Abstract
The intracellular protease from Pyrococcus horikoshii (PH1704) and PfpI from Pyrococcus furiosus are members of a class of intracellular proteases that have no sequence homology to any other known protease family. We report the crystal structure of PH1704 at 2.0-Å resolution. The protease is tentatively identified as a cysteine protease based on the presence of cysteine (residue 100) in a nucleophile elbow motif. In the crystal, PH1704 forms a hexameric ring structure, and the active sites are formed at the interfaces between three pairs of monomers.
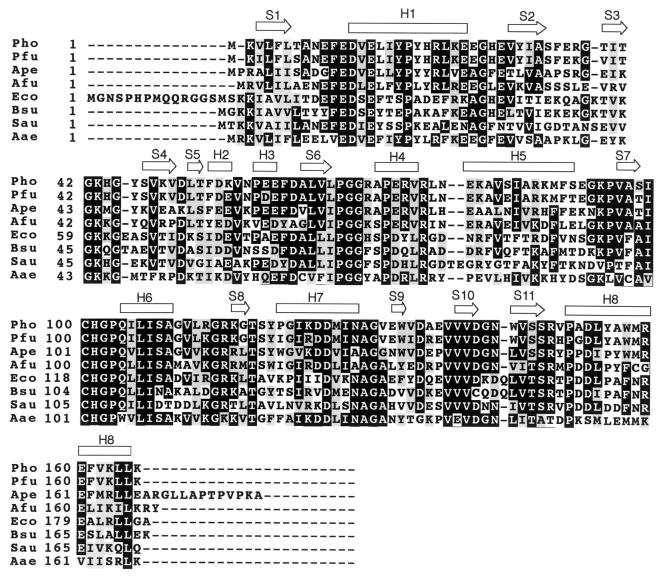
The Pyrococcus horikoshii 1704 gene product (PH1704) has extensive sequence homology (90% identity) to a Pyrococcus furiosus intracellular protease, PfpI. PfpI is characterized by its proteolytic activity and remarkable stability (1, 2). Although PfpI has no detectable sequence homology to any other member of a known protease family, antibodies raised against it crossreact with bovine pituitary proteasome (3). A psi-blast (4) search of databases revealed that PH1704 has homologs in most organisms, although homologs with more than 30% sequence identity are found only in archaea and bacteria. A sequence alignment of PH1704 and several of its homologous proteins is shown in Fig. 1. Only PfpI, among the proteins listed in Fig. 1, has been characterized biochemically, and none have been studied structurally. We have overexpressed the PH1704 gene in Escherichia coli, purified and expressed the protein, and determined its three-dimensional (3D) structure by x-ray crystallography. Here, we report preliminary biochemical data on PH1704 and its 3D crystal structure at 2.0-Å resolution.
Figure 1.
Sequence alignment of PH1704 homologs. Abbreviations are: Pho, P. horikoshii; Pfu, P. furiosus (NCBI access ID, Q51732; percentage identity, 90%); Ape, Aeopyrum pernix (BAA79274.1, 61%); Afu, Archaeoglobus fulgidus (2649300, 53%); Eco, E. coli (P45470, 47%); Bsu, Bacillus subtilis (BAA23403, 43%); Sau, Staphylococcus aureus (Q53719, 39%); Aae, Aquifex aeolicus (2983230, 42%). Secondary structure analysis is based on the crystal structure of PH1704. “H” and “S” refer to helix and strand, respectively.
Methods
Bacterial Expression and Protein Purification.
