Abstract
Background
The Bacteroidetes and Chlorobi species constitute two main groups of the Bacteria that are closely related in phylogenetic trees. The Bacteroidetes species are widely distributed and include many important periodontal pathogens. In contrast, all Chlorobi are anoxygenic obligate photoautotrophs. Very few (or no) biochemical or molecular characteristics are known that are distinctive characteristics of these bacteria, or are commonly shared by them.
Results
Systematic blast searches were performed on each open reading frame in the genomes of Porphyromonas gingivalis W83, Bacteroides fragilis YCH46, B. thetaiotaomicron VPI-5482, Gramella forsetii KT0803, Chlorobium luteolum (formerly Pelodictyon luteolum) DSM 273 and Chlorobaculum tepidum (formerly Chlorobium tepidum) TLS to search for proteins that are uniquely present in either all or certain subgroups of Bacteroidetes and Chlorobi. These studies have identified > 600 proteins for which homologues are not found in other organisms. This includes 27 and 51 proteins that are specific for most of the sequenced Bacteroidetes and Chlorobi genomes, respectively; 52 and 38 proteins that are limited to species from the Bacteroidales and Flavobacteriales orders, respectively, and 5 proteins that are common to species from these two orders; 185 proteins that are specific for the Bacteroides genus. Additionally, 6 proteins that are uniquely shared by species from the Bacteroidetes and Chlorobi phyla (one of them also present in the Fibrobacteres) have also been identified. This work also describes two large conserved inserts in DNA polymerase III (DnaE) and alanyl-tRNA synthetase that are distinctive characteristics of the Chlorobi species and a 3 aa deletion in ClpB chaperone that is mainly found in various Bacteroidales, Flavobacteriales and Flexebacteraceae, but generally not found in the homologs from other organisms. Phylogenetic analyses of the Bacteroidetes and Chlorobi species is also reported based on concatenated sequences for 12 conserved proteins by different methods including the character compatibility (or clique) approach. The placement of Salinibacter ruber with other Bacteroidetes species was not resolved by other phylogenetic methods, but this affiliation was strongly supported by the character compatibility approach.
Conclusion
The molecular signatures described here provide novel tools for identifying and circumscribing species from the Bacteroidetes and Chlorobi phyla as well as some of their main groups in clear terms. These results also provide strong evidence that species from these two phyla (and also possibly Fibrobacteres) are specifically related to each other and they form a single superphylum. Functional studies on these proteins and indels should aid in the discovery of novel biochemical and physiological characteristics that are unique to these groups of bacteria.
Background
The Bacteroidetes and Chlorobi presently comprise two of the main phyla within the Bacteria [1-3]. The bacteria from the Bacteroidetes phylum (previously known as the Cytophaga-Flavobacteria-Bacteroides (CFB) group) exhibit a potpourri of phenotypes including gliding behavior and their ability to digest and grow on a variety of complex substrates such as cellulose, chitin and agar [4-8]. They inhabit diverse habitats including the oral cavity of humans, the gastrointestinal tracts of mammals, saturated thalassic brines, soil and fresh water [9-13]. The Bacteroides species such as B. thetaiotaomicron and B. fragilis are among the dominant microbes in the large intestine of human and other animals [14,15]. These bacteria in the human colon are also important opportunistic pathogens and they are involved in causing abscesses and soft tissue infections of the gastrointestinal tract, as well as diarrheal diseases [15-18]. Other bacteroidetes species, such as Porphyromonas gingivalis and Prevotella intermedia, are major causative agents in the initiation and progression of periodontal disease in humans [12,19,20].
In contrast to wide distribution of Bacteroidetes species in diverse habitats, bacteria from the phylum Chlorobi occupy a narrow environmental niche mainly consisting of anoxic aquatic settings in stratified lakes (chemocline regions), where sunlight is able to penetrate [21-24]. Some of these bacteria also exist as epibionts in phototrophic consortiums with other bacteria, particularly β-proteobacteria [21,25]. The Chlorobi, which are also commonly known as Green Sulfur bacteria, are all anoxygenic obligate photoautotrophs, which obtain electrons for anaerobic photosynthesis from hydrogen sulfide [22,23,26]. Although the Bacteroidetes and Chlorobi are presently recognized as two distinct phyla [1,3], these two groups are closely related in phylogenetic trees based on 16S rRNA as well other gene sequences [27-30]. Conserved indels (i.e. inserts or deletions) in a number of widely distributed proteins (viz. FtsK, UvrB and ATP synthase α subunit), that are uniquely present in species from these two groups, also strongly indicate that these two groups of species shared a common ancestor exclusive of all other bacteria [30].
The species from the Bacteroidetes and Chlorobi phyla are presently distinguished from other bacteria primarily on the basis of their branching in phylogenetic trees [2,3,27]. We have previously described a 4 aa conserved insert in DNA Gyrase B as well as a 45 aa conserved insert in SecA protein that were specific for the Bacteroidetes species [30]. In Chlorobi as well as Chloroflexi species, their light harvesting pigments are located in unique membrane-attached sac-like structures referred to as 'chlorosomes' [22,24,31,32]. A number of genes involved in the synthesis of chlorosomes components in Chlorobi have been identified by genomic and mutational analysis [26] and a few of them, viz. Fenna-Matthew-Olson (FMO) protein [33], are unique for this group [32,34]. However, the number of characteristics that are either unique to species from these two phyla, or are commonly shared by members of these phyla, are very limited. In the past few years, complete genomes of several Bacteroidetes and Chlorobi species have become available (see Table 1). Additionally, sequencing of genomes for many other Bacteroidetes/Chlorobi species is at different stages of completion (see Table 1), but considerable sequence information for these genomes is available in the NCBI database.
Table 1.
General Characteristics of Bacteroidetes/Chlorobi Genomes
| Strain Name | Taxonomic Order | Genome Size (Mb)** | GC Content (%) | Protein Number | Reference# | |
| Bacteroidetes | Porphyromonas gingivalis W83# | Bacteroidales | 2.34 | 48.3 | 1909 | [58] |
| Bacteroides fragilis NCTC 9343# | 5.24 | 44 | 4184 | [16] | ||
| Bacteroides fragilis YCH46# | 5.31 | 33.5 | 4578 | [35] | ||
| Bacteroides thetaiotaomicron VPI-5482# | 6.29 | 42 | 4778 | [15] | ||
| Gramella forsetii KT0803# | Flavobacteriales | 3.8 | 36.6 | 3559 | [59] | |
| Flavobacteria bacterium BBFL7* | 5 | 35.0 | 2592 | a | ||
| Flavobacteriales bacterium HTCC2170* | 5 | 37.0 | 3478 | a | ||
| Flavobacterium johnsoniae UW101* | - | 35.2 | 4985 | DOE-JGI | ||
| Flavobacterium sp. MED217* | 5 | 39.8 | 3735 | a | ||
| Cellulophaga sp. MED134* | 5 | 38.2 | 2944 | a | ||
| Croceibacter atlanticus HTCC2559* | 5 | 33.9 | 2719 | a | ||
| Polaribacter irgensii 23-P* | - | 31.0 | 2557 | a | ||
| Psychroflexus torquis ATCC 700755* | 5 | 32–33.0 | 6751 | a | ||
| Robiginitalea biformata HTCC2501* | 5 | 56.4 | 3228 | a | ||
| Tenacibaculum sp. MED152* | 5 | 30.6 | 2679 | a | ||
| Cytophaga hutchinsonii ATCC 33406# | Sphingobacteriales | 4.43 | 38.8 | 3785 | CP000383 | |
| Salinibacter ruber DSM 13855# | 3.59 | 66.5 | 2801 | [36] | ||
| Chlorobi | Chlorobium chlorochromatii CaD3# | Chlorobia | 2.57 | 44.3 | 2002 | CP000108 |
| Chlorobium limicola DSM 245* | 2.4 | 51.3 | 2435 | DOE-JGI | ||
| Chlorobium phaeobacteroides BS1* | 2. | 45.5 | 3791 | DOE-JGI | ||
| Chlorobium phaeobacteroides DSM 266* | 2.4 | 48.3 | 2789 | DOE-JGI | ||
| Chlorobaculum tepidum formerly Chlorobium tepidum TLS# | 2.15 | 56 | 2252 | [24] | ||
| Chlorobium luteolum formerly Pelodictyon luteolum DSM 273# | 2.36 | 57.3 | 2083 | CP000096 | ||
| Chlorobium clathratiforme formerly Pelodictyon phaeoclathratiforme BU-1* | - | 48.1 | 2762 | DOE-JGI | ||
| Prosthecochloris aestuarii DSM 271* | - | 50.1 | 2313 | DOE-JGI | ||
| Chlorobium phaeovibrioides formerly Prosthecochloris vibrioformis DSM 265* | - | 53.0 | 1747 | DOE-JGI |
#Indicates a completely sequenced genome. The references to the published genomes are provided. For others that are fully sequenced but not published, accession numbers for the genomes are given.
* Indicates that these genomes are at draft assembly stages. The information regarding genome sizes etc. in these cases have been obtained from the NCBI microbial sequence database.
The revised names for a number of Chlorobi species are as proposed in Ref. 45.
# The sequences marked 'a' are being sequenced by Gordon and Betty Moore Foundation Marine Biotechnology Intiative; DOE-JGE, indicates Department of Energy Joint Genome Institute. For some of the completed genomes, which are not published, their accession numbers are provided.
