct costs of the consortium. We hoped that 20% of those institutions would be successful, allowing extended stable support. In fall 2004, Pomona, Grinnell, Swarthmore, and Davidson were each awarded 4-yr educational grants from HHMI, which included funding for GCAT. With this funding, we could purchase microarrays to meet the growing demand of the consortium. For yeast microarrays, we purchased our own whole-genome oligonucleotide sets and contracted with the microarray core facility at Washington University in St. Louis, MO, to produce microarrays for GCAT. Membership in GCAT has continued to grow in two dimensions (Figure 2), beginning with exclusively yeast microarrays in 2001 to 11 different types of microarrays in 2006. In the first 7 yr, GCAT provided ∼5000 DNA microarrays for use by approximately 6000 undergraduates.
Figure 2.
GCAT growth over seven years. GCAT has expanded the number of microarrays distributed (right Y-axis) and the number of faculty (left Y-axis) participating each year.
As GCAT grew, we recognized two new limiting factors: faculty training and appropriate software. Very few GCAT faculty had formal training with microarray techniques or analysis of the data. This need was expressed very clearly by the numerous attendees at an American Society for Microbiology (AMC) symposium chaired by AMC in 2002 (Campbell, 2002). In response, a core group decided to develop student-friendly lab protocols and to offer workshops for faculty training. Free software programs such as ScanAlyze and Cluster (Eisen, 2006) were restricted to the Windows platform, whereas many commercial packages were cumbersome and prohibitively expensive. Therefore, one of us (L.J.H.) worked with several undergraduates to write MAGIC Tool (Heyer et al., 2005) for data analysis. MAGIC Tool is written in Java, and so runs on all major computer platforms; it is freely available, and is open source (Heyer and Campbell, 2004a).
From the outset, GCAT has been guided by a few simple principles:
bring genomic methods into the undergraduate curriculum, primarily through student research;
share resources to make experiments affordable;
be as inclusive as possible so all schools can participate;
create a clearinghouse of information for faculty;
provide all data freely to anyone for pedagogical use;
develop a distributed community to help each other trouble-shoot and develop curriculum;
make assessment a fundamental requirement for participation; and
encourage participants to set their own educational and research goals.
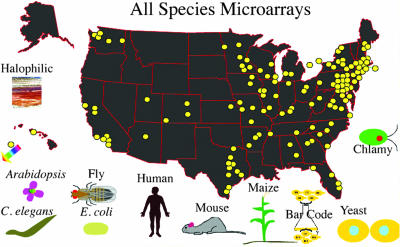
By following these principles, GCAT has reached many campuses, some of which have been overlooked in national educational reform efforts (e.g., small campuses and community colleges), have student populations who are underrepresented in science, or both (Figure 3). Microarrays provided by GCAT have been used by 141 faculty on 134 campuses in 36 states as well as two universities in Canada and one university in Australia. Of the 134 U.S. campuses, 21 (16%) serve large numbers of students from underrepresented minorities (1 in Hawaii, 3 that serve a mixture of ethnic groups, 7 historically black colleges or universities, and 10 Hispanic-serving institutions; U.S. Department of Education, 2006). A majority of the GCAT campuses are small, 4-yr, liberal arts colleges, but GCAT membership includes faculty from three community colleges as well as large universities such as the University of Georgia (25,000 undergraduates), California State University at Sacramento (23,000 undergraduates), University of Louisville (22,000 undergraduates), Boston College (9000 undergraduates), University of Southern Maine (8600 undergraduates), and Georgetown University (7000 undergraduates). In addition to direct support for these schools, GCAT has provided student-friendly protocols, curriculum, and pedagogical advice to research powerhouses such as Massachusetts Institute of Technology (MIT, Cambridge, MA) and University of California at San Diego as they began using microarrays in undergraduate laboratories. GCAT helps faculty overcome some of the common barriers to the introduction of new technologies into undergraduate curricula, and it offers faculty the freedom to adapt the materials to their own research interests and institutions.
Figure 3.
Map of GCAT-participating schools. GCAT is composed of 141 faculty on 134 campuses in 36 states, including two universities in Canada and one in Australia (colored arrow), with nodes serving as hyperlinks to the appropriate departments (GCAT, 2006b). This screen shot is from an interactive map that allows viewers to see the geographical distribution of users for each type of microarray. Contact information for GCAT faculty is available and organized by academic year.
Student Outcomes
Students who work with DNA microarrays tend to be juniors (29%) or seniors (57%), and either biology (74%) or chemistry (14%) majors. Men and women are equally represented and 18% are from underrepresented ethnic groups. Nearly 60% want to pursue medical careers and 33% want to pursue a Ph.D. in cell/molecular biology. Based on these career goals, it is not surprising that most students had already completed introductory biology (93%), organic chemistry (77%), calculus (72%) and first-year physics (67%). Only 5% had completed a course in genomics or bioinformatics before working with the GCAT materials.
