Abstract
Genomic array technologies provide a means for profiling global changes in gene expression under a variety of conditions. However, it has been difficult to assess whether transcriptional or posttranscriptional regulation is responsible for these changes. Additionally, fluctuations in gene expression in a single cell type within a complex tissue like a tumor may be masked by overlapping profiles of all cell types in the population. In this paper, we describe the use of cDNA arrays to identify subsets of mRNAs contained in endogenous messenger ribonucleoprotein complexes (mRNPs) that are cell type specific. We identified mRNA subsets from P19 embryonal carcinoma stem cells by using mRNA-binding proteins HuB, eIF-4E, and PABP that are known to play a role in translation. The mRNA profiles associated with each of these mRNPs were unique and represented gene clusters that differed from total cellular RNA. Additionally, the composition of mRNAs detected in HuB–mRNP complexes changed dramatically after induction of neuronal differentiation with retinoic acid. We suggest that the association of structurally related mRNAs into mRNP complexes is dynamic and may help regulate posttranscriptional events such as mRNA turnover and translation. Recovering proteins specifically associated with mRNP complexes to identify and profile endogenously clustered mRNAs should provide insight into structural and functional relationships among gene transcripts and/or their protein products. We have termed this approach to functional genomics ribonomics and suggest that it will provide a useful paradigm for organizing genomic information in a biologically relevant manner.
Understanding global gene expression at the level of the whole cell will require detailed knowledge of the contributions of transcription, pre-mRNA processing, mRNA turnover, and translation. Although the sum total of these regulatory processes in each cell accounts for its unique expression profile, few methods are available to independently assess each process en masse. DNA arrays are well suited for profiling the steady-state levels of mRNA globally (i.e., the transcriptome). However, because of posttranscriptional events affecting mRNA stability and translation, the expression levels of many cellular proteins do not directly correlate with steady-state levels of mRNAs (1, 2). We have been able to reduce the complexity of gene expression profiling by using mRNA-binding proteins involved in RNA processing and translation to recover mRNA subsets contained in cellular messenger ribonucleoprotein complexes (mRNPs). We report that mRNAs present in mRNP complexes have structural features in common and are dynamic in response to the induction of differentiation by treatment with retinoic acid (RA).
Most mRNAs contain sequences that regulate their posttranscriptional expression and localization (3). These regulatory elements reside in introns and exons of pre-mRNAs as well as in both coding and noncoding regions of mature transcripts (4, 5). An example of a sequence-specific regulatory motif is the AU-rich element (ARE) present in the 3′-untranslated regions of early-response gene (ERG) mRNAs, many of which encode proteins essential for growth and differentiation (6–9). Regulation via the ARE is poorly understood, but the mammalian ELAV/Hu proteins have been shown to bind to ARE sequence elements in vitro and to affect posttranscriptional mRNA stability and translation in vivo (10–14).
There are four ELAV/Hu mammalian homologues of the Drosophila ELAV RNA-binding protein (15, 16). HuA (HuR) is ubiquitously expressed, whereas HuB, HuC, and HuD (and their respective alternatively spliced isoforms) are predominantly found in neuronal tissue but can also be expressed as tumor cell-specific antigens in some small cell carcinomas, neuroblastomas, and medulloblastomas (reviewed in ref. 14). All Hu proteins contain three RNA-recognition motifs (16–18), which confer their binding specificity for AREs (16). The evidence for ARE binding by Hu proteins began with the identification of an AU-rich binding consensus sequence from a randomized combinatorial RNA library that was screened with recombinant HuB (19, 20). These and other studies demonstrated that Hu proteins bind in vitro to several ARE-containing ERG mRNAs including c-myc, c-fos, granulocyte–macrophage colony-stimulating factor, and GTPase-activating protein-43 (12, 19–26). The binding of Hu proteins to ARE-containing mRNAs can result in the stabilization (10–13) and increased translatability of mRNA transcripts (10, 26). The neuron-specific family member, HuB (Hel-N1) is one of the earliest neuronal markers produced in teratocarcinoma cells after RA treatment to induce neuronal differentiation (26, 27). When neuronal Hu proteins are ectopically expressed in various preneuronal cell lines, neurites form spontaneously (26, 28, 29).
Previous attempts to identify subsets of mRNAs bound by RNA-binding proteins used reverse transcription–PCR amplification and iterative selection (20, 30). In this study, we report the direct isolation of subsets of total cell mRNA from endogenous mRNP complexes and the identification of these mRNAs en masse without amplification by using cDNA arrays. We show that the mRNAs present in mRNP complexes share structural features and can vary in response to cellular changes such as induction of differentiation. Specifically, we immunoprecipitated epitope-tagged HuB–mRNP complexes from P19 embryonic carcinoma cell lysates and identified a subset of ARE-containing mRNAs encoding cell-cycle regulators, transcription factors, and other ERG products. In parallel experiments, poly(A)-binding protein (PABP) and 5′-cap-binding protein (eIF-4E) mRNP complexes were found to contain mRNAs whose profiles differed from one another and from the profiles of the HuB–mRNP and the total cellular transcriptome. The profiles of mRNA species detected in these mRNP complexes were highly reproducible and potentially represent structurally and functionally related mRNA subsets. On treatment of these HuB-expressing P19 cells with RA to induce neuronal differentiation, the population of mRNAs detected in the HuB–mRNP complexes changed to include additional ARE-containing mRNAs known to be up-regulated in neurons. The ability to classify the mRNA components of mRNP complexes into distinct subsets should help elucidate the structural and functional networking of gene transcripts and how their expression is regulated posttranscriptionally.