The availability of genomic sequences provide an opportunity to carry out in depth studies to identify novel molecular characteristics that are unique to these groups of bacteria and can be used for their diagnostics as well as biochemical and functional studies. Earlier comparative genomic studies on Bacteroidetes/Chlorobi species have been limited to only a few species and they have focused on specific aspects. The studies by Kuwahara [35] and Cerdeño-Tárraga et al. [16], who sequenced the genomes of B. fragilis strains, revealed that these genomes contained extensive DNA inversions in comparison to B. thetaiotaomicron. These inversion events are indicated to be important in terms of generating cell surface variability in these bacteria to avoid their recognition by the immune system. Large expansion of genes involved in the biosynthesis of cell surface polysaccharides and other antigens was also noted in these genomes [16,35]. A comparative analysis by Eisen et al. [24] of C. tepidum TLS genome identified many probable cases of lateral gene transfers (LGTs) between this species and Archaea; in all about 12% of C. tepidum's proteins were indicated to be most similar to those from the archaea. Similarly, the analysis of S. ruber genome by Mongodin et al. [36] has identified many cases of potential LGT between S. ruber and haloarchaea, particularly involving the rhodopsin genes.
In our recent work, we have used comparative genomics to systematically identify various proteins that are uniquely found in either all members, or particular subgroups, of a number of important groups of prokaryotes. These studies have identified large number of proteins that are specific for alpha proteobacteria [37], chlamydiae [38], Actinobacteria [39], epsilon proteobacteria [40] and Archaea [41]. Such genes and proteins, because of their specificity for different phylogenetic or taxonomic groups, provide novel means for diagnostics and evolutionary studies [38,39,42-44] and for the discovery of important biochemical and physiological characteristics that are unique to these groups of prokaryotes. However, thus far no comparative study has examined different genes/proteins that are uniquely present in species from the Bacteroidetes and Chlorobi phyla or are commonly shared by species from these two groups. In order to identify proteins that are uniquely found in the Bacteroidetes and/or Chlorobi groups of species, we have carried out systematic blast searches on all open reading frames in the genomes of P. gingivalis W83, B. fragilis YCH46, B. thetaiotaomicron VPI-5482, G. forsetii KT0803, C. luteolum DSM 273 and C. tepidum TLS against all available sequences in the NCBI non-redundant (nr) database. This has led to identification of large numbers of proteins that are distinctive characteristics of species from different taxonomic groups within the Bacteroidetes phylum (e.g. specific for the Bacteroides genus, specific for the Bacteroidales and Flavobacteriales orders, or specific for the entire Bacteroidetes phylum). Additionally, large numbers of proteins that are specific for the Chlorobi species as well as some proteins that are uniquely shared by the Bacteroidetes and Chlorobi phyla have also been identified. This work also describes three conserved indels in important housekeeping proteins (viz. alanyl-tRNA synthetase, DNA polymerase subunit III and ClpB) that are distinctive characteristics of either the Chlorobi or the Bacteroidales-Flavobacteriales-Flexebacteraceae groups. Phylogenetic analyses of the Bacteroidetes and Chlorobi were also carried out based on a concatenated sequence alignment for 12 highly conserved proteins and the results of these analyses support the inferences derived from the species distribution of various molecular signatures.
Results
Phylogenetic analyses of Bacteroidetes and Chlorobi species
Table 1 lists some characteristics of various Bacteroidetes and Chlorobi genomes that have been completely sequenced as well as for many others that are currently being sequenced. The taxonomy of the Chlorobi species has undergone significant revision in the past few years leading to name changes and new taxonomic groupings for a number of species [45]. The newly proposed and former names for some of the species that will be discussed in the present work are as follows: Chlorobium tepidum changed to Chlorobaculum tepidum; Pelodictyon luteolum changed to Chlorobium luteolum; Prosthecochloris vibrioformis changed to Chlorobium phaeovibrioides; Pelodictyon phaeoclathratiforme changed to Chlorobium clathratiforme. Although many of these species in the databases are still referred to by their former names, we have used the revised nomenclature in our work to interpret the results of evolutionary and comparative genomic studies.
Prior to undertaking comparative analyses of Bacteroidetes and Chlorobi genomes, phylogenetic analyses on these species were carried out to get an overview of their evolutionary relationships, which can serve as a reference point for comparative genomic analyses. Phylogenetic analysis of Bacteroidetes and Chlorobi species has been carried out previously using 16S rRNA sequences and a few isolated protein sequences [27-30,46-49]. However, recent studies show that analyses based on larger dataset derived from multiple genes/proteins sequences provide a more reliable phylogenetic inference [50]. Hence, phylogenetic analyses for these species was carried out based on concatenated sequences for 12 highly conserved proteins involved in a broad range of functions (see Methods section). The final sequence alignment for phylogenetic analysis contained a total of 6998 aligned positions.
Phylogenetic trees were constructed using the neighbour-joining (NJ), maximum-likelihood (ML) and maximum-parsimony (MP) methods [51]. The results of these analyses for the NJ and ML methods are presented in Fig. 1. The trees were rooted using the sequences for Deinococcus-Thermus species. The tree in both cases consisted of two well-resolved clades (100% bootstrap scores by both treeing methods), one comprising of various Chlorobi species and the other containing various Cytophaga-Flavobacteria-Bacteroides (CFB) species. In these trees, S. ruber appeared as a deep branching outgroup of the Chlorobi clade, but in view of the low bootstrap score of the node indicating this relationship and the long branch that separated them, this relationship was not reliable. The topology of various species in the MP tree was very similar, except that in this tree S. ruber formed the outgroup of Chlorobi as well as various other CFB group of bacteria (results not shown). In addition to the uncertain position of S. ruber, different species belonging to the genus Flavobacterium did not form a coherent phylogenetic group (Fig. 1). For most other Bacteroidetes species, sequence information for multiple species from the same genera was not available.
Figure 1.
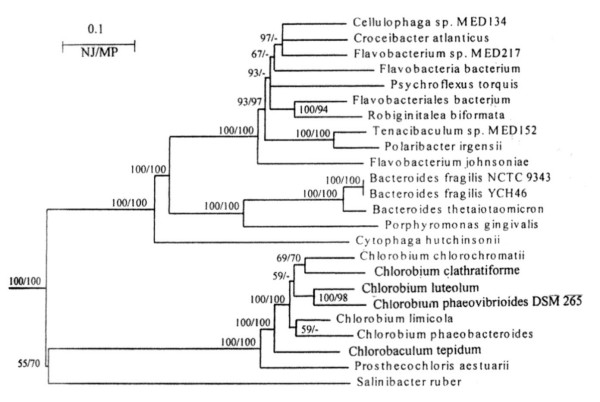
Neighbour-joining tree based on concatenated sequences for 12 highly conserved proteins. The tree was rooted using sequences for Deinococcus-Thermus species and numbers on the nodes indicate bootstrap scores in the NJ and maximum-likelihood analyses (NJ/MP). The branching position of G. forsetii, which became available after this analysis was completed, is not shown. However, our analysis of a smaller dataset indicates that it exhibits closest affinity for the flavobacteria Psychrobacter torquis (results not shown).
Phylogenetic analysis on the concatenated dataset was also performed employing the character compatibility approach [52]. In this approach, all sites in the sequence alignment where only two amino acid states are found, with each state present in at least two species, are examined for mutual compatibility to find the largest clique of mutually compatible characters [52-56]. By removing all homoplasic as well as fast-evolving characters from dataset, this approach provides a powerful means for obtaining correct topology in difficult to resolve cases [56,57]. Our concatenated dataset for the 12 proteins contained 832 positions where only two amino acids were found, with each amino acid present in a minimum of two species. The mutual compatibility of these binary state sites was determined as described in the Methods section.
The compatibility analysis identified two largest sets of compatible characters (referred to as cliques) each containing 410 characters. These cliques were identical in all respects except that the relative branching positions of Chlorobaculum tepidum and Chlorobium chlorochromatii, which differed by a single character, were interchanged. A composite of these cliques, in which the branching positions of these two species are not resolved, is shown in Fig. 2. A large number of characters (i.e. 200) in this clique distinguished the Chlorobi-Bacteroidetes species from the two Deinococcus-Thermus species, which were included to serve as outgroup. The clique is comprised of two main clades, one consisting of various species belonging to the Bacteroidetes group and the other of different Chlorobi species. The species from each of these clades were distinguished by a large numbers of characters. In the Bacteroidetes clade, S. ruber was found to be the deepest branching lineage and its specific association with other Bacteroidetes species was supported by 21 uniquely shared characters, which is a highly significant result [57]. Additionally, the two main orders within the Bacteroidetes viz. Bacteriodales and Flavobacteriales, for which sequence information is available from multiple species, were clearly distinguished based upon multiple characters. However, these analyses detected no uniquely shared character between the C. hutchinsonii and S. ruber. Although, these two species are currently placed in the order Sphingobacteriales, phylogenetic trees do not support a specific grouping of them (see Fig. 1). Different Flavobacterium species again did not group together indicating that this genus does not constitute a phylogenetically coherent taxon. Within the Chlorobi clade, Prosthecochloris aesturaii was found to be the deepest branching lineage, but branching order of other Chlorobi species was not resolved.
Figure 2.

Character compatibility tree (or the largest clique of mutually compatible characters) based on two states sites in the concatenated sequence alignment for the 12 proteins. The clique consisted of 410 mutually compatible characters. The numbers of characters that distinguished different clades are indicated on the nodes. Rooting was done using the sequences for Deinococcus-Thermus species.
Comparative Genomic Studies on Bacteroidetes and Chlorobi Species
To identify proteins that are uniquely present in species from the Bacteroidetes and Chlorobi phyla at various taxonomic levels (genus and above), systematic Blastp searches were performed on each protein or ORF in the genomes of the following species: Bacteroides (P. gingivalis W83 [58], B. fragilis YCH46 [35], B. thetaiotaomicron VPI-5482 [15]); Flavobacteria (G. forsetii str. KT0803 [59]); Chlorobi (C. luteolum DSM 273 and C. tepidum TLS [24]). These analyses have identified a large number of proteins that are specific for different taxonomic groups. A brief description of these signature proteins and their evolutionary significances are discussed below.