Nearly 80% of the students were able to progress through the experimental procedure far enough to have their microarrays scanned, although only 54% reported that they obtained usable data. Of faculty surveyed, 70% reported that some of their students obtained usable data. Among these 34 faculty, the average success rate (scanned microarrays with usable data) of their students was 81%, with 22 faculty reporting that 100% of their students produced usable data. Therefore, the overall student success rate as reported by faculty is estimated to be ∼56% (0.70 × 0.81). By comparison, 212 (76%) of the 277 microarrays scanned at Davidson College between June 17, 2005, and July 28, 2006, produced usable results. This is a very impressive success rate for a method that early skeptics thought was impossible for undergraduates to use. Notably, some faculty whose students at first obtained no usable data are now getting very good results due to improved methods for RNA isolation and cDNA production. As faculty determine the best way to produce labeled probes for their system, the success rate should continue to climb above 70%.
Knowledge Gains.
Eleven questions to test knowledge were presented in identical form on the two surveys (pre- and postsurveys) taken many weeks apart; in total, 409 students responded to both sets of questions (see Supplemental Material B–D for the 2 surveys and 11 knowledge questions). Students were instructed to answer questions without the use of notes or consultation with friends. Those questions presented hypothetical scenarios pertaining to gene expression and microarray experimentation techniques. The questions were not focused on details or specific facts, but they were designed to be very challenging and to emphasize problem solving and data analysis. With the exception of the final question, correct response rates for each question in the presurvey were below 50%. On average, students were not knowledgeable about microarray experimentation relating to either DNA or RNA at the outset of their GCAT experiences. The average percentage of correct responses across all test items before GCAT training was 30.5%. Item 5 was particularly difficult for student participants; only 5.0% of students answered it correctly on the preprogram survey. Correct response rates for each item and students' knowledge gains are found in Table 1.
Table 1.
Scores on 11 knowledge questions for pre- and postsurveysa (n = 409)
| Question | Subject matter | Correct pre-GCAT (%) | Correct post-GCAT (%) | Increase (%) |
|---|---|---|---|---|
| 1 | Microarray experimental error–dye bias | 23.1 | 59.3 | 36.2 |
| 2 | Microarrary experimental error–gradient | 32.7 | 43.2 | 10.5 |
| 3 | Microarray negative controls | 28.9 | 39.2 | 10.3 |
| 4 | Microarray experimental design | 33.9 | 72.1 | 38.2 |
| 5 | Gene expression ratios using a graph | 5.0 | 10.8 | 5.8 |
| 6 | Gene expression–probability | 20.9 | 21.1 | 0.2 |
| 7 | Gene expression–gene clusters | 30.0 | 52.3 | 22.3 |
| 8 | Gene expression–regulatory cascade | 28.4 | 43.3 | 14.9 |
| 9 | Gene expression–gene circuit graphs | 37.9 | 49.7 | 11.8 |
| 10 | Interpreting microarray results | 40.0 | 59.0 | 19.0 |
| 11 | Diagnosis with microarrays | 54.9 | 67.4 | 12.5 |
a Performance increased significantly (p < 0.05) on all questions except item 6.
Knowledge scores improved substantially after the GCAT program; the average percentage of correct responses on the post-GCAT survey was 47.1%. Correct responses for each item increased on average by 16.5%. All gains were statistically significant, with the exception of item 6. Questions 1 and 4 showed particularly large improvements, and both specifically pertain to microarray experimentation. Knowledge gains and final performance were lowest on items 5 (10.8% correct) and 6 (21.1% correct); the subject matter for these two questions relates to gene expression ratios and probability.
Although the pre- and postsurveys showed significant gains for students who worked with GCAT materials, it would be interesting to know whether similar gains were possible for students who learned about microarrays in a lecture-only course. Fortunately, one GCAT faculty member volunteered to have her genomics lecture course of 18 students take the pre- and postsurveys as a control group. Lectures and reading assignments were congruent with other classes that used GCAT materials, but the control class did not conduct laboratory experiments. Students in the control group gained an average of 3.5% correct responses at the end of the semester, and this increase was not statistically significant. Conversely, the remaining students, who implemented GCAT materials in their laboratories (n = 377), showed significant increases on knowledge questions (p < 0.01); the average student increased by 16.4%. This improvement is roughly equivalent to two additional correct answers on the 11-item quiz. There was significant correlation with time spent working with the microarrays and use of GCAT materials (p < 0.05). Students who conducted microarray experiments improved significantly in knowledge assessments over the course of a semester, whereas students who did not participate in laboratory activities did not show significant knowledge gains over the same amount of time.