Materials and Methods
P19 Cells, Transfection, and RA Treatment.
Murine P19 embryonal carcinoma cells were obtained from American Type Culture Collection and were maintained as suggested. They were stably transfected with a simian virus 40 promoter-driven pα2-gene 10-HuB plasmid that expressed a gene 10-tagged neuron-specific HuB protein (Hel-N2) (20). The plasmid was maintained with 0.2 mg/ml G418 (Sigma). Although it lacks 13 amino acids from the hinge region connecting RNA-recognition motifs (RRMs) 2 and 3 of Hel-N1, the RRMs are identical and in vitro-binding experiments have indicated no differences in the AU-rich RNA-binding properties of Hel-N1 and Hel-N2 (refs. 20 and 24; unpublished observations).
Chemical treatment with RA was used to induce neuronal differentiation by treating 5 × 105 P19 cells placed in a 60-mm Petri dish (Fisher 8–757-13A) with 0.5 μM retinoic acid (Sigma R2625) as previously described (20, 26). The RA-treated HuB (Hel-N2) stably transfected P19 cells grew neurites and displayed characteristic neuronal markers and morphology but did not terminally differentiate and remained susceptible to being killed by mitotic inhibitors.
Sources of Antibodies.
Monoclonal anti-g10 antibodies were produced as previously described (20, 26). Polyclonal sera reactive with HuA were produced as previously described (19, 31). An antibody reactive with PABP was kindly provided by N. Sonenberg, McGill University. Antibody against eIF-4E was obtained from Transduction Laboratories, San Diego, CA.
Immunoprecipitation of Endogenous mRNP Complexes from Cell Lysates.
Cells were removed from tissue culture plates with a rubber scraper and washed with cold PBS. The cells were resuspended in approximately two pellet volumes of polysome lysis buffer containing 100 mM KCl, 5 mM MgCl2, 10 mM Hepes, pH 7.0, and 0.5% Nonidet P-40 with 1 mM DTT, 100 units/ml RNase OUT (GIBCO/BRL), 0.2% vanadyl ribonucleoside complex (GIBCO/BRL), 0.2 mM PMSF, 1 mg/ml pepstatin A, 5 mg/ml bestatin, and 20 mg/ml leupeptin added fresh at time of use. The lysed cells were then frozen and stored at −100°C. At the time of use, the cell lysate was thawed and centrifuged at 16,000 × g in a tabletop microfuge for 10 min at 4°C. The mRNP cell lysate contained approximately 50 mg/ml total protein.
For immunoprecipitations, Protein-A Sepharose beads (Sigma) were swollen 1:5 V/V in NT2 Buffer (50 mM Tris, pH 7.4/150 mM NaCl/1 mM MgCl2/0.05% Nonidet P-40) supplemented with 5% BSA. A 300-μl aliquot of the 1:5 V/V preswollen protein-A bead slurry was used per immunoprecipitation reaction and incubated overnight at 4°C with excess immunoprecipitating antibody (typically 5–20 μl depending on the reagent). The antibody-coated beads were washed with ice-cold NT2 buffer and resuspended in 900 μl of NT2 buffer supplemented with 100 units/ml RNase OUT/0.2% vanady/ribonucleoside complex/1 mM DTT/20 mM EDTA. The beads were briefly vortexed, and 100 μl of the mRNP cell lysate was added and immediately centrifuged and a 100-μl aliquot removed to represent total cellular RNA. The immunoprecipitation reactions were tumbled at room temperature for 2 hours and then washed 4 times with ice-cold NT2 buffer followed by 2 washes with NT2 buffer supplemented with 1 M urea. Washed beads were resuspended in 100 μl NT2 buffer supplemented with 0.1% SDS and 30 μg proteinase K incubated for 30 min in a 55°C water bath, phenol-chloroform-isoamylalcohol extracted and ethanol precipitated.
RNase Protection Assay.
Immunoprecipitated RNA was assayed by RNase protection by using the PharMingen Riboquant assay according to the manufacturer's suggestions (45014K). Myc-related gene and cyclin template sets were used (45356P and 45620P, respectively). Protected riboprobe fragments were visualized on a phosphorimaging screen (Molecular Dynamics) after 24 h of exposure. Phosphorimages were scanned by using the Molecular Dynamics storm 860 system at 100 μm resolution and analyzed by using Molecular Dynamics image quant software (ver. 1.1).
Probing of cDNA Arrays.
cDNA array analysis was performed by using Atlas Mouse Arrays (CLONTECH) that contain a total of 597 cDNA segments spotted in duplicate side by side on a nylon membrane. Probing of cDNA arrays was performed as described in the CLONTECH Atlas cDNA Expression Arrays User Manual (PT3140–1). Briefly, RNA was extracted from HuB stably transfected P19 cells and used to produce reverse-transcribed probes. A pooled set of primers complementary to the genes represented on the array (CLONTECH) was used for the reverse transcription probe synthesis, which was radiolabeled with P32 α-dATP and purified by passage over CHROMA SPIN-200 columns (CLONTECH). After hybridization, the array membrane was washed and visualized by using a phosphorimaging screen (Molecular Dynamics).