Proteins (or ORFs) that are Specific for the Species within the Bacteroidetes Phylum
The blast searches on various ORFs from P. gingivalis, B. fragilis YCH46 and B. thetaiotaomicron genomes have identified 27 proteins that are present in most of the species from the Bacteroidetes phylum (Table 2). In addition to most fully sequenced Bacteroidetes genomes, the homologs of these proteins are also present in most Bacteroidetes species whose genomes have been partially sequenced. One of these proteins, PG0448, is present in all of the Bacteroidetes species, whose genomes have been either partially or fully sequenced. Two other proteins, PG1850 and PG2092, are present in all fully as well as partially sequenced genomes, except those from the Bacteroides genus. The absence of these proteins in only the species from this genus is very likely due to selective gene loss from this lineage and their genes also likely originated in a common ancestor of the Bacteroidetes phylum. Similarly, four proteins viz. PG0449, PG0779, PG1679 and PG2066, are present in virtually all other Bacteroidetes genomes, but they are missing in C. hutchinsonii. Their absence again is very likely due to selective gene loss from C. hutchinsonii. Eight additional proteins viz. PG0202, PG0362, PG0399, PG0482, PG0621, PG01281, PG1367 and PG1394, are present in all other fully as well as partially sequenced Bacteroidetes genomes, but they are only missing in S. ruber. The absence of these proteins in S. ruber can also be explained by selective gene loss. However, in view of the fact that S. ruber branches very deeply in comparison to all other Bacteroidetes species (Figs. 1 and 2), it is also possible that their genes evolved in a common ancestor of the other Bacteroidetes species after the divergence of S. ruber.
Table 2.
Proteins that are Specific for the Phylum Bacteroidetes
| Protein Name | Accession No. | Length | Possible/Predicted Function | Comments |
| PG0202 | NP_904537 | 165 | Uroporphyrinogen-III synthase HemD, putative; COG1587, HemD; pfam02602, HEM4 | Missing in S. ruber+ |
| PG0362 | NP_904673 | 722 | Hypothetical | Missing in S. ruber |
| PG0399 | NP_904705 | 156 | Putative lipoprotein | Missing in S. ruber |
| PG0448 | NP_904748 | 434 | Toluene × outer membrane/transport protein (OMPP1/FadL/TodX); pfam03349 | All species present |
| PG0449 | NP_904749 | 441 | TPR domain protein; cd00189, TPR; COG3071, HemY; COG4783, Putative Zn-dependent protease | Not found in F. johnsoniae and C. hutchinsonii |
| PG0482 | NP_904777 | 143 | Hypothetical protein | Missing in S. ruber |
| PG0621 | NP_904906 | 182 | Hypothetical protein | Missing in S. ruber |
| PG0779 | NP_905041 | 157 | ExbD, Biopolymer transport protein; COG0848 | Not found in F. bacterium HTC, F. johnsoniae and C. hutchinsonii |
| PG1281 | NP_905462 | 387 | Putative DNA mismatch repair protein; pfam01713, Smr; COG1193, Mismatch repair ATPase | Not found in C. atlanticus and S. ruber+ |
| PG1367 | NP_905532 | 200 | Hypothetical protein | Missing in S. ruber |
| PG1394 | NP_905555 | 165 | Putative trans-membrane | Missing in S. ruber |
| PG1626 | NP_905755 | 554 | Putative hemin receptor | Missing in C. hutchinsonii; also present in C. phaeobacteriodes. |
| PG1679 | NP_905797 | 464 | Putative trans-membrane | Not found in P. ruminicola and C. hutchinsonii |
| PG1850 | NP_905940 | 302 | Hypothetical protein | Missing in Bacteroides species+ |
| PG2066 | NP_906128 | 351 | Putative lipoprotein | Missing in P. ruminicola and C. hutchinsonii |
| PG2092 | NP_906153 | 419 | Hypothetical protein | Missing in Bacteroides species |
| BF0296 | YP_097579 | 988 | Outer membrane assembly protein | Missing in P. gingivalis and S. ruber+ |
| BF0439 | YP_097722 | 565 | Putative outer membrane protein probably involved in nutrient binding | Missing in P. gingivalis, and Tenacibaculum |
| BF0534 | YP_097817 | 192 | Putative acetyl-transferase | Missing in P. gingivalis, C. atlanticus and S. ruber |
| BF0665 | YP_097947 | 531 | Putative exported protein | Missing in P. gingivalis, and C. hutchinsonii |
| BF0751 | YP_098036 | 577 | Putative exported protein | Missing in P. gingivalis, P. torquis, R. biformata and C. hutchinsonii |
| BF1057 | YP_098341 | 506 | Putative exported protein | Missing in P. gingivalis, and C. hutchinsonii |
| BF1254 | YP_098538 | 507 | Putative exported protein | Missing in P. gingivalis, and C. hutchinsonii |
| BF1327 | YP_098610 | 514 | Putative exported protein | Missing in P. gingivalis, and C. hutchinsonii |
| BF3185 | YP_100464 | 490 | Putative exported protein | Missing in P. gingivalis, and C. hutchinsonii |
| BF3612 | YP_100889 | 542 | Putative exported protein | Missing in P. gingivalis and C. hutchinsonii |
| BF4330 | YP_101602 | 538 | Putative exported protein | Missing in P. gingivalis,, R. biformata and C. hutchinsonii |
For the proteins listed here, all significant blast hits are from various Bacteroidetes species, except as noted below. These proteins are present in all of the Bacteroidetes species listed in Table 1, except as noted in the comments.
+For the protein PG0202, a significant hit is also observed from C. phaeobacteroides; For BF0296 significant hits also observed from P. aestuarii and C. phaeobacteroides.
Of the proteins listed here, the following proteins are homologous to each other: BF0751, BF1057, BF1327, BF3185; BF1254, BF3612, BF4330.
We have also come identified a 3 aa deletion in a conserved region of ClpB protease that is present in all other Bacteroidetes species, except S. ruber (Additional file 1). Similar to the genes for the above 8 proteins, this deletion likely occurred in a common ancestor of the other Bacteroidetes species after the branching of S. ruber. Besides Bacteroidetes species, this indel is also present in the ClpB homologs from C. phaebacteroidetes (only Chlorobi species containing this protein) and the archaeum Methanospirillum hungatei, which is likely due to LGT. The remaining proteins in Table 2, of which 7 (BF0751, BF1057, BF1327, BF3185; BF1254, BF3612, BF4330) are homologous to each other, are missing in 1 or 2 sequenced species (e.g. P. gingivalis, B. fragilis, C. hutchinsonii or S. ruber) and their distribution pattern can also be explained by similar mechanisms as discussed above. Except for a few proteins that show limited similarity to conserved domains (CDs) found in other proteins [60], most of the proteins in Table 2 are of unknown function.
These searches have also identified several proteins that at present appear unique for the species from the Bacteroidales and Flavobacteriales orders. These proteins are listed in Table 3. Of these, the first 4 proteins viz. PG0336, PG1302, PG1537, PG2030 are present in nearly all complete as well as partially sequenced species from the above two orders, but they are not found in S. ruber as well as C. hutchinsonii. The latter two species, which show much deeper branching than all other Bacteroidales species (Figs. 1 and 2), are currently placed in the order Sphingobacteriales. The genes for the proteins listed in Table 3 have thus likely originated in a common ancestor of the Bacteroidales and Flavobacteriales after the divergence of Sphingobacteria. Thirty-seven additional proteins in Table 3 are also uniquely present in either all or many of the sequenced Bacteroidales species and a small number of flavobacteria species including G. forsetii. A large number of these proteins are only missing in P. gingivalis, which is likely due to gene loss. Of the proteins listed in Table 3, seven are indicated to be conjugative transposon proteins: TraJ, TraN, TraK, TraF, TraE and TraB (PG1251 and PG1479, PG1475, PG1478, PG1482, PG1483 and BF0127, respectively). Four of them are present in two clusters very close to each other (PG1478-PG1479, PG1482-PG1483), supporting their involvement in related functions [61,62]. The genes for these proteins have also likely evolved in a common ancestor of the Bacteroidales and Flavobacteriales, followed by gene losses in various species.
Table 3.
Proteins that are Specific for the Bacteroidales and Flavobacteriales Orders
| Genome ID No. [Accession No.] | Possible/Predicted Function | Genome ID No. [Accession No.] | Possible/Predicted Function |
| PG0336 [NP_904650] | Hypothetical protein | PG1276 [NP_905457] | DNA-binding protein, histone-like family |
| PG1302 [NP_905476] | Hypothetical protein | PG1389 [NP_905551] | DNA-binding protein, histone-like family |
| PG1537 [NP_905677] | Hypothetical protein | PG1444 [NP_905595] | Hypothetical protein |
| PG1251 [NP_905435] | Conjugative transposon protein TraJ | PG1475 [NP_905621] | Conjugative transposon protein TraN |
| PG2030 [NP_906097] | Hypothetical protein | PG1478 [NP_905624] | Conjugative transposon protein TraK |
| PG1492 [NP_905638] | Hypothetical protein | PG1479 [NP_905625] | Conjugative transposon protein TraJ |
| PG0302 [NP_904618] | Hypothetical protein | PG1482 [NP_905628] | Conjugative transposon protein TraF |
| PG0330 [NP_904645] | DNA-binding protein, histone-like family; smart00411, BHL | PG1483 [NP_905629] | Conjugative transposon protein TraE |
| PG0555 [NP_904845] | DNA-binding protein, histone-like family | PG1488 [NP_905634] | Hypothetical protein |
| PG0829 [NP_905084] | Hypothetical protein | PG1494 [NP_905640] | Hypothetical protein |
| PG0870 [NP_905118]* | Hypothetical protein | PG2040 [NP_906106] | DNA-binding protein, histone-like family |
| PG0875 [NP_905123] | TnpA; DNA replication, recombination & repair, COG2452 | PG2127 [NP_906183] | Hypothetical protein |
| PG1206 [NP_905397]c | Mobilizable transposon, tnpC protein | BF1773 [YP_099054]* | Probable truncated integrase; cd01185, INT_Tn4399 |
| Missing in Porphyromonas gingivalis W83 | |||
| BF0127 [YP_097410] | TraB | BF1727 [YP_099008] | Putative outer membrane protein maybe involved in nutrient binding |
| BF0136 [YP_097419] | Tetracycline resistance element mobilization: RteC | BF1860 [YP_099142] | Hypothetical protein |
| BF0146 [YP_097429] | Hypothetical protein | BF1926 [YP_211559] | Hypothetical protein |
| BF0319 [YP_097602]* | Putative exported protein | BF2130 [YP_099411] | Hypothetical protein |
| BF0342 [YP_097625] | Putative exported protein | BF2214 [YP_099495] | Hypothetical protein |
| BF1067 [YP_098351] | Hypothetical protein | BF3164 [YP_100443] | Putative lipoprotein |
| BF1422 [YP_098707] | Hypothetical protein | BF4258 [YP_101533] | Hypothetical protein |
| BF1567 [YP_098851] | Hypothetical protein | ||
The first five proteins in this table (viz. PG0336, PG1302, PG1537, PG1626, PG2030) are present in all Bacteroidales and Flavobacteriales species listed in Table 1; The other proteins are present in many of the Bacteroidales and Flavobacteriales species.