The cloning of PH1704 from P. horikoshii genomic DNA was carried out according to the “sticky-end PCR” method (5). Two pairs of primers were used to produce two PCR products, which were mixed, heated, and cooled to produce a DNA fragment with NdeI and BamHI sticky ends, which was in turn inserted into pET21a (6). A selenomethionine derivative of the protein was expressed in a methionine auxotroph, E. coli strain B834 (DE3)/pSJS1244 (7, 8) grown in M9 medium supplied with selenomethionine. In the purification process, the cell lysate was subjected to heating (80°C for 30 min), anion exchange (HiTrap-Q), and size-exclusion column chromatography (Superdex 75). To avoid potential oxidation of the protein, 10 mM DTT was used in all buffers. The yield of protein was typically 15 mg of pure protein/liter of culture. Initial crystallization conditions were screened by using the sparse matrix method (ref. 9; Hampton Research, Laguna Niguel, CA) at room temperature. One microliter of 20 mg/ml PH1704 in 20 mM Tris⋅HCl, pH 7.5, and 1 mM EDTA was mixed with 1 μl of 0.1 M trisodium citrate dihydrate, pH 5.6, 0.2 M potassium tartrate tetrahydrate, 2.0 M ammonium sulfate, and equilibrated with 0.5 ml of the same solution in the reservoir by the vapor diffusion sitting drop method. Diffraction-quality crystals were obtained 2 days after setup.
Data Collection and Reduction.
X-ray diffraction data sets were collected at three wavelengths at the Macromolecular Crystallography Facility beamline 5.0.2 at the Advanced Light Source at Lawrence Berkeley National Laboratory. The crystal was soaked in a drop of mother liquor with 30% glycerol (about 50 μl) for about 40 s before being flash-frozen in liquid nitrogen and exposed to x-ray. All data sets were processed with denzo (10) and reduced with scalepack (10) and programs in the CCP4 package (11). The statistics of the data collection and reduction are shown in Table 1 and Table 2, respectively.
Table 1.
Data collection
| Measurement | Value
|
||
|---|---|---|---|
| Peak | Edge | Remote | |
| Wavelength, Å | 0.97938 | 0.9796 | 0.9686 |
| Resolution, Å | 2.0 | 2.0 | 2.0 |
| No. of unique data | 129,941 | 130,210 | 129,727 |
| Redundancy | 5.2 | 5.3 | 5.3 |
| Overall | |||
| completeness | 98.9 (95.6) | 98.9 (95.8) | 98.9 (95.7) |
| Rmerge, % | 7.3 (34.6) | 6.6 (31.8) | 6.4 (35.3) |
| Average I/σ(I) | 21.9 (4.9) | 23.3 (5.2) | 19.3 (3.7) |
Space group P41212. Cell: a = b = 124.7 Å, c = 129.0 Å. Each protein monomer has four selenomethionines. Values in parentheses are for highest-resolution shell, from 2.03 to 2.00 Å.
Table 2.
Refinement statistics
| Measurement | Value |
|---|---|
| Resolution, Å | 20–2.0 |
| No. of unique reflections | 65,700 |
| No. of parameters to fit | 12,660 |
| σ cutoff | None |
| R factor, % | 18.4 |
| Rfree, % | 20.0 |
| rmsd bonds, Å | 0.005 |
| rmsd angles, ° | 1.2 |
| rmsdNCS,* Å | 0.12 |
| Average B factor of protein | 25.2 |
| Average B factor of water | 32.7 |
| No. of water molecules | 279 |
| Residues in | |
| Ramachandran plot, % | |
| Most favored | 90.6 |
| Additional | 8.7 |
| Disallowed† | 0.7 |
Three models were built. The rmsd is an average of the rmsd values between AB, BC, and AC.
The only residue in a disallowed main chain conformation is Cys-100. This is consistent with its proposed role as the nucleophile (see text).
Model Building and Refinement.
The program solve (12) was used to locate the selenium sites in the crystal and to calculate initial phases. The initial multiwavelength anomalous dispersion phases (13) were further improved by solvent flattening and histogram matching with the dm program in the CCP4 package (11). The map calculated by using the improved phases was of excellent quality. Three models were built by using the o program (14) and refined by using cns (15). No noncrystallographic symmetry (NCS) constraints or restraints were applied in the last few cycles of refinement. The refinement statistics are shown in Table 2. The atomic coordinates and structure factors have been deposited into the Brookhaven Protein Data Bank with the accession number 1G2I.
Results
Biochemical Characterization of PH1704.