Analysis of cDNA Arrays.
Phosphorimages were scanned by using the Molecular Dynamics STORM 860 System at 100 μm resolution and stored as .gel files. Images were analyzed by using atlasimage 1.0 and 1.01 software (CLONTECH). The signal for any given gene was calculated as the average of the signals from the two duplicate cDNA spots. A default external background setting was used in conjunction with a background-based signal threshold to determine gene signal significance. The signal for a gene was considered significantly above background if the adjusted intensity (total signal minus background) was more than 2-fold the background signal. Comparisons of multiple cDNA array images were performed by using an average of all of the gene signals on the array (global normalization) to normalize the signal intensity between arrays. Changes in the mRNA profile of HuB–mRNP complexes in response to RA treatment were considered significant if they were 4-fold or greater. cDNA array images and overlays were prepared by using Adobe photoshop 5.0.2 (Adobe Systems, Mountain View, CA).
Results
Identifying mRNA Subsets Associated with RNA-Binding Proteins by Using a Multiprobe RNase Protection Assay.
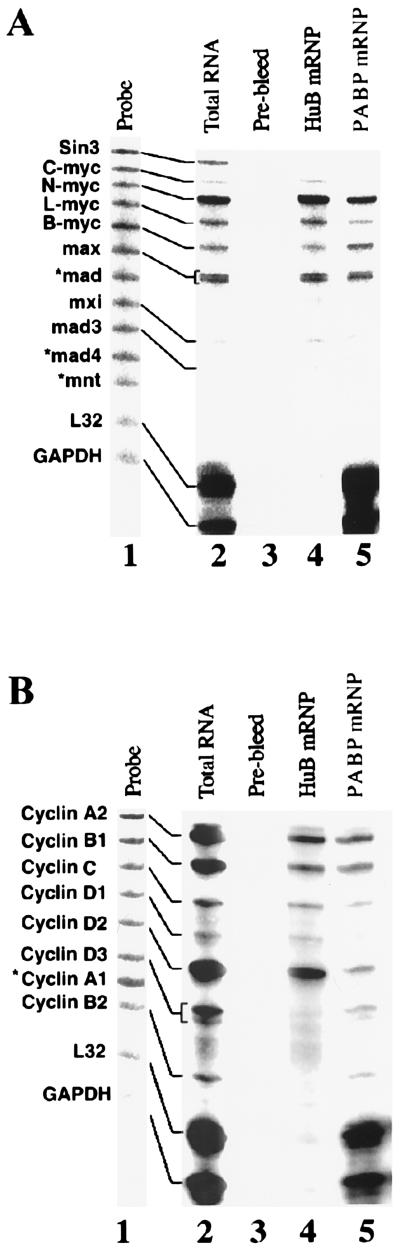
Previously we reported the ability to immunoprecipitate HuB (Hel-N1) by using a monoclonal anti-g10 epitope tag and identified an mRNA encoding NF-M protein by using reverse transcription–PCR (10). In this study, we expanded this approach by using a multiprobe RNase protection assay to rapidly optimize the immunoprecipitation of several endogenous mRNP complexes containing different mRNA-binding proteins. By using a multiprobe system, we assayed mRNP pellets containing many mRNAs in a single lane of a polyacrylamide gel. On the basis of our previous work (19, 31), we chose multiprobe template sets specific for several myc and cyclin mRNA targets as well as two control housekeeping genes, GAPDH and L32. As shown in Fig. 1, we immunoprecipitated HuB– and PABP–mRNP complexes from extracts of murine P19 cells stably transfected with g10-HuB cDNA. No mRNAs were detected in pellets immunoprecipitated with polyclonal prebleed rabbit sera (Fig. 1 A and B, lane 3) or with many other rabbit, mouse, and normal human sera tested with this assay (data not shown). The profiles of mRNAs associated with HuB–mRNP complexes included n-myc, l-myc, b-myc, max, and cyclins A2, B1, C, D1 and D2, but not sin3, cyclin D3, cyclin B2, L32, or GAPDH mRNAs (Fig. 1 A and B, lane 4). In contrast, the profiles of mRNAs extracted from PABP–mRNP complexes resembled the profiles of total RNA but showed enriched levels of L32 and GAPDH and decreased levels of sin3 mRNA (Fig. 1 A and B, lane 5). We conclude that antibodies reactive with these cellular RNA-binding proteins can be used to immunoprecipitate mRNP complexes and to recover mRNAs with which they are specifically associated. This is consistent with the postulated role of Hu proteins in regulating posttranscriptional gene expression during cell growth and differentiation (10–14, 16, 19–29, 31).
Figure 1.