*Also present in C. phaeobacteroides.
The following proteins are homologous to each other: PG0330, PG0555, PG1276, PG1389, PG2040; PG0829, PG1444, PG1488; PG1251, PG1479; BF1422, BF1860; BF1926, BF4258.
Proteins that are specific for the order Bacteroidales or the genus Bacteroides
There are 4 genomes for the Bacteroidales species (P. gingivalis W83, B. thetaiotaomicron VPI-5482 and B. fragilis strains: NCTC 9343 and YCH46) that have been fully sequenced. Additionally, sequence information for a large number of genes/proteins from Prevotella intermedia 17 and Prevotella ruminicola 23, which belong to this order is available in the in NCBI database (see Table 1). Our blast searches on proteins from P. gingivalis genome have identified 52 proteins that are uniquely shared by either all or most of the sequenced Bacteriodales species and whose homologs are not found in any other species, except where noted (Table 4). Thirty-nine of these 52 proteins are uniquely found in all 4 fully sequenced Bacteroidales species. These species also form a strongly supported clade in phylogenetic trees (Figs. 1 and 2). Thus, it is likely that the genes for these proteins evolved in a common ancestor of this order. The remaining 13 proteins are lacking in at least one of the Bacteroides species (noted in Table 4), which is likely due to gene loss. In addition to the sequenced Bacteroidales species, high scoring homologs for many of the above proteins were also found in the two Prevotella species. These latter homologs were detected via genomic blasts against partially sequenced genomes from these species (see Methods).
Table 4.
Proteins Unique to the Bacteroidales Order
| Genome ID No. [Accession No.] | Possible/Predicted Function | Genome ID No. [Accession No.] | Possible/Predicted Function |
| PG0018 [NP_904375]x | hypothetical protein | PG1133 [NP_905341] | conserved hypothetical protein |
| PG0082 [NP_904431]+ | putative exported protein | PG1139 [NP_905347] | conserved hypothetical protein; cd02966, Tlp_A_like_family |
| PG0125 [NP_904468] | conserved hypothetical protein | PG1214 [NP_905405] | hypothetical protein |
| PG0179 [NP_904515]+ | putative exported protein | PG1301 [NP_905475]+ | conserved hypothetical protein |
| PG0188 [NP_904523]x | lipoprotein, putative | PG1333 [NP_905502]x | putative exported protein |
| PG0216 [NP_904548]+ | conserved hypothetical exported protein | PG1352 [NP_905517] | putative conserved hypothetical protein |
| PG0217 [NP_904549]+ | cons. hypothetical exported protein | PG1388 [NP_905550]+ | conserved hypothetical protein |
| PG0218 [NP_904550]+ | conserved hypothetical exported protein | PG1441 [NP_905593]x | lysozyme-related protein; cd00737, endolysin_autolysin; COG3772, phage-related lysozyme |
| PG0246 [NP_904573]+ | putative DNA-binding protein | PG1442 [NP_905594] | TraB |
| PG0312 [NP_904628]+ | putative transmembrane protein | PG1458 [NP_905606]x | hypothetical protein |
| PG0326 [NP_904641] | hypoth; COG3637, Opacity protein & related surface antigens | PG1473 [NP_905619]+ | conjugative transposon protein TraQ |
| PG0366 [NP_904677]+# | hypothetical protein | PG1621 [NP_905750] | conserved hypothetical exported protein |
| PG0434 [NP_904735]+ | putative transmembrane protein | PG1757 [NP_905859]x | hypothetical protein |
| PG0541 [NP_904834]+ | conserved hypothetical protein | PG1881 [NP_905968]+ | putative lipoprotein |
| PG0574 [NP_904862] | hypothetical protein | PG1889 [NP_905974]x | hypothetical protein |
| PG0717 [NP_904988]+ | lipoprotein, putative | PG1945 [NP_906027]+ | conserved hypothetical protein |
| PG0781 [NP_905043]+ | putative membrane protein | PG2006 [NP_906077]+ | conserved hypothetical membrane protein |
| PG0816 [NP_905074]x | hypothetical protein | PG2079 [NP_906141] | conserved hypothetical protein |
| PG0831 [NP_905085] | Cons. protein maybe related to TraB | PG2083 [NP_906145]+ | conserved hypothetical protein |
| PG0843 [NP_905095]x | conserved hypothetical protein | PG2116 [NP_906174]x | transposase |
| PG0851 [NP_905101] | Toprim domain protein | PG2130 [NP_906186] | FimX |
| PG0910 [NP_905150] | FHA domain protein, cd00060 | PG2131 [NP_906187]+ | 60 kDa protein; OmpA, COG2885.2 |
| PG0937 [NP_905172] | putative exported protein | PG2133 [NP_906189]x | lipoprotein, putative |
| PG0961 [NP_905192]+ | conserved hypothetical protein | PG2149 [NP_906203] | putative conserved exported protein |
| PG1050 [NP_905267]x | putative lipoprotein | PG2168 [NP_906219]+ | FimX |
| PG1125 [NP_905334]+ | conserved hypothetical protein | PG2224 [NP_906265]x | hypothetical protein |
All significant hits for these proteins are observed from the following Bacteroidales species, for which sequence information is available in the NCBI or TIGR microbial sequence database. P. gingivalis W83, B. fragilis NCTC 9343, B. fragilis YCH46 , B. thetaiotaomicron VPI-5482, Prevotella intermedia 17, P. ruminicola 23. Unless noted below, these proteins are present in all of the above-mentioned species. Of these proteins, the following are homologous to each other: PG0179, PG2133; PG0217, PG0218; PG0816, PG1458; PG0831, PG1442; PG2130, PG2168.
+Not found at present in either 1 or both Prevotella species.
xMissing in one of the Bacteroides species as well as Prevotella species.
#Also present was C. phaeobacteroides BS1.
The majority of the Bacteroidales-specific proteins are hypothetical and some of them are indicated as putative exported proteins (Table 4). Some interesting proteins in this list include the FimX proteins (PG2130, PG2168) that are involved in fimbriae production, which is necessary for adhesion to host surfaces [63]. Also of interest is the conserved hypothetical protein, PG1139, which shows slight but significant similarity to the conserved domain in the TlpA-like family, responsible for cytochrome maturation. One of the proteins PG0366 also had a significant hit from C. phaeobacteroides, which could be due to LGT [64]. There are seven proteins in Table 4 (PG0216-PG0218, PG1441-PG1442, PG2130-PG2131) that are present in clusters of two or three, suggesting that they could form functional units. Further, a number of proteins in this table (viz. PG0179, PG2133; PG0217, PG0218; PG0816, PG1458; PG0831, PG1442; PG2130, PG2168) are homologous to each other, indicating that they resulted from gene duplication events.
The Bacteroides genus contains three fully sequenced genomes corresponding to B. thetaiotaomicron VPI-5482 and B. fragilis strains NCTC 9343 and YCH46. The blast searches on B. fragilis YCH46 genome have identified 185 proteins that are mainly specific for these species (Additional file 2) and their homologs are not found in P. gingivalis. For 10 of these proteins, significant similarity was also observed for at least one of the two Prevotella species, suggesting that within the order Bacteriodales, species from the Bacteroides and Prevotella genera may be more closely related to each other in comparison to P. gingivalis. Most of these proteins are of unknown functions, however, some have been annotated as transmembrane or lipoproteins. Thirty-nine of these proteins are present in clusters of two to four in the genome indicating that they could be involved in related functions. A number of proteins in this table are homologous to each other indicating that they have likely resulted from gene duplication events.
Proteins that are specific for the order Flavobacteriales
The complete genome of the first flavobacteria species viz.G. forsetii KT0803 became available very recently [59]. However, sequence information for a large number of other Flavobacteriales species, whose genomes are being sequenced (see Table 1), is available in the NCBI database. Our blastp and PSI-blast searches on different ORFs in the G. forsetii genome have identified 38 proteins that are uniquely present in virtually all of the Flavobacteriales species (Table 5). Twenty-six of these proteins are present in all Flavobacteriales species listed in Table 1, whereas the remaining 12 are missing in only one of the species. An additional group of 146 proteins are also specific for the Flavobacteriales, but they are missing in some flavobacteria species or limited to only a small numbers of flavobacteria (Additional file 3). Because the genomes for most of the Flavobacteriales species are not complete at the present time, these proteins were not separated into different groups.