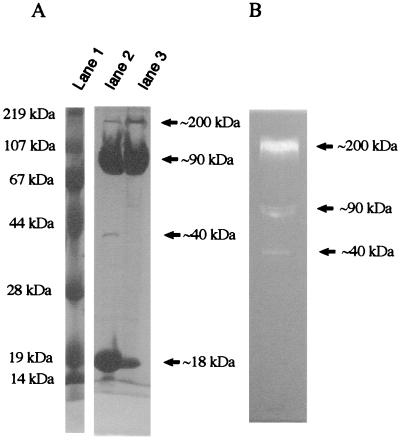
The ORF of gene PH1704 codes for a polypeptide chain of 166 amino acids. The purified protein appears as four bands on an SDS/PAGE gel, with apparent molecular masses of 18, 40, 90, and 200 kDa (Fig. 2A). Prolonged heating in the presence of SDS increases the fraction of monomeric protein (18 kDa; Fig. 2A, lane 2). A gelatin-SDS/PAGE protease assay was carried out according to the procedure established for PfpI (1) to determine the proteolytic activity of the individual protein bands. The 200-kDa band showed the highest activity in the SDS/PAGE gelatin overlay assay (Fig. 2B), although it makes up only a small fraction of the total protein. Weaker activities were shown by the 90-kDa and 40-kDa bands (Fig. 2B). N-terminal sequencing of the blotted protein from the 200-kDa band identified it unambiguously as PH1704 (data not shown). These results suggest that the most active form of the protein is a partially SDS-resistant, multimeric complex corresponding to the 200-kDa band.
Figure 2.
Electrophoretic migration of PH1704 and gelatin protease assay. (A) 10% SDS-PAGE gel of purified PH1704. Lane 1, molecular markers; lane 2, PH 1704 boiled for 20 min in sample buffer; lane 3, PH1704 in sample buffer without boiling. (B) Gelatin overlay of 8% SDS-PAGE gel of purified PH1704. Proteolytic activity is manifested by cleared zones in the dark background. Molecular weights indicated are based on molecular markers in gelatin-SDS-PAGE gel.
Structure Determination.
The PH1704 crystals belong to the space group P41212 with cell dimensions of a = b = 124.7 Å, and c = 129 Å. There are three monomers in an asymmetric unit. Three models, A, B, and C for the three chains, were built by using the o program (14). The numbering of the residues is from 1 to 166, 201 to 366, and 401 to 566, for monomers B, A, and C, respectively.
Monomer Structure.
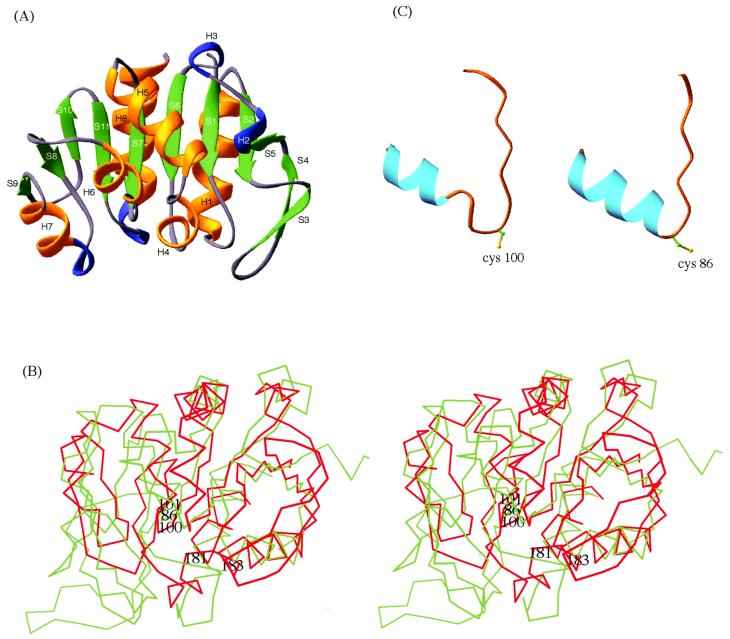
Each monomer consists of an α/β sandwich. A secondary structure analysis and ribbon diagram of the structure are shown in Fig. 1 and Fig. 3A, respectively. There are 11 β strands and eight helices as determined by the Database of Secondary Structure of Proteins (DSSP; ref. 16). The central β sheet consists of six β strands (S2, S1, S6, S7, S11, and S10). It is flanked by helices H8 and H1, and strands S3 and S4 on one side, and by helices H2, H3, H4, H5, H6, H7, and strands S9 and S8 on the other side.