Attitude Changes.
After their GCAT experiences, students rated their change in interest and understanding of genomics, biology, and research on a 7-point scale where 1 is decreased a lot and 7 is increased a lot. On average, students' interest and understanding of all three areas increased over the course of the GCAT program (Table 2). Students also rated the effectiveness of various GCAT activities on a 7-point scale, where 1 is not effective at all and 7 is highly effective. Table 3 presents the percentage of students who rated these activities at least 4.00 and at least 5.00 on the 7-point scale. The average effectiveness value students assigned to all of the activities was 5.20, and mean scores on individual activities ranged from 5.06 to 5.32. On average, students did not judge any activity to be drastically more or less effective than others. All average ratings are above 5.0 on the 7-point scale, indicating that students judged all of the activities to be effective.
Table 2.
Student attitude change (on a 7-point scale where 1 is decreased a lot and 7 is increased a lot) in interest and understanding of subject areas (n = 409)
| Area | Mean | SD |
|---|---|---|
| Genomics | 5.5 | 1.1 |
| Biology | 5.5 | 1.1 |
| Research | 5.4 | 1.2 |
Table 3.
Student responses (on a 7-point scale where 1 is not effective at all and 7 is highly effective) measuring satisfaction with methods used in lab
| GCAT activity | % of students who rated the activity with at least 4.00 or 5.00 |
|||
|---|---|---|---|---|
| Mean | ≥ 4.00 | ≥ 5.00 | Na | |
| Practicing data analysis before I began analyzing my own data | 5.25 | 93.6 | 67.1 | 313 |
| Isolating RNA or genomic DNA to produce probe | 5.32 | 94.1 | 70.0 | 323 |
| Producing the fluorescently labeled probe | 5.22 | 94.4 | 68.9 | 306 |
| Hybridizing the probe with the spotted DNA | 5.20 | 92.8 | 70.1 | 334 |
| Designing my own experiment | 5.13 | 87.3 | 64.3 | 244 |
| Analyzing data from public domain source | 5.22 | 94.7 | 65.8 | 325 |
| Reading papers that used DNA microarrays | 5.06 | 88.9 | 62.4 | 343 |
a Number of students who did not rate the activity ″not applicable.″
Faculty Outcomes
2005–2006 Academic Year Responses.
Faculty estimated the number of weeks allocated for each of the activities performed by their students (i.e., isolate mRNA, make cDNA probes, make total genomic DNA probes, hybridize probes to microarray, analyze students' own data, analyze data from public sources, and (students) design their own experiments). We evaluated both the time devoted to each activity (e.g., 2.6 wk) and the frequency with which faculty members reported a particular activity (e.g., 80% of the faculty may have reported doing a particular activity) but not the frequency with which each activity was done at each institution (i.e., 80% does not mean an activity was done 80% of the time at an institution). Hybridizing probes to a microarray (80.0%) was the activity reported most often by the faculty, whereas only a small percentage asked that students make total genomic DNA probes (12.2%). Excluding the lecture-only control group, 90.6% of the professors reported that their students performed at least three of the GCAT activities during the semester, using an average of 1.8 wk per task. Students were given the most time to make total genomic DNA probes (2.6 wk) and to analyze their own data (2.5 wk). The least amount of time was allotted for making cDNA probes and analyzing public domain data (1.12 wk for each). Professors were asked how they measured student performance when they used GCAT materials (Table 4). The most common assessment tool used by GCAT professors was informal feedback (62.2%), but term papers and lab reports were nearly as popular (51.1%). Other methods for assessment included tests (42.2%) and poster presentations (33.3%). About 24% of the professors reported “other” techniques (e.g., three faculty used lab notebooks, whereas single responses were recorded for honors thesis, constant discussion with the student, constructive participation in course discussion [graded daily], laboratory work, and quizzes). A small number of faculty (8.8%) assessed students through preparation of a manuscript for publication.
Table 4.
Faculty assessment methods from 2005 to 2006 academic year
| Assessment method | Professors who used each assessment method (%) |
|---|---|
| Test | 42.2 |
| Term paper/lab report | 51.1 |
| Poster presentation | 33.3 |
| Oral presentation | 26.6 |
| Manuscript for publication | 8.8 |
| Course evaluation | 33.3 |
| Informal feedback | 62.2 |
| Other | 24.4 |
Faculty received funding to support use of GCAT resources from a variety of sources, with most support coming from departmental funds (89.0%). Institutional and extramural funds each supported 20% of the participating faculty. Only 4.4% of professors indicated that they received no funding for using the materials provided by GCAT. Although most professors (61.7%) did not feel that their implementation of GCAT materials was limited by computer resources, 38.3% indicated that they experienced such limitations.