Table 5.
Proteins that are Specific for Species from the Flavobacteriales Order
| orf No. [Accession No.] | Possible/Predicted Function | Genome ID No. [Accession No.] | Possible/Predicted Function |
| orf89 [CAL65078]b | membrane protein | orf1826 [CAL66812] | hypothetical protein |
| orf92 [CAL65081] | secreted protein | orf1872 [CAL66858] | hypothetical protein |
| orf107 [CAL65096] | membrane protein | orf2280 [CAL67264]c | hypothetical protein |
| orf110 [CAL65099] | conserved hypothetical protein | orf2667 [CAL67651] | conserved hypothetical protein |
| orf191 [CAL65180] | membrane or secreted protein | orf2698 [CAL67682] | secreted protein |
| orf403 [CAL65392]b | hypothetical protein | orf2700 [CAL67684] | hypothetical protein |
| orf509 [CAL65498]c | hypothetical protein | orf2718 [CAL67702] | hypothetical protein |
| orf612 [CAL65599]b | secreted protein | orf2731 [CAL67715]b | secreted protein |
| orf983 [CAL65970]a | hypothetical protein | orf2756 [CAL67740] | secreted protein |
| orf995 [CAL65982] | phospholipid/glycerol acyltransferase; smart00563, PlsC | orf2825 [CAL67809] | secreted protein |
| orf998 [CAL65985] | membrane protein | orf2844 [CAL67828] | membrane protein |
| orf1059 [CAL66046] | membrane protein | orf2917 [CAL67899] | secreted protein |
| orf1078 [CAL66065]a | membrane or secreted protein | orf2939 [CAL67921] | hypothetical; COG1577, ERG12 |
| orf1453 [CAL66440] | secreted protein | orf3043 [CAL68025] | hypothetical protein |
| orf1469 [CAL66456]a | secreted protein | orf3076 [CAL68058]e | hypothetical protein |
| orf1555 [CAL66542] | secreted protein | orf3240 [CAL68223] | membrane protein |
| orf1618 [CAL66605]d | secreted protein | orf3266 [CAL68249]a | conserved hypothetical protein |
| orf1766 [CAL66752] | hypothetical protein | orf3313 [CAL68296] | hypothetical protein |
| orf1776 [CAL66762] | hypothetical protein | orf3501 [CAL68484] | conserved hypothetical protein |
Unless otherwise indicated, all significant hits for the proteins listed in this table are from the following Flavobacteriales species, for which sequence information is presently available. G. forestii KT0803, F. bacterium BBFL7, F. bacterium HTCC2170, F. johnsoniae UW101, Flavobacterium sp. MED217, Cellulophaga sp. MED134, C. atlanticus HTCC2559, P. irgensii 23-P, P. torquis ATCC 700755, R. biformata HTCC2501, Tenacibaculum sp. MED152. The proteins are present in all of these species except as noted below:
aMissing in Polaribacter irgensii 23-P
bMissing in Flavobacterium johnsoniae UW101
cMissing in Flavobacteria bacterium BBFL7
dMissing in Psychroflexus torquis ATCC 700755
eMissing in Robiginitalea biformata HTCC2501
Proteins that are unique for the Chlorobi Phylum
The genomes of three Chlorobi species viz. C. luteolum DSM 273,C. tepidum TLS, and C. chlorochromatii CaD3, have been fully sequenced. Our blast analyses on the C. tepidum and C. luteolum genomes have identified 51 proteins that are uniquely shared by species from this phylum (Table 6). In addition to the 3 completely sequenced genomes, homologs of these proteins are also present in six others Chlorobi species (see Table 1), for which sequence information is available in the NCBI database. The genes for these proteins likely originated in a common ancestor of various Chlorobi species, which form a distinct, strongly supported, clade in phylogenetic trees (see Figures 1 and 2). The vast majority of these proteins are of hypothetical or unknown functions. However, 5 of them are indicated to be involved in functions related to photosynthesis. Of these, Plut_0264 and Plut_0265 are clustered in the genome and they correspond to chlorosome envelope proteins C and A, respectively. The protein Plut_1500, which is indicated as bacteriochlorophyll A protein, corresponds to the FMO protein that is involved in the attachment of chlorosomes to the cytoplasmic membrane [34]. The other two photosynthesis-related proteins, Plut_0620 and Plut_1628, are annotated as photosystem P840 reaction centre protein PscD and the photosystem P840 reaction centre cytochrome c-551, respectively [24,32]. Three additional chlorobi-specific proteins, Plut_1714-Plut_1716, are clustered together in the genome indicating that they may form a functional unit [61]. There are 65 additional proteins that are also specific for the Chlorobi species (Additional file 4). However, unlike the proteins in Table 6, these proteins are missing in a number of the Chlorobi species and their species distribution does not show any clear pattern and these could involve gene loss or LGTs [65]. However, the first 8 proteins listed in this additional file (viz. Plut_0107, Plut_0759, Plut_0762, Plut_0981, Plut_0985, Plut_1092, Plut_1145, Plut_1858) are only found in C. luteolum and C. phaeovibrioides. These two species form a strongly supported clade in various phylogenetic trees (Figs. 1 and 2) and a specific relationship between them is further supported by the unique presence of these shared proteins. For one of the proteins in this table, Plut_1345, a significant hit is also observed from C. hutchinsonii, which could be due to LGT [64,65].
Table 6.
Proteins that are Specific for the Chlorobi Species
| Genome ID No. [Accession No.] | Possible/Predicted Function | Genome ID No. [Accession No.] | Possible/Predicted Function |
| Plut_0059 [YP_373992] | Hypothetical protein | Plut_1225 [YP_375130] | Hypothetical protein |
| Plut_0074 [YP_374007] | orfCR | Plut_1238 [YP_375143] | Hypothetical protein |
| Plut_0111 [YP_374044] | Hypothetical protein | Plut_1332 [YP_375234] | TPR repeat |
| Plut_0145 [YP_374078] | Hypothetical protein | Plut_1409 [YP_375311] | Hypothetical protein |
| Plut_0160 [YP_374093] | Hypothetical protein | Plut_1465 [YP_375367] | Hypothetical protein |
| Plut_0264 [YP_374195] | chlorosome envelope protein C | Plut_1469 [YP_375371] | Hypothetical protein |
| Plut_0265 [YP_374196] | chlorosome envelope protein A; Bac_chlorC, pfam02043 | Plut_1491 [YP_375393] | Hypothetical protein |
| Plut_0278 [YP_374209] | Hypothetical protein | Plut_1500 [YP_375402] | FMO, BchlA protein |
| Plut_0282 [YP_374213] | Hypothetical protein | Plut_1517 [YP_375417] | Hypothetical protein |
| Plut_0295 [YP_374226] | Hypothetical protein | Plut_1608 [YP_375505] | Srm |
| Plut_0325 [YP_374256] | Hypothetical protein | Plut_1625 [YP_375522] | Hypothetical protein |
| Plut_0409 [YP_374340] | Hypothetical protein | Plut_1628 [YP_375525] | Photosystem P840 reaction center cytochrome c-551 |
| Plut_0422 [YP_374353] | Hypothetical protein | Plut_1682 [YP_375579] | Hypothetical protein |
| Plut_0489 [YP_374420] | Hypothetical protein | Plut_1714 [YP_375611] | Hypothetical protein |
| Plut_0499 [YP_374430] | Hypothetical protein | Plut_1715 [YP_375612] | Hypothetical protein |
| Plut_0540 [YP_374467] | Hypothetical protein | Plut_1716 [YP_375613] | Hypothetical protein |
| Plut_0572 [YP_374498] | Hypothetical protein | Plut_1725 [YP_375622] | Hypothetical protein |
| Plut_0620 [YP_374546] | Photosystem P840 reaction center protein PscD | Plut_1742 [YP_375639] | Hypothetical protein |
| Plut_0666 [YP_374587] | Hypothetical protein | Plut_1743 [YP_375640] | Hypothetical protein |
| Plut_0713 [YP_374634] | Hypothetical protein | Plut_1746 [YP_375643] | Hypothetical protein |
| Plut_0779 [YP_374695] | Hypothetical protein | Plut_1933 [YP_375818] | Hypothetical protein |
| Plut_0950 [YP_374855] | Hypothetical protein | Plut_2003 [YP_375888] | orfCR |
| Plut_1012 [YP_374917] | Hypothetical protein | Plut_2041 [YP_375926] | Hypothetical protein |
| Plut_1195 [YP_375100] | Hypothetical protein | Plut_2100 [YP_375985] | Hypothetical protein |
| Plut_1217 [YP_375122] | Hypothetical protein | Plut_2117 [YP_376002] | Hypothetical protein |
| Plut_1223 [YP_375128] | Hypothetical protein |
All significant Blast hits for these proteins are observed only from the following Chlorobi species for which sequence information is presently available: C. luteolum DSM 273, C. tepidum TLS, C. chlorochromatii CaD3, C. limicola DSM 245, C. phaeobacteroides BS1, C. phaeobacteroides DSM 266, C. clathratiforme BU-1, P. aestuarii DSM 271 and C. phaeovibrioides DSM 265. Of these proteins, the following proteins are homologous to each other:Plut_0059, Plut_1491; Plut_0074, Plut_2003.
In addition to the proteins that are uniquely found in various Chlorobi species, we have also identified two large conserved inserts in two widely distributed proteins that are distinctive characteristics of the Chlorobi phylum. The first of these signatures is a 28 aa insert in the DNA polymerase III alpha subunit encoded by the dnaE gene that is required for chromosomal replication in bacteria [66]. The large insert in DnaE is present in all of the Chlorobi homologs but it is not found in any other species (Fig. 3). A smaller insert of 3–4 aa is probably also present in this regions in some Bacteroidales species, but based on their different sizes and sequence characteristics, these inserts are of independent origin. The other Chlorobi-specific signature consists of a 12–14 aa insert in alanyl-tRNA synthetase (Fig. 4), which plays an essential role in protein synthesis. This insert is again present in all Chlorobi homologs but not found in any other species indicating that it provides a reliable molecular marker for this group.