Figure 3.
Structural features of PH1704. (A) Ribbon diagram of PH1704. “H” and “S” refer to helix and strand, respectively. Green, gold, and blue code for β-strand, α-helix, and 310 helix, respectively. (B) The superposition of the Cα trace of amidotransferase domain of GMP synthetase (green) and that of PH1704 (red). The three members of the catalytic triad, Cys-86, His-181, and Glu-183 of the amidotransferase and Cys-100 and His-101 of PH1704 are labeled. (C) The “nucleophile elbows” in PH1704 (Left) and amidotransferase domain of GMP synthetase (Right).
The structure was compared with protein structures in the Protein Data Bank (PDB) by the dali program (17). The two proteins with the highest Z scores are the noncatalytic domain of E. coli catalase HPII which has no known function (18), and the glutamine amidotransferase (GA) domain of GMP synthetase (19). Fig. 3B shows the Cα trace of GA aligned with that of PH1704. These two enzymes hydrolyze chemically related substrates, an amide bond in the case of GA and a peptide bond in the case of PH1704. In addition to the overall similarity in the folding topology between PH1704 and GA, there is a “nucleophile elbow” motif in both structures. The nucleophile elbow is a distinctive strand–nucleophile–helix motif that was first recognized in α/β hydrolases (20). The nucleophile of a catalytic triad in an α/β hydrolase, either a cysteine or a serine, resides in a sharp turn that connects the strand and the helix. Residues 96–109 (S7 and H6) of PH1704 form a nucleophile elbow-like motif (Fig. 3C). The Cα traces of this fragment can be aligned with the nucleophile elbow in amidotransferase with an rms deviation (rmsd) of 1.2 Å. The φ/ψ angles for the potential nucleophile Cys 100 of PH1704 fall in an unfavorable region in the Ramachandran plot; this is characteristic of the nucleophile in a nucleophile elbow. The sequence around Cys-100, S-I-C-H-G-P, is also consistent with the consensus sequence small-x-Nu-x-small-small for α/β hydrolases.
Quaternary Structure.
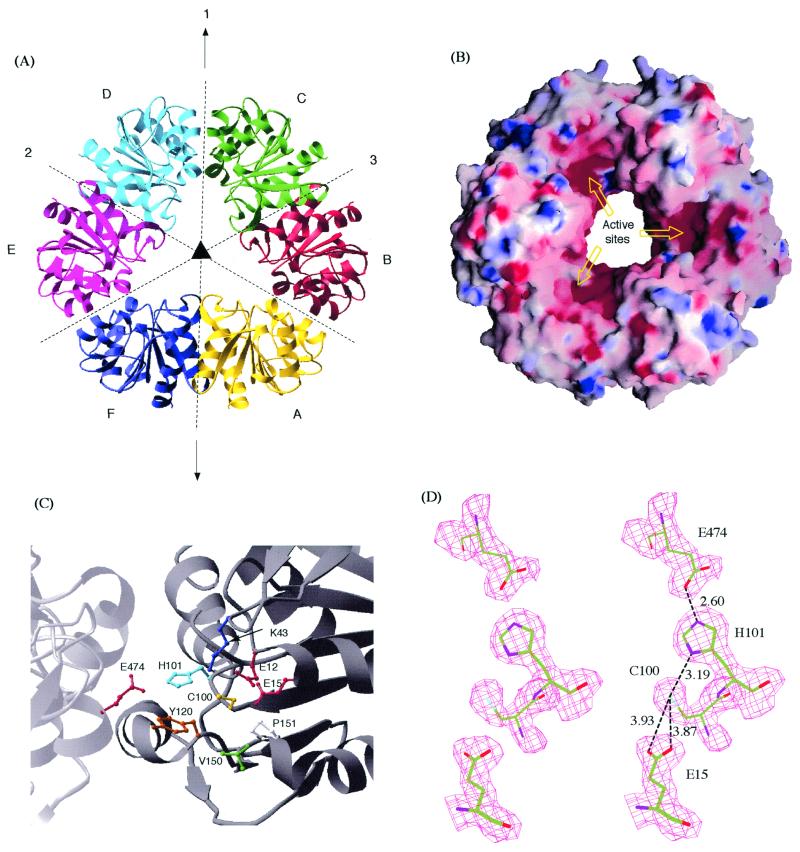
Because only oligomeric forms of the protein showed activity, we looked closely into the quaternary structure. There are six kinds of intermolecular contacts in the crystal (Fig. 4A). The contacts between monomer B and C as well as that between A and B are symmetric and bury a significant amount of surface areas (1689 Å2 and 1431 Å2, respectively). The areas buried by the other four contacts are much smaller, and vary from 675 Å2 to 348 Å2.
Figure 4.