GCAT faculty rated their agreement with statements describing their experiences with GCAT (Table 5). Most faculty responded that they would not have access to microarray technology without GCAT, and they reported a positive overall GCAT experience. Faculty participants generally agreed that the online protocols and e-mail distribution list (GCAT-L) were helpful. Working with DNA microarrays is inherently an interdisciplinary effort, as illustrated by two unsolicited faculty comments. A biology faculty member commented about GCAT,
“You have awakened parts of my brain that have been dormant since my last stats course. The only reason I have gone over the manual so carefully is that this is my first time teaching microarrays, or even using them, for that matter. GCAT has been remarkably helpful to me. In fact I don't think I would have undertaken this new module in my lab course without the tools GCAT makes available.”
Conversely, a mathematics professor remarked (with identifiers removed for anonymity),
“I am working with a student who is trying to do some serious data analysis on [Dr. X's] chips – we are having great fun learning and thinking about how to understand and analyze all of this data – we are going back to basics – and have already found some interesting things – we are excited that our mathematical results seem to be synching up with [Dr. X's] biological results/insights. I hope we are not the first on board with the GCAT project that are primarily data analysis oriented folks – but I daresay, if we are, we won't be the last! This project provides a great area of study for undergrad students interested in data analysis but not necessarily the actually bench work (but of course they need to understand what happened on the bench to understand the data!). Also, it is a fantastic opportunity for math/stats and bio majors (and professors!) to interact! Hmmm… looks like your project may be expanding to us lab phobic (but data loving!) types!”
These quotes illustrate the power of providing stimulating opportunities to faculty who otherwise would not venture out of their comfort zones.
Table 5.
Faculty responses from 2005 to 2006 academic year by using a 5-point scale, where 1 is strongly disagree and 5 is strongly agree
| Mean | SD | |
|---|---|---|
| I would have access to microarray technology without GCAT. | 1.5 | 0.75 |
| The online protocols available on the GCAT website were useful. | 4.4 | 0.69 |
| The GCAT-Listserv was helpful. | 4.2 | 1.0 |
| The collection of other GCAT members as a support network was a significant factor in launching microarray technology on my campus. | 4.2 | 0.79 |
| Overall, I had a positive experience using GCAT. | 4.6 | 0.60 |
| I would use GCAT again in the future. | 4.7 | 0.63 |
2004 GCAT Workshop Outcomes.
Immediately following the 2004 workshop, all faculty indicated the workshop was very good. One year later, 67% of the respondents said that, overall, the workshop they attended was excellent (80% of the wet lab attendees and 57% of the dry lab attendees). The remainder reported the workshop was very good (29%) or good (4%); none reported that it was fair or poor. When asked to select the aspects of the workshops that, in retrospect, were most valuable in preparing for and teaching during the 2004–2005 academic year, participants consistently indicated that the handouts and notebook were critical (70%). Additionally, 48% found the protocols for data analysis valuable, and 39% found the protocols for hybridization valuable. In open-ended responses, two participants wrote that gaining confidence to use the tools was important, and one wrote that doing the data analysis in the workshop was useful. Importantly, 61% of the respondents indicated that networking with instructors and other participants via GCAT was among the most important aspects of the workshop. This finding is consistent with the 2004 on-site evaluation, which indicated that participants felt the collaborative nature of the workshops was among the most valuable aspect of the workshop.
Upon completion of the workshop, respondents to the 2004 evaluation indicated that they intended to alter existing courses to include data analysis with MAGIC Tool, expected to add a wet lab in upper-division courses, and that they planned to emphasize microarrays in several courses across the curriculum. The 2005 survey asked the 2004 workshop participants whether these courses had in fact been altered to accommodate what they had learned (Table 6). Nineteen of the 24 respondents (79%) who attended the workshop used the materials in at least one course (including independent study) during the 2004–2005 academic year, as did the one instructor who could not attend the workshop but received the written materials. The others indicated that they were still in the curricular planning phases or had committed to using the materials in a class scheduled for the 2005–2006 academic year. Of the 20 respondents who reported using the workshop-supplied information during 2004–2005, 18 said they met at least one goal that they had proposed before taking the workshop. Many participants used the materials in more than one class; two participants indicated they altered three courses to use what they learned in the workshop. Two respondents added a dry lab, and two added a wet lab. The average number of courses modified by wet lab workshop attendees was 1.6 (SD = 0.84); the average number of modified courses for dry lab attendees was 0.86 (SD = 0.77). These values differ significantly (t = −2.2, p < 0.05), suggesting that attendance at the wet lab workshop may yield better preparation or more confidence for using the microarray tools.