Multiprobe RNase protection analysis of mRNAs associated with mRNP complexes. mRNP complexes from P19 cell lysates were immunoprecipitated and the pelleted RNA extracted and quantitated by RNase protection as described in Materials and Methods by using the PharMingen Riboquant assay. (A) mMyc MultiProbe template set; (B) mCyc-1 MultiProbe template set. Lanes: 1, undigested riboprobe (slightly larger than RNase digested product because of riboprobe plasmid template); 2, total cellular RNA; 3, rabbit prebleed serum control; 4, mRNAs extracted from HuB mRNPs; 5, mRNAs extracted from PABP mRNPs. *, mRNA species not detected in total RNA.
Identifying mRNA Subsets Associated with RNA-Binding Proteins en Masse by Using cDNA Arrays.
We further expanded our ability to identify the mRNAs associated in endogenous mRNP complexes by using cDNA array filters containing 597 known murine genes. Like the multiprobe RNase protection assay, this approach provided a highly specific and sensitive method for detecting mRNAs without amplification or iterative selection.
After assessing the overall gene expression profile of HuB-transfected P19 cells (the transcriptome), HuB– and PABP–mRNP complexes as well as eIF-4E–mRNP complexes were separately immunoprecipitated and the captured mRNAs identified on cDNA arrays. mRNAs extracted from immunoprecipitated pellets showed no reactivity with the genomic DNA spots normally used to orient the perimeter of the array membrane, further indicating selectivity for specific mRNAs in these mRNP complexes. The initial alignment of these arrays was facilitated by spiking the hybridization reaction with radiolabeled λ-phage markers that hybridized with six DNA spots on the bottom of the array membrane. Once the alignment register was established, subsequent blots did not require the use of spiked λ markers for orientation.
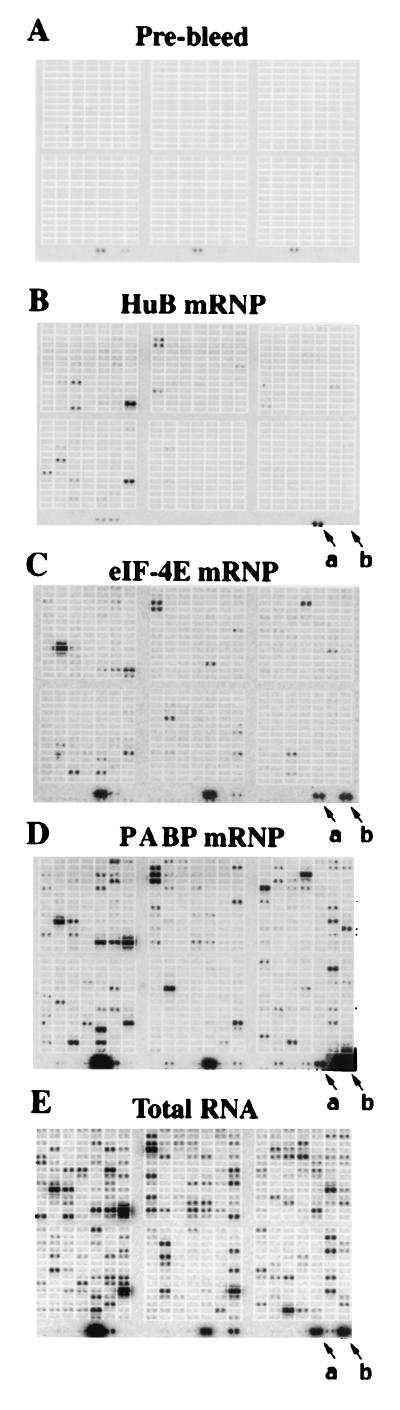
Arrays generated from immunoprecipitations with rabbit prebleed sera were essentially blank with the exception of the spiked λ markers observed at the bottom of the array (Fig. 2A). Immunoprecipitated HuB–mRNP and eIF-4E–mRNP complexes each contained slightly more than 10% of the mRNAs detected in the total RNA but differed considerably from one another (Fig. 2 B, C, and E). In addition to the data presented here, the phosphorimages for the cDNA arrays described in this paper can be obtained as supplementary Figs. 5–13 at http://bioinformatics.duke.edu/pubs/keene.
Figure 2.
Profiles of mRNAs associated with mRNP complexes by using cDNA arrays. As described in Materials and Methods, RNA was extracted from immunoprecipitated mRNPs or total cell lysates and used to make reverse-transcribed probes for Atlas Mouse Arrays containing 597 double-spotted cDNA segments (CLONTECH). (A) Prebleed; (B) HuB–mRNP complexes; (C) eIF-4E–mRNP complexes; (D) PABP–mRNP complexes; (E) total cellular RNA. An example of the quality of enrichment between mRNA profiles is demonstrated in the relative abundance of β-actin and ribosomal protein S29 mRNAs (Fig. 2, arrows a and b, respectively).
Like HuB and eIF-4E, PABP has been implicated in facilitating mRNA stabilization and translation (32–35). Not surprisingly, PABP mRNPs contained many more detectable mRNAs than those observed in the HuB or eIF-4E mRNPs (Fig. 2D). The profile of the mRNAs in the PABP mRNPs from these cells closely resembled that of the transcriptome. However, as was seen in HuB and eIF-4E mRNPs, some mRNAs were enriched or depleted in the PABP mRNPs as compared with the total RNA (Fig. 2 D and E). The profiles and relative abundance of mRNAs detected in these mRNP complexes were highly reproducible, but the absolute number of mRNA species detectable on the phosphorimages occasionally varied as a result of differences in the specific activity of the probe.