Hexameric structures and putative catalytic sites of PH1704. (A) Ribbon diagram of the hexamer with the threefold NCS axis (perpendicular to the plane of paper) and the three twofold axes (in the plane of paper) are shown. Axis 1 coincides with a crystallographic symmetry axis. Axis 2 and 3 are NCS symmetry axes. (B) Surface representation of the hexamer color-coded by electrostatic potential. The diameter of the center opening is ≈24 Å. (C) Ball and stick model of the putative active site. All of the residues surrounding Cys-100 are shown. (D) A stereogram of the proposed catalytical triad and Glu-15 with the corresponding solvent-flattened experimental map. The distances shown are in Å.
In the case of the AB contact, there are a pair of salt bridges between Arg-22 and Glu-225 and the symmetry-related Arg-222 and Glu-25. Otherwise, the interactions are mainly hydrophobic. Residues that are buried in the interface include Tyr-18, Tyr-218, Val-14, Val-214, Leu-154, and Leu-354. Several other hydrophobic residues, including Tyr-46, Tyr-246, Ile-17, and Ile-217, are partially buried at the interface. In contrast, contact BC is primarily ionic in nature. There is hydrogen-bonding between His-101 and Glu-474, Ser-108 and Asp-525, and their symmetry-related counterparts, as well as electrostatic interactions between Arg-77 and Asp-526 and its symmetric pair Asp-126 and Arg-477. In addition, Ile-123, Ile-523, Ile-107, and Ile-527 are also buried in this contact. Both contacts, BC and AB, are likely to be biologically relevant instead of fortuitous crystal packing. In addition to the fact that these two buried surface areas are considerably larger than that of the other four intermolecular contacts, many of the residues involved in the ionic interactions and most of the hydrophobic residues involved in these two contacts are conserved among close homologs (Fig. 1).
Six monomers in the crystal are connected through the two major contacts described above to form a closed ring. The ribbon diagram and the surface of the hexamer are shown in Fig. 4 A and B, respectively. Monomer A, B, and C are crystallographically determined whereas D, E, and F are generated by a crystallographic twofold symmetry operation (around axis 1). Thus, the complex can be considered as a dimer of trimers, because there are two NCS twofold axes (axis 2 and 3) and one crystallographic twofold axis (axis 1) perpendicular to the threefold axis. Axis 2 relates trimer BCD to trimer AFE, and axis 3 relates trimer CDE to trimer BAF, with rmsd values of 0.42 and 0.42 Å, respectively. The complex can be considered also as a trimer of dimers because there is an NCS threefold axis, which relates dimer BC to DE, DE to FA, and FA to BC, with an rmsd of 0.63 Å.