Table 6.
Faculty goals prior to 2004 workshop and percentage who accomplished these goals
| Proposed change | % Participants (n = 20) |
|---|---|
| Proposed to use in specific lectures and used the material in those lectures | 35 (7) |
| Proposed to use in specific labs and used the material in those labs | 60 (12) |
| Proposed to use in research and did so | 15 (3) |
| Proposed to use in specific lectures but used the materials in other ways | 20 (4) |
| Proposed to use in specific labs but used the materials in other ways | 15 (3) |
The GCAT workshop materials were used in 10 different types of courses. MAGIC Tool was used in genetics classes by 25% of respondents. The software was used by other respondents in Biochemistry (2), Introductory Biology (2), Bioinformatics (2), Molecular Biology (1), Advanced Molecular/Cell Biology (1), Data Analysis (1), Biotechnology (1), Cell Physiology (1), and Microbiology (1). Twenty-five percent of the respondents indicated that they used the microarray tools for independent research with students. Interestingly, only one faculty member developed a wet lab component but did not use MAGIC Tool software, which reveals the intense need for free software that is student friendly. An additional benefit faculty identified was their increased collaboration as a result of the workshop (Table 7). This result addresses important recommendations in the BIO2010 report that call for increasing faculty development opportunities and building communities with a shared commitment to educational reform.
Table 7.
Faculty-perceived collaborative benefits from attending 2004 GCAT workshop
| Collaborations after returning to home campus | % Participants (n = 23) |
|---|---|
| Talked with GCAT faculty via GCAT-L | 48 |
| Collaborated with faculty at my home institution to assist in curriculum/course development | 48 |
| Worked with teaching assistants | 9 |
| Discussed material at department/faculty meetings | 48 |
| Shared with colleagues at other institutions | 9 |
| Other | 13 |
Twenty faculty reported that a total of approximately 800 students participated in a course or in research that used workshop materials in some way. Individual faculty reports ranged from engagement of 2–220 students, with an average of 39.5 and median of 20 per faculty member. Sixteen students were involved in advanced tutorials or independent research using the microarray tools; most had successful experiences. Five students made presentations at their respective colleges or universities or at the regional Sigma Xi conference; one received a grant for an honors proposal using the microarray technique. Four students were conducting research for the first time. However, one respondent indicated that his two students had a less than optimal experience because the data were not readable and the term allowed no time for replication.
Open-ended faculty comments included the following:
“… the presentation of this subject makes [students] realize and practice the close interaction biology/genetics has with other fields like mathematics. They enjoyed [being] introduced to a novel genetic technique. They said they can understand better and related more [of] their class to real life, like when they watch health news and advances in science.”
“Because [this course] was an absolutely introductory exposure to using microarrays for faculty and students, exposure was limited. I anticipate a strong uptick in activity in the next year as new molecular faculty become involved.”
“Many students have come back and said they got jobs or were assigned or allowed to do special projects in graduate schools because of their experience with microarrays. Many others come back and tell how helpful what they learned in the class was with job experiences or graduate school and how they feel ahead of many others attending classes.”
“I think students were extremely excited to have exposure to microarray technology and data analysis.”
“The students said this made them think about what they were doing more critically and it made the whole process seem less ‘magical.’”
“I didn't have quite as much time as I had hoped for data analysis. I found that it took longer than I anticipated for students to grasp the analysis.”
“… none of our arrays worked. Unfortunately, I think a lot of it was lost on the students. Negative results tend to confuse them, they are not yet appreciative of the fact that experiments don't always work.”
DISCUSSION
Students
The main purpose of GCAT has been to use DNA microarrays as a vehicle to bring genomics into the undergraduate curriculum. The NRC recommends undergraduate curricula should blend mathematics with cell/molecular biology and laboratory experiences that are research-based and interdisciplinary (NRC, 2003, 2005). GCAT provides ready access to an exciting area of interdisciplinary research that is moving into clinical applications—DNA microarrays. Analyzing real microarray data requires students to understand the complexities of genomics and use quantitative methods such as bioinformatics to understand their data and statistical analysis to interpret their results. Students enjoy working with cutting-edge techniques, and they see the value of an integrative approach to science. GCAT helps teachers provide students with valuable skills and train them to think in ways that are critical to the future success of research scientists (Hartwell et al., 1999).