Although some of the mRNAs detected in the immunoprecipitated mRNPs correspond to messages that are highly abundant in the total RNA profile, many other mRNAs that were readily detectable in the total mRNA profile were not observed in the pelleted mRNPs. Additionally, the relative abundance of mRNA species differed among the various mRNP complexes in comparison with the total cellular RNA. Because the cDNA arrays from total RNA were generated by using one-tenth the quantity of lysate used for mRNP immunoprecipitations, we did not compare the absolute quantities of each mRNA detected in mRNP complexes with those observed in the total RNA. It is more accurate to compare the relative abundance of each mRNA species to each other within each microarray. For example, the relative abundance of the mRNAs encoding β-actin and ribosomal protein S29 (Fig. 2, arrows a and b, respectively) is approximately equal in total cellular RNA but varied dramatically among each of the mRNP complexes. Many other examples of this distinction are readily apparent in Fig. 2. These findings indicate that the mRNA profiles detected in HuB–, eIF-4E–, and PABP–mRNP complexes are distinct from one another and from those of the transcriptome.
Although the mRNA profile of HuB mRNPs was not expected to resemble that of PABP or eIF-4E mRNPs, we were surprised that the mRNA profile of eIF-4E mRNPs differed so extensively from that observed for PABP–mRNP complexes. One possible explanation for the observed differences is that the antibody used to immunoprecipitate eIF-4E–mRNP complexes interacts only with a subset of mRNPs in which the eIF-4E 5′ cap-binding protein is accessible. An alternative explanation for the differences in mRNA profiles of PABP and eIF-4E mRNPs is that they reflect distinct functional subsets of mRNAs whose expression is regulated differently at the level of translation (36).
RA Treatment of P19 Cells Changes the Profile of mRNAs in HuB–mRNP Complexes.
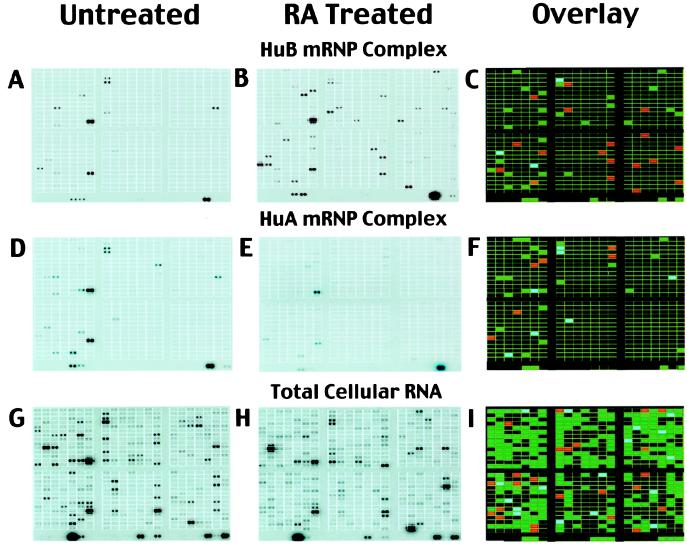
Because HuB is predominantly a neuronal protein believed to play a role in regulating neuronal differentiation, we investigated whether the mRNA population found in HuB–mRNP complexes changes in response to RA, a chemical inducer of neuronal differentiation. We treated HuB-transfected P19 cells with RA to induce the onset of neuronal differentiation and then immunoprecipitated HuB–mRNP complexes followed by identification of the mRNAs on cDNA arrays. Comparison of the mRNA profiles extracted from the HuB mRNPs before and after RA treatment revealed that 18 mRNAs were either exclusively present or greatly enriched (4-fold or greater) in RA-treated HuB mRNPs (Fig. 3 A–C, red bars). In addition, three mRNAs (T lymphocyte-activated protein, DNA-binding protein SATB1 and HSP84) decreased in abundance by 4-fold or greater in response to RA treatment (Fig. 3C, blue bars). To determine whether the changes observed in the mRNA profile of the HuB–mRNP complexes were unique, we immunoprecipitated the ubiquitously expressed ELAV family member HuA (HuR) from these same RA-treated cells. Although there were a few changes to the HuA mRNP profile after treatment with RA, they were minor in comparison with HuB and were represented predominantly by 4-fold or greater decreases in the presence of certain mRNAs (Fig. 3 D–F, blue bars).
Figure 3.
Comparison of the mRNA profiles from HuB mRNPs before and after treatment with RA. As described in Materials and Methods, phosphorimages were scanned and stored as gel files and then analyzed by using atlasimage 1.01 software (CLONTECH). A default external background setting was used in conjunction with a background-based signal threshold to determine gene signal significance. Comparisons of multiple cDNA array images were performed by using an average of all of the gene signals on the array (global normalization) to normalize the signal intensity between arrays. Changes in the mRNA profile of HuB–mRNP complexes in response to RA treatment were considered significant if they were 4-fold or greater. (A) Untreated p19 HuB mRNPs; (B) RA-treated p19 HuB mRNPs; (C) comparison of A and B; (D) untreated p19 HuA (HuR) mRNPs; (E) RA-treated p19 HuA mRNPs; (F) comparison of D and E; (G) untreated p19 total RNA; (H) RA-treated p19 total RNA; (I) comparison of G and H. Red bars represent mRNAs that increased 4-fold or greater after RA treatment, and blue bars represent mRNAs that decreased 4-fold or greater. Green bars indicate mRNAs of approximately equal abundance.