Discussion
The activity of an intracellular protease must be strictly regulated to prevent cytoplasmic proteins from unwanted proteolytic degradation. This is ensured by cells in several different ways. Some proteases, like calpains and caspases, are highly specific (21, 22). Others are confined in endosomal and lysosomal compartments by a lipid membrane (cathepsins; refs. 23 and 24). ATP-dependent proteases, including proteosomes (25, 26), CLpP (27), Lon, and HslV (28), employ a different mechanism. They all form barrel-like oligomeric structures with the active sites sequestered inside the barrel. The ATPase-containing regulatory complexes cover the outer port of the proteolytic chamber and regulate the entry of the substrates into the chamber. This feature of compartmentalization was also recognized in crystal structures of two ATP-independent intracellular proteases, Gal6 (29) and leucine aminopeptidase (LAP) (30). In each case, the active site resides in a cavity formed by a hexamer, and the access to the cavity is restricted by a small opening. Because the ATPase domain, which is thought to be involved in substrate unfolding and locomotion, is absent in these two structures, it was suggested that this class of compartmentalized proteases may specialize in the hydrolysis of small peptides that can freely permeate the small opening (31).
PH1704 and PfpI can be categorized into the same class as Gal6 and LAP. PfpI and PH1704 have been identified as ATP-independent proteases in a previous assay (2) and our gelatin-overlay protease assay described earlier, respectively. Although the overall structure of the PH1704 complex is more open, the active sites of PH1704 are located in hindered positions in the complex (Fig. 4B) and are not accessible to even the smallest globular protein. The active sites of PH1704 also lack the cleft that defined specificity and that binds to peptides of certain lengths, further suggesting that the protease may have a broad specificity. Interestingly, it was proposed that the physiological function of PfpI, a protein of high homology to PH1704, is to hydrolyze small peptides to provide a nutritional source for P. furiosus (3).
The presence of the nucleophile elbow in the PH1704 structure suggests that Cys-100 may be the active site nucleophile (Fig. 4C). In the crystal structure, Cys-100 is surrounded by Glu-12, Glu-15, Lys-43, His-101, Tyr-120, Val-150, and Pro-151 (Fig. 4C). Cys-100 also forms a “catalytic triad” with His-101 and Glu-474 (from an adjacent monomer). The catalytic triad in PH1704 shares the same handedness with the triads in papain type cysteine proteases, namely the cysteine interacts with δN, whereas the glutamate hydrogen-bonds with ɛN of the histidine (Fig. 4D). An aspect of the triad is its formation on a dimer interface, which is consistent with the experimental observation that the activity was found only for the oligomeric forms of the protein.
Acknowledgments
We thank Dr. David King for characterizing PH1704 with electrospray mass spectrometry and Dr. Gerry McDermott for assistance in data collection. We also thank Dr. Edward Berry, Zhaolei Zhang, Dr. Alfonso Martinez, Dr. Dong-Hae Shin, and Dr. Luhua Lai for their help throughout the project. The work was supported by the Director, Office of Science, Office of Biological and Environmental Research, of the U.S. Department of Energy under Contract no. DE-AC03-76SF00098.