Figure 3.
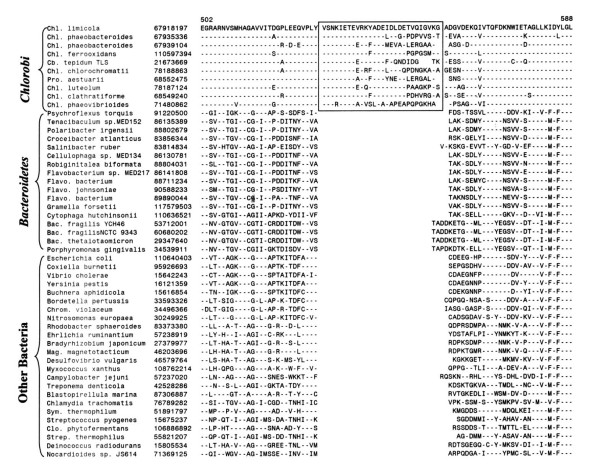
Partial sequence alignments of the DnaE protein showing a large insert of about 28 aa that is uniquely present in Chlorobi homologs. The dashes (-) denote identity with the amino acid on the top line. Except for the Chlorobi species, this insert is not found in any other organism. Sequence information for only representative species from other groups of bacteria is shown. Abbreviations in the species names are: Bac., Bacteroides; Cb., Chlorobaculum; Chl., Chlorobium; Chrom., Chromobacterium; Clo., Clostridium; Pro., Prosthecochloris; Sym., Symbiobacterium; Flavo., Flavobacterium; Strep., Streptococcus.
Figure 4.
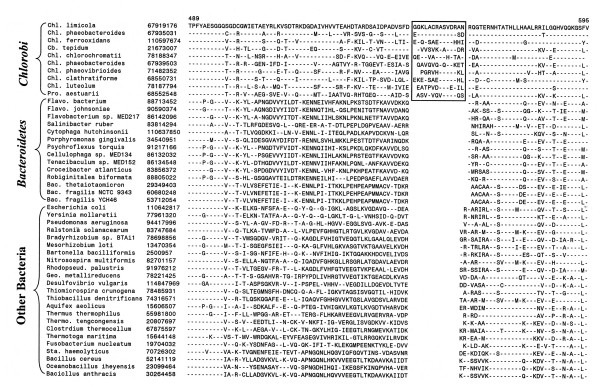
Partial sequence alignments of alanyl-tRNA synthetase showing a conserved insert of about 12–14 aa that is a distinctive characteristic of Chlorobi homologs and not found in other bacteria. The dashes (-) denote identity with the amino acid on the top line. Additional abbreviations: Geo., Geobacter; Sta., Staphylococcus; Thermo., Thermoanaerobacter.
Proteins that are uniquely shared by the Bacteroidetes and Chlorobi species
The Bacteroidetes and Chlorobi species generally branch very close to each other in phylogenetic trees [27-30]. However, there are very few characteristics known that are uniquely shared by species from these two phyla. Our analysis has identified 3 proteins (PG0081, PG0649 and PG2432 in Table 7) that are uniquely found in virtually all of the fully as well as partially sequenced Bacteroidetes and Chlorobi genomes. These results are significant because Bacteroidetes or Chlorobi species do not share any protein in common with different species from any other group of bacteria. Of these proteins, the protein PG0081 is also found in Fibrobacter succinogenes. A close and specific relationship of F. succinogenes to the Bacteroidetes and Chlorobi groups was strongly suggested by our earlier work based on conserved indels in different proteins [30]. This inference is reinforced by the unique presence of this protein in these different groups. The absence of the protein PG0649, which is present in all other Bacteroidetes and Chlorobi species, in S. ruber, is probably due to gene loss. Three other proteins, PG1818, PG1977 and BF2465 although they appear unique to the Bacteriodales and Chlorobi, their homologs are not detected in most Flavobacteria including G. forsetii. It is likely that the genes for these proteins also evolved in a common ancestor of the Bacteroidetes and Chlorobi phyla followed by gene losses in particular Bacteroidetes lineages. All of these proteins are of unknown functions except PG1818, which is annotated as a putative transmembrane protein with significant similarity to the conserved domain of the ResB-like family.
Table 7.
Proteins that are Uniquely Shared by the Bacteroidetes and Chlorobi Species
| Protein I.D. No. [Accession No.] | PG0081 [NP_904430] | PG0649 [NP_904929] | BF2432 [YP_099715] | PG1818 [NP_905917] | PG1977 [NP_906051] | BF2465 [YP_099748] |
| Length (aa) | 725 aa | 194 aa | 1478 | 238 aa | 668 aa | 93 aa |
| Possible Function | Hypoth. | Hypoth. | Hypoth. | Putative transmembrane | Hypoth. | Hypoth. |
| Bacteroidetes | ||||||
| P. gingivalis | * | * | * | * | * | * |
| B. fragilis NCT | * | * | * | * | * | * |
| B. fragilisYCH | * | * | * | * | * | * |
| B. thetaiotaomi. | * | * | * | * | * | * |
| Prev. intermedia | * | * | * | * | * | * |
| Prev. ruminicola | * | * | * | * | * | |
| G. forestii | * | * | * | |||
| F. bacterium BBF | * | * | * | * | ||
| F. bacterium HTC | * | * | * | |||
| F. johnsoniae | * | * | * | |||
| Flavobacterium | * | * | * | |||
| Cellulophaga | * | * | * | |||
| C. atlanticus | * | * | * | |||
| Polibacter irgensii | * | * | * | |||
| Psychro. torquis | * | * | * | |||
| Rob. biformata | * | * | ||||
| Tenacibaculum | * | * | * | |||
| Cyto. hutchinsonii | * | * | * | * | * | |
| Salinibacter ruber | * | * | * | |||
| Chlorobi | ||||||
| Cb. tepidum | * | * | * | * | ||
| C. chlorochrom. | * | * | * | * | * | * |
| C. luteolum | * | * | * | * | * | * |
| C. limicola | * | * | * | * | * | |
| C. phaeobac BS1 | * | * | * | * | * | |
| C. phaeobac DSM | * | * | * | * | * | |
| C. clathratiforme | * | * | * | * | * | * |
| Prosthec. aesturii | * | * | * | * | * | * |
| C. phaeovibrioides | * | * | * | * | * |
All significant blast hits for these proteins are from the indicated (marked by *) Bacteroidetes and Chlorobi species. For the protein PG0081, a homolog is also present in Fibrobacter succinogenes subsp. succinogenes S85 (identified by blast search against the partial sequence). The blank space indicates that no hit showing significant similarity was detected at the present time.
Discussion
This work has identified a large number of proteins that are specific for Bacteroidetes and Chlorobi species at various taxonomic levels. Homologs exhibiting significant similarity to these proteins are not found in any other bacteria, except in a few isolated cases. Among the proteins that are specific for the Bacteroidetes, 27 proteins are specific for the entire phylum as their homologs are present in species from all three main orders within this phylum. Many other proteins are limited to various clades within the Bacteroidetes phylum. These include 41 proteins that are common to the Flavobacteriales and Bacteroidales orders; 53 and 38 proteins that are specific for the Bacteriodales and Flavobacteriales orders, respectively; and 185 proteins that are specific for the Bacteriodes genus. We have also identified 51 proteins that are specific for the Chlorobi species and 6 proteins that are uniquely shared by the Bacteroidetes and Chlorobi phyla. Two large conserved inserts in the DnaE and AlaRS proteins that are distinctive characteristics of the Chlorobi species were also discovered in this work. In addition, a deletion in ClpB protein that is mainly specific for the Bacteriodales, Flavobacteriales and Flexibacteraceae was also identified. In earlier work, a number of conserved inserts that are specific for either the Bacteroidetes phylum (viz. SecA and Gyrase B) or commonly shared by the Bacteroidetes and Chlorobi species were also described. Based upon their specificity for the Bacteroidetes and Chlorobi species, these molecular markers provide novel and more definitive means for identifying and circumscribing species from these groups.
The species distribution patterns of these signature proteins and conserved indels strongly suggest that they or the genes for them have evolved at various stages in the evolution of these bacteria (Fig. 5). However, subsequent to their evolution or introduction in these genes, these genomic characteristics are stably retained in various descendents of these lineages with minimal gene loss or LGTs, as has also been found in other related studies [37-41,44,67,68]. The evolutionary relationship among the Bacteroidetes species as deduced from these signature proteins is in complete agreement with their branching pattern in phylogenetic trees (Figs. 1 and 2). The unique presence of several signature proteins as well as conserved indels in a number of essential proteins (viz. FtsK, UvrB and ATP synthase alpha subunit) by different Bacteroidetes and Chlorobi species provides compelling evidence that species from these two groups shared a common ancestor exclusive of all other bacteria. In earlier studies, a close relationship of Fiborbacteres to the Bacteroidetes and Chlorobi was also observed [2,30]. The species from all these three groups were found to contain large conserved indels in RNA polymerase β ' subunit and serine hydroxymethyl transferase, that were not found in any other bacteria [30]. The species from these three groups also branched in the same position based on distribution profiles of signature sequences in a number of other proteins and in different phylogenetic trees [30,69]. The unique shared presence of the protein PG0081 by all sequenced Chlorobi and Bacteroidetes species as well as Fibrobacter succinogenes, provides further evidence that species from these three groups form a single superphylum and that they shared a common ancestor exclusive of all other bacteria [30].
Figure 5.