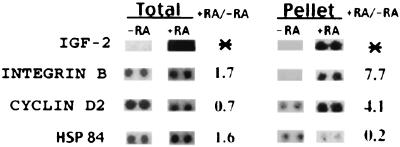
The changes in the HuB-associated mRNA profile in response to RA treatment did not merely reflect changes in the total cellular mRNA (Fig. 3 G–I). However, in some cases, changes in the HuB mRNA profile reflected global changes in the total RNA profile. Numerous examples of differentially enriched or depleted mRNAs detected in HuB–mRNP complexes are evident by comparing Fig. 3 C and I. For comparative purposes, this is depicted in Fig. 4 by realignment and enlargement of representative spots. For example, IGF-2 mRNA was detectable only in total RNA and HuB–mRNP complexes from RA-treated cells (Fig. 4). However, other HuB mRNP-bound mRNAs, such as integrin β, cyclin D2, and Hsp 84, increased or decreased in abundance disproportionately to their changes in the total RNA profile after RA treatment (Fig. 4). The disparity between changes in the mRNA profiles of total RNA and HuB mRNPs possibly results from changes in compartmentalization of mRNAs that flux through mRNP complexes in response to RA treatment. We conclude that the mRNA profiles derived from these mRNP complexes are dynamic and can reflect the state of growth, as well as changes in the cellular environment in response to a biological inducer like RA.
Figure 4.
Comparison of ribonomic profiling with global gene profiling. Selected examples of mRNAs that demonstrate differences between the total RNA profile in comparison with HuB-bound mRNAs before and after RA treatment. *, mRNAs detected only in RA-treated cells.
By using GenBank and Expressed Sequence Tag databases, we identified the 3′ UTR sequences for mRNAs enriched in RA-treated HuB–mRNP complexes (Table 1). Many of the mRNAs for which 3′ UTR sequences were available contained uridylate-rich motifs similar to those previously found to bind to Hu proteins in vitro (19–22, 37, 38). Many of these mRNAs encode proteins that are expressed in neuronal tissues or are known to be up-regulated after RA-induced neuronal differentiation. The sequence alignment shown in Table 1 is consistent with the previous results of Levine et al. (19) and Gao et al. (20), who used in vitro selection to derive a consensus RNA-binding sequence for HuB. On the basis of the direct mRNP analytical methods described in this paper, it may be possible to discern in vivo target sequence preferences for other RNA-binding proteins.
Table 1.
Examples of consensus sequences found in the 3′ UTRs of neuronal HuB target mRNAs
| Gene (GenBank 3′ UTR accession no.) consensus sequence |
|---|
| CD44 (M27129)----AUUUUCUAUUCCUUUUUUAUUUUAUGUCAUUUUUUUA |
| IGF-2 (M14951)------UAAAAAACCAAAUUUGAUUGGCUCUAAACA |
| HOX 2.5 (M34857)--------UCACUCUUAUUAUUUAU |
| AAAUUUUAUUAAGUUA |
| HSP47 (J05609)----------------AUUUUAUCUGUUA |
| UUUUUCUCCCUUUUUUAGUUUUUUCAAA |
| GADD45 (L28177)----UUUUCUUUUUUUUUUUUGGUCUUUAU |
| AAAUUCUCAGAAGUUUUAUUAUAAAUCUU |
| PN-1 (X70296)-----UUCUGUUAAAUAUUUUUAUAUACUGCUUUCUUUUUU |
| AUUUUAUAGUAGUUUUUAUGUUUUUAUGGAAAA |
| AUUUGCCUUUUUAAUUCUUUUU |
| KROX24, EGR1 (M20157)---UUUGUGGUUUUAUUUUACUUUGUACUU |
| UUUUGUUUUCCUU |
| N-cadherin (M31131)----------UUUUUUAUUUUCUGUAUUUUUU |
| UUUUUUUAAAUUUUUUUAUUUUCUUUUU |
| UUUUUAAUUUUUUAAUUUUUUUU |
| CD51 (U14135)--------------AAUGGUUUAUAUUUAUGAU |
| UUGUUUAUAUCUUCAAU |
| CF2R (L03529)-------UGCAUCGAUCCGUUGAUUUACUACU |
| ITGB1 (Y00769)----------UAUAAUUUUUAAUUUUUUAUUAUUUU |
| UUUUUUUUUUUUCUUUAAUUCCUGGU |
| CTCF (U51037)---------UUAUGAAUGUUAUAUUUGU |
| UCUUAAUUUUUUCUCUUUUUUUU |
| TGF-β 2(X57413)--------UUUUUUCCUUUUAAUUGUAAAUGGUUCUUU |
| UCAGAUUGUAUAUAUUUGUUUCCUUU |
| UUCAAUUUUUUUUAUAUACUAUCUU |
| UUUUUCUUUAAUUGGUUUUUUU |
| MTRP (U34259)-----UCUUGUTCUGAGCAUUUAUUUUCAAA |
| UUCUCGUCUUGUUUAUUUUACAA |
| In vitro sequence (ref. 19) UUUAUUU |
Discussion
This paper describes the use of mRNA-binding proteins to purify endogenous mRNP complexes and to identify their associated mRNAs en masse by using cDNA array analysis. Earlier attempts to identify mRNA targets of the HuB protein by using high-throughput methods required reverse transcription–PCR amplification and in vitro iterative selection and identified several structurally related ERG mRNAs from neuronal tissues (20, 39). Most of these mRNAs contained ARE-like sequences in their 3′ untranslated regions, which is a characteristic of ERG mRNAs (14, 19–21). It has been demonstrated that Hu proteins can bind ERG mRNAs and affect their stability and/or translational activation (10–14, 19–22, 24–29, 37, 38, 40). The in vitro approach of Gao et al. (20) yielded a distinct mRNA subset from human brain and medulloblastoma cells with ERG sequence characteristics. Because of the extreme sensitivity associated with PCR amplification and the iterative in vitro selection procedures needed to enrich the population, this method was limited. As described in this paper, the more direct in vivo approach obviates the need for in vitro binding and PCR amplification and has allowed the identification of mRNA transcripts with linked structural and perhaps functional properties. In addition, recognizable HuB protein RNA-binding sequences were identified within the in vivo captured mRNA subset (Table 1).