Based on the knowledge surveys, students have made significant gains in many areas (Table 1). Although we provide here some preliminary evidence that GCAT offers learning benefits over a control group, the conclusions one can draw from these data are limited by the small size of the control group. As a result, we are expanding our evaluation efforts to include more control classes from a variety of institutions in an attempt to determine more concretely the learning gains associated with the wet lab portion of GCAT. The high percentage of microarrays with usable data is a tribute to the student-friendly protocols and faculty support network. Students attending a wide range of institutions have been able to perform microarray experiments, because the costs of microarrays are low and the software is free. Only 25% of GCAT faculty have access to extramural funding of some kind (including HHMI educational grants), which explains why affordability is so critical to GCAT's success. Furthermore, student interest and understanding in genomics and appreciation of research increased (Table 2) in part because they felt the methods were beneficial (Table 3).
Based on student learning gains, GCAT faculty and students should devote more time to gene expression ratios and probability, because these topics are essential to understanding gene expression data. Student weakness in these topics reflects the traditional lack of effective integration of mathematics in biology programs. Working with microarrays creates an opportunity for faculty to integrate math and biology, as recommended in BIO2010. The next area of concern is to make sure students fully understand the experimental method and how to troubleshoot. Fewer than half of student participants were able to answer items 2, 3, 8, and 9 correctly after their GCAT experience, and all of these items pertain to microarray experimentation methods. Although significant gains were observed for these questions, there is room for additional improvement. Professors might want to emphasize a wider range of microarray techniques in their future implementations of GCAT activities.
Faculty
Faculty development is an ongoing concern for every campus, and BIO2010 recognized this as a critical issue (NRC, 2003). GCAT provides an easy way for faculty to learn a new method with their students. The GCAT protocols and MAGIC Tool software minimize the risk to faculty of trying this method for the first time. Often, faculty do not have colleagues on campus who can help them. GCAT's network of supportive colleagues encourages faculty to learn a new and intimidating method. The workshops are efficient and effective, based on the number of courses altered and the number of students affected after 1 yr. NSF has provided funding for three more summers of workshops, projected to involve a total of 120 faculty. If we multiply the number of faculty trained each year (40) by the number of students affected based on the 2004 workshop (39.5), then by 2009, ∼9480 undergraduates will have been provided with access to microarrays as a direct result of future GCAT workshops. This number does not include the current number of GCAT faculty (141 to date) and all the students they will reach. Furthermore, because many GCAT faculty teach at minority-serving institutions, another BIO2010 goal is being supported—diversification of future researchers.
No program is perfect, and there are areas where GCAT could improve. Because faculty indicated that interactions with other GCAT members were very significant factors when they launched microarray technology on their own campuses, additional networking resources such as online curriculum workshops or electronic communication could potentially enhance GCAT faculty training and success rates. Workshop participants from 2004 indicated that some additional information or materials would have facilitated increased use of microarray data analysis in the curriculum. Four primary suggestions were clear from open-ended comments:
1. Help with course planning.
Faculty particularly sought additional instructor guidelines (perhaps lesson plans), especially focused on how best to explain and present the experimental design, and, critically, requested information regarding the prep time needed to incorporate the materials into the course with confidence. The need for a course guide for teaching the analysis component was noted by several respondents.
Response: In the future, GCAT may sponsor curriculum development workshops, but currently there is no funding for this. At this time, the best option for new faculty is comparing notes with other GCAT faculty on GCAT-L.
2. Help with analysis.
Faculty requested comparisons with other software packages and more instruction on related analysis programs (e.g., ScanAlyze).
Response: The workshops do help with data analysis but due to the high costs of commercial software, we support only free programs. ScanAlyze is free, but does not work on Macintosh, and does only part of the data analysis, whereas MAGIC Tool does the full analysis in a single program. MAGIC Tool was designed to be student-friendly and to help users understand the consequences of various actions, such as background subtraction. Most other programs were designed for researchers and do not readily lend themselves to instructional applications. Faculty may decide to use other programs, and GCAT does not place any constraints on the software programs its members use. The former company Silicon Genetics provided free access to GeneSpring, a Windows-compatible analysis program, to GCAT members. Agilent Technologies is continuing to provide free access to GeneSpring during research-style classes, after reviewing the laboratory syllabus, but no longer permits publications to use their graphics, even undergraduate research projects.
3. Documentation.
Suggestions included developing help files, distributing slide sets (PowerPoint) of the lectures, creating a detailed handout for MAGIC Tool explaining why certain tasks are performed, and publishing a troubleshooting guide.
Response: NSF has provided funds for a helpdesk staffed by students as well as for the production of tutorials that contain movies to teach users how to use MAGIC Tool. We hope this will address the needs of many faculty and students.
4. Networking.
Faculty want contact with others using similar protocols.