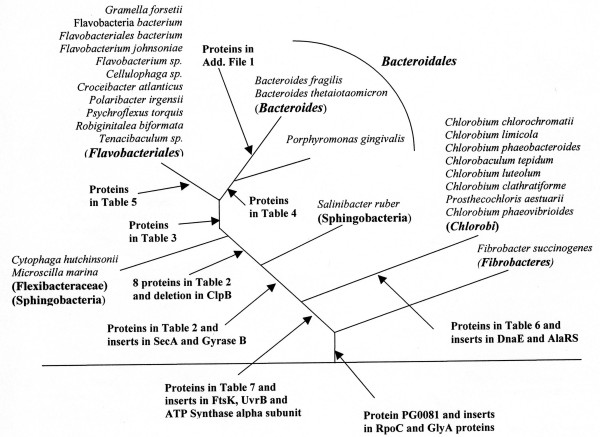
A summary diagram showing the evolutionary stages where different signature proteins and conserved indels that are specific for the Bacteroidetes and Chlorobi species have likely evolved or originated. Some of the conserved inserts that are specific for these groups or indicate their branching position relative to other bacterial phyla have been described in earlier work [30,43,69].
This paper also reports phylogenetic analyses of Bacteroidetes and Chlorobi species based on a concatenated alignment of 12 highly conserved proteins. The branching order of various species in the trees obtained using different phylogenetic methods were in general very similar with the clades corresponding to the Chlorobi species and the Cytophaga-Flavobacteria-Bacteroides species, well resolved from each other with 100% bootstrap scores. The species corresponding to the two main groups within the Bacteroidetes phylum (viz. the Bacteriodales and Flavobacteriales orders) were also clearly resolved. However, in the trees constructed by traditional phylogenetic methods such as NJ, ML and MP, the phylogenetic placement of S. ruber was not resolved. In all of these trees, S. ruber appeared either as a very deep branch of the Chlorobi clade (i.e. in NJ and ML trees) or as outgroup of both the Chlorobi and the CFB clades (MP tree). In contrast to these trees, when the same dataset was analyzed by means of the character compatibility or clique approach, S. ruber formed the deepest branch of the Bacteroidetes species and its specific association with this group was supported by 21 uniquely shared characters, indicating strongly that this affiliation was reliable [57]. These results provide evidence that the character compatibility approach, which removes all fast-evolving as well as homoplasic sites from a given dataset, provides a powerful means for obtaining correct topology in cases, such as that for S. ruber, whose phyletic affinity has proven difficult to establish by traditional phylogenetic methods [13,52,56,57].
The cellular functions of most of the Bacteroidetes or Chlorobi-specific proteins identified in the present work are not known. A few of the Chlorobi-specific proteins are involved in chlorosome- or photosynthesis-related functions, which is expected as Chlorobi is one of the few bacteria phyla that possesses photosynthetic ability [22,31,32,34,70]. A number of other proteins exhibit weak sequence similarity to conserved domains in certain other proteins, but considering that the overall sequence similarity is not significant, the actual functions of these proteins could be quite different. Therefore, an important task for the future is to determine the cellular functions of these Bacteroidetes or Chlorobi specific proteins. Likewise, it is also of much interest to determine the functional significance of the conserved indels in SecA, Gyrase B and ClpB proteins that are distinctive characteristics of the Bacteroidetes [30], and of the inserts in DnaE and AlaRS proteins that are specific for the Chlorobi species. The retention of these signature proteins and conserved indels by all species from these groups strongly suggests that they are functionally important for these bacteria. Hence, further studies on these molecular signatures should lead to the discovery of novel biochemical and physiological characteristics that are unique to these bacteria. The primary sequences of many of these genes/proteins that are specific for the Bacteroidetes or Chlorobi species are highly conserved and they provide novel means for identification of both known as well as novel species belonging to these groups by means of PCR-based and immunological methods. Several Bacteroidetes species play central role in the initiation and progression of periodontal diseases in humans [12,18,19,58]. Hence, the proteins that are specific to these bacteria also provide important potential targets for development of therapeutics and vaccines for treatment and prevention of periodontal diseases.
Methods
Identification of Proteins that are Specific for Bacteroidetes and Chlorobi
The blastp searches were carried out on each ORF in the genomes of P. gingivalis W83 [58], B. fragilis YCH46 [35], B. thetaiotaomicron VPI-5482 [15], G. forsetii KT080 [59], Chlorobium (Pelodictyon)luteolum DSM 273 and C. tepidum TLS [24], to identify proteins that are specific for the Bacteroidetes and Chlorobi phyla at different taxonomic levels. The blastp searches were performed against all organisms (i.e. using the NCBI non-redundant (nr) database) with default settings except that the low complexity filter was not used [71]. The proteins that were of interest were those where either all significant hits were from these groups of species or which involved a large increase in E values from the last Bacteroidetes-Chlorobi hit to the first hit from any other organism and the E values for the latter hits in most cases > 10-4, which indicates a weak similarity that could occur by chance. However, higher E values were sometimes acceptable particularly for smaller proteins as the magnitude of the E value depends upon the length of the query sequence. All promising proteins were further analyzed using the position-specific iterated (PSI)-blast program [71]. This program creates a position-specific scoring matrix from statistically significant alignments produced by the blastp program and then searches the database using this matrix. The PSI-blast is more sensitive in identifying weak but biologically relevant sequence similarity as compared to the blastp program [71]. In the present work, a protein was considered to be specific for a given group if all hits producing significant alignments were from that group of species. However, we have also retained a few proteins where 1 or 2 isolated species from other groups of bacteria also had acceptable E values, as they provide possible cases of lateral gene transfer. For all of the Bacteroidetes or Chlorobi-specific proteins identified in the present work, their protein ID's, accession numbers and any information regarding cellular functions (such as COG number or the presence of any conserved domain) are presented here. Preliminary sequence information regarding the presence of a homolog of a query protein in the partially sequenced genomes of P. intermedia, P. ruminicola and F. succinogenes subsp. succinogenes S85 were obtained via genomic blasts against The Institute for Genomic Research database for unfinished microbial genomes [72]. In describing various proteins in the text, "PG," "BF," "BT," "orf", "Plut" and "CT" indicate the identification numbers of the proteins in the genomes of P. gingivalis W83, B. fragilis YCH46, B. thetaiotaomicron VPI-5482,G. forsetii KT080, C. (Pelodictyon) luteolum DSM and C. tepidum TLS, respectively.
Phylogenetic Analysis
The amino acid sequences for the 12 conserved proteins viz. RNA polymerase β subunit, RNA polymerase β ' subunit (RpoC), alanyl-tRNA synthetase (AlaRS), arginyl-tRNA synthetase, phenylalanyl-tRNA synthetase, elongation factor-Tu, elongation factor G, RecA protein, DNA gyrase subunit A, DNA gyrase subunit B, Hsp60 or GroEL protein and DnaK or Hsp70 protein, for different species were downloaded from the NCBI database and aligned using the ClustalX (1.83) program using the default settings [73]. The sequences for two deep-branching species, D. radiodurans and T. aquaticus [27], were included in this dataset for rooting purposes. The sequence alignments for all 12 proteins were concatenated into a single large alignment containing 8899 positions. Poorly aligned regions from this alignment were removed with the Gblocks 0.91b program [74], using the default settings, except that allowable gap position was selected to half. This resulted in a final sequence alignment of 6998 sites, which was used for phylogenetic analyses. A NJ tree based on this alignment (bootstrapped 1000 time) was constructed based on Kimura's model [75] using the TREECON programs [76]. Maximum-likelihood and MP trees were computed using the WAG+F model plus a gamma distribution with four categories [77] using the TREE-PUZZLE [78] and Mega 3.1 program [79], respectively. All trees were bootstrapped 100 times [80], unless otherwise indicated.
The character compatibility analysis on the concatenated alignment was carried out as described recently [57]. Using the program "DUALSITE" [57], all sites in the alignments where only two amino acid states were found, with each state present in at least two species, were selected. All columns with any gaps were omitted. The sites where one of the states is present in only a single species are not useful for compatibility analysis. All useful two state sites were converted into a binary file of "0, 1" characters using the DUALSITE program and this file was used for compatibility analysis [57]. The compatibility analysis was carried out using the CLIQUE program from the PHYLIP (ver. 3.5c) program package [81] to identify the largest clique(s) of compatible characters. The cliques were drawn and the numbers of characters that distinguished different nodes were indicated. The sequence information for G. forsetii, which became available after these analyses were completed [59] is not included in these trees.
Identification of conserved indels
Multiple sequence alignments for large numbers of proteins have been created in our earlier work [82-84]. These alignments were visually inspected to search for any indels in a conserved region that was uniquely present in C. tepidum (the only Chlorobi species present in these groups). The specificity of any potential indel for these groups was evaluated by carrying out by blastp searches on a short segment of the sequence (between 80–120 aa) containing the indel and the flanking conserved regions against the nr database. The purpose of these blast searches was to obtain information from all available homologs to determine the specificities of the indels.
Abbreviations
CD, conserved domain; CFB, Cytophaga-Flavobacteria-Bacteroides; Indel, insert or deletion; ORF, open reading frame; ORFans, open reading frames of unknown functions; AlaRS, alanyl-tRNA synthetase; RGC, rare genetic change; RpoC, RNA polymerase β '-subunit.
Authors' contributions
The initial blastp searches on various genomes were carried out by RSG with the computer assistance provided by Venus Wong. EL analyzed the results of these blast searches to identify various group-specific proteins and confirmed their specificities by means of PSI-blast and genomic blasts. RSG carried out the phylogenetic analyses and identified the conserved indels described here. RSG was also responsible for conceiving and directing this study and for the final evaluation of results. RSG was responsible for the preparation of the final manuscript. All authors have read and approved the submitted manuscript.