Interestingly, some of the mRNAs detected by these methods may not be directly associated with the targeted RNA-binding protein yet would still be considered bone fide components of the mRNP complex. Therefore, the methods described here represent a potentially useful approach to mapping the mRNA–protein infrastructure of cells with implications for functional outcomes. Indeed, HuB, eIF-4E, and PABP are mRNA-binding proteins that have been implicated in mRNA stabilization and translational activation (5, 26, 34, 35). Several investigators have suggested that mRNAs that are regulated in developmental or temporal patterns, or whose protein products participate in the same regulatory pathway, should be considered functionally linked (41, 42). We suggest that organizing subsets of mRNAs into mRNP complexes during a process such as differentiation may provide the cell with a means to regulate the expression of multiple genes posttranscriptionally.
We have termed ribonomics the identification and analysis of linked mRNA subsets by using RNA-associated proteins. Ribonomics is distinct from transcriptomics, which is used to assess the total mRNA complement of the genome. The characterization of structurally and/or functionally related subsets of mRNAs by using ribonomics or other partitioning methods may facilitate our understanding of gene products that are expressed simultaneously or sequentially for a specific outcome. For example, if mRNAs such as CD44, Egr-1/Zif 268, or neuronal cadherin (Table 1 and Fig. 3) are regulated posttranscriptionally in Hu–mRNP complexes, their protein products may act correspondingly to help activate neuronal differentiation (41). Additionally, cell-specific mRNA-binding proteins like neuronal HuB can be used to capture cell-type-specific mRNA subsets from extracts of complex tissues or tumors without the need for microdissection. In this manner, a ribonomic approach should provide a better understanding of how cells form dynamic mRNA networks and regulate information flow during gene expression.
Acknowledgments
We thank Drs. Ning Lu, Dragana Antic, William Miller, and Ulus Atasoy for intellectual contributions. This work was supported by research grants CA60083, CA79907, and AI46451 from the National Institutes of Health (NIH) (J.D.K.). S.A.T. was supported by NIH Viral Oncology training grant CA09111.
Abbreviations
- mRNPs
messenger ribonucleoprotein complexes
- ARE
AU-rich element
- ERG
early-response gene
- PABP
poly(A)-binding protein
- RA
retinoic acid
References
- 1.Gygi S P, Rochon Y, Franza B R, Aebersold R. Mol Cell Biol. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Futcher B, Latter G I, Monardo P, McLaughlin C S, Garrels J I. Mol Cell Biol. 1999;19:7357–7368. doi: 10.1128/mcb.19.11.7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Richter J D. In: Translational Control. Hershey J W B, Mathews M B, Sonenberg N, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 481–504. [Google Scholar]
- 4.Jacobson A, Peltz S W. Annu Rev Biochem. 1996;65:693–739. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]
- 5.Wickens M, Anderson P, Jackson R J. Curr Opin Genet Dev. 1997;7:220–232. doi: 10.1016/s0959-437x(97)80132-3. [DOI] [PubMed] [Google Scholar]
- 6.Caput D, Beutler B, Hartog K, Thayer R, Brown-Shimer S, Cerami A. Proc Natl Acad Sci USA. 1986;83:1670–1674. doi: 10.1073/pnas.83.6.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shaw G, Kamen R. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- 8.Schiavi S C, Belasco J G, Greenberg M E. Biochim Biophys Acta. 1992;1114:95–106. doi: 10.1016/0304-419x(92)90009-n. [DOI] [PubMed] [Google Scholar]
- 9.Chen C Y, Shyu A B. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 10.Jain R G, Andrews L G, McGowan K M, Pekala P H, Keene J D. Mol Cell Biol. 1997;17:954–962. doi: 10.1128/mcb.17.2.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levy N S, Chung S, Furneaux H, Levy A P. J Biol Chem. 1998;273:6417–6423. doi: 10.1074/jbc.273.11.6417. [DOI] [PubMed] [Google Scholar]