Response: In addition to electronic community building via GCAT-L, GCAT faculty attend many professional workshops and may seek each other out during these meetings. As stated in number 1 above, GCAT may organize curriculum workshops, but currently does not have funding to do this.
No matter how much the GCAT community offers to faculty, some problems are institutional and cannot be solved by GCAT. Faculty challenges include the following:
Faculty may not be able to predict their teaching assignments into future years, so long-term planning for curricular change can be difficult.
Although many faculty have a great desire to use these new materials, they require extensive time to prepare. In some cases, admirable goals cannot be met within the existing time constraints.
The need for unusually large amounts of computer RAM must be considered in advance; some labs are not adequately equipped.
Faculty found it hard to imagine and develop productive lesson plans to incorporate the tools.
Future Directions for GCAT
By focusing on microarrays as a tool for understanding gene expression and functional genomics, GCAT has accomplished many of the BIO2010 goals; but not all faculty want to work with microarrays. Exploration of other routes may help faculty bring genomics into their courses while remaining consistent with our goals. Two additional efforts are underway currently. The first is a collaboration with Dr. Sarah Elgin at Washington University who is working with college faculty on actual, authentic DNA sequencing projects. She has collected resources and protocols so that undergraduates can learn to finish and annotate genome sequences (Elgin, 2005). The other project is a collaboration with Randy Rettburg and Drew Endy at MIT, working in the field of synthetic biology (Rettburg and Endy, 2006). Synthetic biology blends mathematics, computer science, and engineering with molecular and cell biology (SyntheticBiology, 2006). Furthermore, to extend the pipeline of students who can work in genomics as undergraduates, we have developed microarray wet lab simulations and paper activities for high school students (GCAT, 2006a; Campbell et al., 2006b). These tools allow teachers to use hands-on learning activities to blend mathematics with biology in a way that students enjoy and retain.
Accessing Materials
Any faculty member may join GCAT and there is no fee for joining. All you need to do is sign up for GCAT-L to receive e-mail announcements (http://www.bio.davidson.edu/projects/GCAT/GCAT-L.html), including the free summer workshops for faculty (http://www.bio.davidson.edu/projects/GCAT/gcat.html#workshops). Only undergraduates can use the DNA microarrays, though anyone can analyze data with MAGIC Tool or use any of the other resources on the GCAT or MAGIC Tool websites. The microarray simulation kit is available for anyone (http://www.bio.davidson.edu/projects/GCAT/HSChips/HSchips.html). GCAT invites faculty who teach undergraduates to participate in synthetic biology (http://www.bio.davidson.edu/projects/GCAT/Synthetic/synthetic.html) or to contact The International Genetically Engineered Machine (iGEM) leaders directly (http://parts2.mit.edu/wiki/index.php/Main_Page).
ACKNOWLEDGMENTS
GCAT gratefully thanks several agencies for financial contribution: The Waksman Foundation for Microbiology, National Science Foundation, Howard Hughes Medical Institute (grants 52005120, 52005137, 52005202, and 52005328), Associated Colleges of the South, Davidson College, Pomona College, and Missouri Western State University. Many investigators have donated time and resources to GCAT: Patrick Brown, Barbara Dunn, Leroy Hood, Michael Katz, Krassen Dimitrov, John Aitchison, Steve Proper, Richard Bookman, Jef Boeke, Anna Ballew, Corey Nislow, Fred Blatner, Patrick Schnable, David Galbraith, Elaine Mardis, Sarah C. R. Elgin, Seth Crosby, and Chris Sawyer. We thank Sally O'Connor, Diane Okamuro, Mary Clutter, Machi Dilworth, Gerald Selzer, Rob Last, and Angela Klaus at NSF for their vision, guidance, and encouragement. We thank the Institute for Systems Biology, Georgetown University, and Morehouse College for hosting our first three workshops. We also thank Peggy Maiorano for invaluable support at Davidson and all the GCAT faculty and students who have willingly ventured into uncharted educational territory. GCAT faculty workshops were funded by four NSF grants: DBI-0627478 (2006–2010), DBI-0520908 (2005), DBI-0408386 (2004), and DBI-0305176 (2003). NSF also funded the first microarray scanner for GCAT via grant DBI-0099720.