Abbreviations
- PH1704
Pyrococcus horikoshii 1704 gene product
- PfpI
Pyrococcus furiosus intracellular protease
- NCS
noncrystallographic symmetry
- rmsd
rms deviation
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1G2I).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.260503597.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.260503597
References
- 1.Blumentals I I, Robinson A S, Kelly R M. Appl Env Microbiol. 1990;56:1992–1998. doi: 10.1128/aem.56.7.1992-1998.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Halio S B, Blumentals I I, Short S A, Merrill B M, Kelly R M. J Bacteriol. 1996;178:2605–2612. doi: 10.1128/jb.178.9.2605-2612.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Snowden L, Blumentals I I, Kelly R. Appl Env Microbiol. 1992;58:1134–1141. doi: 10.1128/aem.58.4.1134-1141.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zeng G. BioTechniques. 1998;25:206–208. doi: 10.2144/98252bm05. [DOI] [PubMed] [Google Scholar]
- 6.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 7.Leahy D J, Hendrickson W A, Aukhil I, Erickson H P. Science. 1992;258:987–991. doi: 10.1126/science.1279805. [DOI] [PubMed] [Google Scholar]
- 8.Kim R, Sandler S J, Goldman S, Yokota H, Clark A J, Kim S-H. Biotech Lett. 1998;20:207–210. [Google Scholar]
- 9.Jancarik J, Kim S-H. J Appl Crystallogr. 1991;24:409–411. [Google Scholar]
- 10.Otwinowski Z, Minor W. Methods Enzymol. 1997;277:307–327. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 11.Dodson E J, Winn M, Ralph A. Methods Enzymol. 1997;277:620–633. doi: 10.1016/s0076-6879(97)77034-4. [DOI] [PubMed] [Google Scholar]
- 12.Terwilliger T C, Berendzen J. Acta Crystallogr D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hendrickson W A, Horton J R, LeMaster D M. EMBO J. 1990;9:1665–1672. doi: 10.1002/j.1460-2075.1990.tb08287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones A, Kleywegt G. Methods Enzymol. 1997;277:173–208. doi: 10.1016/s0076-6879(97)77012-5. [DOI] [PubMed] [Google Scholar]
- 15.Brünger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 16.Kabsch W, Sander C. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 17.Holm L, Sander C. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 18.Bravo J, Mate M J, Schneider T, Switala J, Wilson K, Loewen P C, Fita I. Proteins. 1999;34:155–166. doi: 10.1002/(sici)1097-0134(19990201)34:2<155::aid-prot1>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 19.Tesmer J J, Klem T J, Deras M L, Davisson V J, Smith J L. Nat Struct Biol. 1996;3:74–86. doi: 10.1038/nsb0196-74. [DOI] [PubMed] [Google Scholar]
- 20.Ollis D L, Cheah E, Cygler M, Dijkstra B, Frolow F, Franken S M, Harel M, Remington S J, Silman I, Schrag J, et al. Protein Eng. 1992;5:197–211. doi: 10.1093/protein/5.3.197. [DOI] [PubMed] [Google Scholar]
- 21.Wang K K. Trends Neurosci. 2000;23:20–26. doi: 10.1016/s0166-2236(99)01479-4. [DOI] [PubMed] [Google Scholar]
- 22.Thornberry N A, Lazebnik Y. Science. 1998;281:1312–1316. doi: 10.1126/science.281.5381.1312. [DOI] [PubMed] [Google Scholar]
- 23.Allen P M, Babbitt B P, Unanue E R. Immunol Rev. 1987;98:171–187. doi: 10.1111/j.1600-065x.1987.tb00524.x. [DOI] [PubMed] [Google Scholar]
- 24.Puri J, Factorovich Y. J Immunol. 1988;141:3313–3317. [PubMed] [Google Scholar]
- 25.Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik H D, Huber R. Nature (London) 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 26.Löwe J, Stock D, Jap B, Zwickl P, Baumeister W, Huber R. Science. 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- 27.Wang J, Hartling J A, Flanagan J M. Cell. 1997;91:447–456. doi: 10.1016/s0092-8674(00)80431-6. [DOI] [PubMed] [Google Scholar]
- 28.Rohrwild M, Pfeifer G, Santarius U, Muller S A, Huang H C, Engel A, Baumeister W, Goldberg A L. Nat Struct Biol. 1997;4:133–139. doi: 10.1038/nsb0297-133. [DOI] [PubMed] [Google Scholar]
- 29.Joshua-Tor L, Xu H E, Johnston S A, Rees D C. Science. 1995;269:945–950. doi: 10.1126/science.7638617. [DOI] [PubMed] [Google Scholar]
- 30.Burley S K, David P R, Taylor A, Lipscomb W N. Proc Natl Acad Sci USA. 1990;87:6878–6882. doi: 10.1073/pnas.87.17.6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Larsen C N, Finley D. Cell. 1997;91:431–434. doi: 10.1016/s0092-8674(00)80427-4. [DOI] [PubMed] [Google Scholar]