- 12.Fan X C, Steitz J A. EMBO J. 1998;17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peng S S, Chen C Y, Xu N, Shyu A B. EMBO J. 1998;17:3461–3470. doi: 10.1093/emboj/17.12.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keene J D. Proc Natl Acad Sci USA. 1999;96:5–7. doi: 10.1073/pnas.96.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Good P J. Proc Natl Acad Sci USA. 1995;92:4557–4561. doi: 10.1073/pnas.92.10.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Antic D, Keene J D. Am J Hum Genet. 1997;61:273–278. doi: 10.1086/514866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kenan D J, Query C C, Keene J D. Trends Biochem Sci. 1991;16:214–220. doi: 10.1016/0968-0004(91)90088-d. [DOI] [PubMed] [Google Scholar]
- 18.Burd C G, Dreyfuss G. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 19.Levine T D, Gao F, King P H, Andrews L G, Keene J D. Mol Cell Biol. 1993;13:3494–3504. doi: 10.1128/mcb.13.6.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao F B, Carson C C, Levine T, Keene J D. Proc Natl Acad Sci USA. 1994;91:11207–11211. doi: 10.1073/pnas.91.23.11207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.King P H, Levine T D, Fremeau R T, Jr, Keene J D. J Neurosci. 1994;14:1943–1952. doi: 10.1523/JNEUROSCI.14-04-01943.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu J, Dalmau J, Szabo A, Rosenfeld M, Huber J, Furneaux H. Neurology. 1995;45:544–550. doi: 10.1212/wnl.45.3.544. [DOI] [PubMed] [Google Scholar]
- 23.Ma W J, Cheng S, Campbell C, Wright A, Furneaux H. J Biol Chem. 1996;271:8144–8151. doi: 10.1074/jbc.271.14.8144. [DOI] [PubMed] [Google Scholar]
- 24.Abe R, Yamamoto K, Sakamoto H. Nucleic Acids Res. 1996;24:2011–2016. doi: 10.1093/nar/24.11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chung S, Eckrich M, Perrone-Bizzozero N, Kohn D T, Furneaux H. J Biol Chem. 1997;272:6593–6598. doi: 10.1074/jbc.272.10.6593. [DOI] [PubMed] [Google Scholar]
- 26.Antic D, Lu N, Keene J D. Genes Dev. 1999;13:449–461. doi: 10.1101/gad.13.4.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao F B, Keene J D. J Cell Sci. 1996;109:579–589. doi: 10.1242/jcs.109.3.579. [DOI] [PubMed] [Google Scholar]
- 28.Wakamatsu Y, Weston J A. Development (Cambridge, UK) 1997;124:3449–3460. doi: 10.1242/dev.124.17.3449. [DOI] [PubMed] [Google Scholar]
- 29.Akamatsu W, Okano H J, Osumi N, Inoue T, Nakamura S, Sakakibara S, Miura M, Matsuo N, Darnell R B, Okano H. Proc Natl Acad Sci USA. 1999;96:9885–9890. doi: 10.1073/pnas.96.17.9885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keene J D. Methods Enzymol. 1996;267:367–383. doi: 10.1016/s0076-6879(96)67023-2. [DOI] [PubMed] [Google Scholar]
- 31.Atasoy U, Watson J, Patel D, Keene J D. J Cell Sci. 1998;111:3145–3156. doi: 10.1242/jcs.111.21.3145. [DOI] [PubMed] [Google Scholar]
- 32.Ross J. Microbiol Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ross J. Trends Genet. 1996;12:171–175. doi: 10.1016/0168-9525(96)10016-0. [DOI] [PubMed] [Google Scholar]
- 34.Wickens M, Kimble J, Strickland S. In: Translational Control. Hershey J W B, Mathews M B, Sonenberg N, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 411–450. [Google Scholar]
- 35.Sachs A B, Sarnow P, Hentze M W. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 36.Lazaris-Karatzas A, Montine K S, Sonenberg N. Nature (London) 1990;345:544–547. doi: 10.1038/345544a0. [DOI] [PubMed] [Google Scholar]
- 37.Ma W J, Chung S, Furneaux H. Nucleic Acids Res. 1997;25:3564–3569. doi: 10.1093/nar/25.18.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fan X C, Myer V E, Steitz J A. Genes Dev. 1997;11:2557–2568. doi: 10.1101/gad.11.19.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andrews L G, Keene J D. Methods Mol Biol. 1999;118:233–244. doi: 10.1385/1-59259-676-2:233. [DOI] [PubMed] [Google Scholar]
- 40.Aranda-Abreu G E, Behar L, Chung S, Furneaux H, Ginzburg I. J Neurosci. 1999;19:6907–6917. doi: 10.1523/JNEUROSCI.19-16-06907.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wen X, Fuhrman S, Michaels G S, Carr D B, Smith S, Barker J L, Somogyi R. Proc Natl Acad Sci USA. 1998;95:334–339. doi: 10.1073/pnas.95.1.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zong Q, Schummer M, Hood L, Morris D R. Proc Natl Acad Sci USA. 1999;96:10632–10636. doi: 10.1073/pnas.96.19.10632. [DOI] [PMC free article] [PubMed] [Google Scholar]