REFERENCES
- Campbell A. M. Genomics in the Undergraduate Curriculum: Rocket Science or Basic Science? [accessed 3 October 2006];2002 doi: 10.1187/cbe.02-06-0014. www.bio.davidson.edu/people/macampbell/ASM/ASM.html. [DOI] [PMC free article] [PubMed]
- Campbell A. M., Eckdahl T. T., Fowlks E., Heyer L. J., Hoopes L.L.M., Ledbetter M. L., Rosenwald A. G. Collaborative programs. Genome Consortium for Active Teaching (GCAT) Science. 2006a;311:1103–1104. doi: 10.1126/science.1121955. [DOI] [PubMed] [Google Scholar]
- Campbell A. M., Zanta C. A., Heyer L. J., Kittinger B., Gabric K. M., Adler L. DNA microarray wet lab simulation brings genomics into the high school curriculum. CBE Life Sci. Educ. 2006b;5:108–115. doi: 10.1187/cbe.06-07-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRisi J. L., Iyer V. R., Brown P. O. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- Eisen M. B. Eisen Lab Software. [accessed 10 September 2006];2006 http://rana.lbl.gov/EisenSoftware.htm.
- Elgin S.C.R. [accessed 10 August 2006];Genomics in Education. 2005 http://www.nslc.wustl.edu/elgin/genomics/index.html.
- Genome Consortium for Active Teaching. GCAT-Listserv (GCAT-L) [accessed 10 August 2006];2003 www.bio.davidson.edu/projects/GCAT/GCAT-L.htm.
- GCAT. Two GCAT Best Practices Workshops. [accessed 10 August 2006];2005 www.bio.davidson.edu/projects/GCAT/workshop3.html.
- GCAT. GCAT DNA Chip Simulations: Dry Lab and Wet Lab Curricula. [accessed 10 August 2006];2006a http://www.bio.davidson.edu/projects/GCAT/HSChips/HSchips.html.
- GCAT. GCAT Faculty Members Using DNA Microarrays with Undergraduate Students. [accessed 12 August 2006];2006b http://www.bio.davidson.edu/projects/GCAT/members/main.html.
- Handelsman J., et al. Scientific teaching. Science. 2004;304:521–522. doi: 10.1126/science.1096022. [DOI] [PubMed] [Google Scholar]
- Hartwell L. H., Hopfield J. J., Leibler S., Murray A. W. From molecular to modular cell biology. Nature. 1999;402(suppl):C47–C52. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- Heyer L. J., Campbell A. M. [accessed 10 August 2006];MicroArray Genome Imaging and Clustering Tool. 2004a www.bio.davidson.edu/MAGIC.
- Heyer L. J., Campbell A. M. Exploring Diauxic Shift Microarray Data with MAGIC Tool. [accessed 4 September 2006];2004b http://gcat.davidson.edu/GCAT/workshop2/derisi_lab.html.
- Heyer L. J., Moskowitz D. Z., Abele J. A., Karnik P., Choi D., Campbell A. M., Oldham E. E., Akin B. K. MAGIC tool: integrated microarray data analysis. Bioinformatics. 2005;21:2114–2115. doi: 10.1093/bioinformatics/bti247. [DOI] [PubMed] [Google Scholar]
- Kuldell N. H. How golden is silence? Teaching undergraduates the power and limits of RNA interference. CBE Life Sci. Educ. 2006;5:247–254. doi: 10.1187/cbe.05-11-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A. Teaching systems biology: an active-learning approach. Cell Biol. Educ. 2005;4:323–329. doi: 10.1187/cbe.04-12-0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Research Council. Washington, DC: National Academies Press; 2003. BIO 2010: Transforming Undergraduate Education for Future Research Biologists. [PubMed] [Google Scholar]
- NRC. Washington, DC: National Academies Press; 2005. Facilitating Interdisciplinary Research. [Google Scholar]
- Pfund C., et al. The merits of training mentors. Science. 2006;311:473–474. doi: 10.1126/science.1123806. [DOI] [PubMed] [Google Scholar]
- Project Kaleidoscope. [accessed 10 August 2006];Investing in Faculty. 2001 http://www.pkal.org/documents/index.cfm?page=3080.
- Rettburg R., Endy D. iGEM—The International Genetically Engineered Machine competition. [accessed 10 August 2006];2006 http://parts2.mit.edu/wiki/index.php/Main_Page.
- Steen L. A., editor. Washington, DC: The Mathematical Association of America; 2005. Math & Bio 2010, Linking Undergraduate Disciplines. [Google Scholar]
- SyntheticBiology. SyntheticBiologyis. [accessed 10 August 2006];2006 http://syntheticbiology.org/FAQ.html.
- Tonidandel S. GCAT Assessment Online. [accessed 10 August 2006];2004 www.bio.davidson.edu/projects/GCAT/assessment/assess.html.
- U.S. Department of Education. United States Department of Education List of Postsecondary Minority Institutions. [accessed 20 July 2006];2006 www.ed.gov/about/offices/list/ocr/edlite-minorityinst.html.