Supplementary Material
A conserved indel (3 aa deletion) in ClpB protein that is mainly specific for the Bacteroidales, Flavobacteriales and Flexibacteraceae species. Partial sequence alignment of the ClpB protein containing this indel region is shown. The boxed region is missing in the Bacteroidales, Flavobacteriales and Flexibacteraceae species. Dashes in the alignment show identity with the amino acid on the top line. The ClpB homologs from C. phaebacteroidetes and Methanospirillum hungatei also lack the boxed region, which could be due to LGT. The beta and gamma proteobacteria contain a larger insert in this region, which has likely occurred independently.
Proteins that are specific for the Bacteroides Genus. All significant hits for these proteins are from the following sequenced Bacteroides species unless otherwise indicated: B. thetaiotaomicron VPI-5482, B. fragilis NCTC 9343 and YCH46.
Proteins specific for Flavobacteria that are missing in several species. All significant hits for these proteins are also from Flavobacteriales species. However, unlike the proteins listed in Table 5, these proteins are either present in only a small number of Flavobacteria or are missing from many species.
Chlorobi-specific proteins that are missing in some species. All significant hits for these proteins are also from Chlorobi species. However, unlike the proteins listed in Table 6, these proteins are not present in all Chlorobi species.
Acknowledgments
Acknowledgements
We thank Venus Wong and Yan Li for providing computer support for blast analyses and Beile Gao for help with some phylogenetic analysis. We thank the investigators at US DOE Joint Genome Institute and Gordon and Betty Moore Foundation Marine Biotechnology Initiative for making the genome data for various Bacteroidetes and Chlorobi species available in public databases prior to publication, which were of central importance in these studies. Blast searches against preliminary sequence data for the P. intermedia, P. ruminicola and F. succinogenes were performed at The Institute for Genomic Research website [72]. This work was supported by a research grant from the Canadian Institute of Health Research.
Contributor Information
Radhey S Gupta, Email: gupta@mcmaster.ca.
Emily Lorenzini, Email: lorenze@muss.cis.mcmaster.ca.
References
- Garrity GM, Bell JA, Lilburn TG. The Revised Road Map to the Manual. In: Brenner DJ, Krieg NR and Staley JT, editor. Bergey's Manual of Systematic Bacteriology, Volume 2, Part A, Introductory Essays. New York, Springer; 2005. pp. 159–220. [Google Scholar]
- Woese CR. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W, Klenk HP. Overview: A phylogenetic backbone and taxonomic framework for prokaryotic systamatics. In: Boone DR and Castenholz RW, editor. Bergey's Manual of Systematic Bacteriology. 2nd. Berlin, Springer-Verlag; 2001. pp. 49–65. [Google Scholar]
- Shah HN. The Genus Bacteroides and Related taxa. In: Balows A, Truper HG, Dworkin M, Harder W and Schleifer KH, editor. The Prokaryotes. 2. Vol. 196. New York, Springer-Verlag; 1992. pp. 3593–3607. [Google Scholar]
- Reichenbach H. The Order Cytophagales. In: Balows A, Truper HG, Dworkin M, Harder W and Schleifer KH, editor. The Prokaryotes. 2. Vol. 199. New York, Springer-Verlag; 1992. pp. 3631–3675. [Google Scholar]
- Paster BJ, Dewhirst FE, Olsen I, Fraser GJ. Phylogeny of bacteroides, prevotella, and porphyromonas spp.and related bacteria. J Bacteriol. 1994;176:725–732. doi: 10.1128/jb.176.3.725-732.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdeman LV, Kelley RW, Moore WEC. Family I. Bacteroidaceae Pribam 1933. In: Krieg NR and Holt JG, editor. Bergey's Manual of Systematic Bacteriology. Ist. Baltimore, Williams and Wilkins; 1984. pp. 602–662. [Google Scholar]
- Shah HN, Gharbia SE, Olsen I. Bacteroides, Prevotella, and Porphyromonas. In: Borrelio SP, Murray PR and Funke G, editor. Topley & Wilson's Microbiology and Microbial Infections, 10th. Vol. 80. London, Hodder Arnold; 2005. pp. 1913–1944. [Google Scholar]
- Ohkuma M, Noda S, Hongoh Y, Kudo T. Diverse bacteria related to the bacteroides subgroup of the CFB phylum within the gut symbiotic communities of various termites. Biosci Biotechnol Biochem. 2002;66:78–84. doi: 10.1271/bbb.66.78. [DOI] [PubMed] [Google Scholar]
- O'Sullivan LA, Weightman AJ, Fry JC. New degenerate Cytophaga-Flexibacter-Bacteroides-specific 16S ribosomal DNA-targeted oligonucleotide probes reveal high bacterial diversity in River Taff epilithon. Appl Environ Microbiol. 2002;68:201–210. doi: 10.1128/AEM.68.1.201-210.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anton J, Oren A, Benlloch S, Rodriguez-Valera F, Amann R, Rossello-Mora R. Salinibacter ruber gen. nov., sp. nov., a novel, extremely halophilic member of the Bacteria from saltern crystallizer ponds. Int J Syst Evol Microbiol. 2002;52:485–491. doi: 10.1099/00207713-52-2-485. [DOI] [PubMed] [Google Scholar]
- Duncan MJ. Genomics of oral bacteria. Crit Rev Oral Biol Med. 2003;14:175–187. doi: 10.1177/154411130301400303. [DOI] [PubMed] [Google Scholar]
- Oren A. The Genera Rhodothermus, Thermonema, Hymenobacter and Salinibacter. In: Dworkin M and al , editor. The Prokaryotes: An Evolving Electronic Resource for the Microbiological Community. 3rd, Release 3.3. New York, Springer-Verlag; 2000. [Google Scholar]
- Salyers AA. Bacteroides of the human lower intestinal tract. Annu Rev Microbiol. 1984;38:293–313. doi: 10.1146/annurev.mi.38.100184.001453. [DOI] [PubMed] [Google Scholar]
- Xu J, Bjursell MK, Himrod J, Deng S, Carmichael LK, Chiang HC, Hooper LV, Gordon JI. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science. 2003;299:2074–2076. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- Cerdeno-Tarraga AM, Patrick S, Crossman LC, Blakely G, Abratt V, Lennard N, Poxton I, Duerden B, Harris B, Quail MA, Barron A, Clark L, Corton C, Doggett J, Holden MT, Larke N, Line A, Lord A, Norbertczak H, Ormond D, Price C, Rabbinowitsch E, Woodward J, Barrell B, Parkhill J. Extensive DNA inversions in the B. fragilis genome control variable gene expression. Science. 2005;307:1463–1465. doi: 10.1126/science.1107008. [DOI] [PubMed] [Google Scholar]
- Durmaz B, Dalgalar M, Durmaz R. Prevalence of Enterotoxigenic Bacteroides fragilis in patients with diarrhea: A controlled study. Anaerobe. 2005;11:318–321. doi: 10.1016/j.anaerobe.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Shah HN, Gharbia SE, Duerden BI. Bacteroides, Prevotella and Porphyromonas. In: Balows A and Duerden BI, editor. Topley &Wilson's Microbiology and Microbial Infections vol 2 Systematic Bacteriology. 9th. Vol. 58. London, Arnold; 1998. pp. 1305–1330. [Google Scholar]
- Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, Levanos VA, Sahasrabudhe A, Dewhirst FE. Bacterial diversity in human subgingival plaque. J Bacteriol. 2001;183:3770–3783. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ximenez-Fyvie LA, Haffajee AD, Socransky SS. Microbial composition of supra- and subgingival plaque in subjects with adult periodontitis. J Clin Periodontol. 2000;27:722–732. doi: 10.1034/j.1600-051x.2000.027010722.x. [DOI] [PubMed] [Google Scholar]
- Overmann J, Garcia-Pichel F. The Phototrophic way of Life. In: Dworkin M and al , editor. The Prokaryotes: An Evolving Electronic Resource for the Microbiological Community. 3rd, Release 3.3. New York, Springer-Verlag; 2000. http://141.150.157.117:8080/prokPUB/chaprender/jsp/showchap.jsp?chapnum=239 [Google Scholar]
- Overmann J. The Prokaryotes. 3rd edition 2003. The Family Chlorobiaceae. [Google Scholar]
- Truper HG, Pfennig N. The Family Chlorobiaceae. In: Balows A, Truper HG, Dworkin M, Harder W and Schleifer KH, editor. The Prokaryotes. 2. Vol. 195. New York, Springer-Verlag; 1992. pp. 3583–3592. [Google Scholar]
- Eisen JA, Nelson KE, Paulsen IT, Heidelberg JF, Wu M, Dodson RJ, DeBoy R, Gwinn ML, Nelson WC, Haft DH, Hickey EK, Peterson JD, Durkin AS, Kolonay JL, Yang F, Holt I, Umayam LA, Mason T, Brenner M, Shea TP, Parksey D, Nierman WC, Feldblyum TV, Hansen CL, Craven MB, Radune D, Vamathevan J, Khouri H, White O, Gruber TM, Ketchum KA, Venter JC, Tettelin H, Bryant DA, Fraser CM. The complete genome sequence of Chlorobium tepidum TLS, a photosynthetic, anaerobic, green-sulfur bacterium. Proc Natl Acad Sci U S A. 2002;99:9509–9514. doi: 10.1073/pnas.132181499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frostl JM, Overmann J. Phylogenetic affiliation of the bacteria that constitute phototrophic consortia. Arch Microbiol. 2000;174:50–58. doi: 10.1007/s002030000172. [DOI] [PubMed] [Google Scholar]
- Bryant DA, Frigaard NU. Prokaryotic photosynthesis and phototrophy illuminated. Trends Microbiol. 2006;14:488–496. doi: 10.1016/j.tim.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Olsen GJ, Woese CR, Overbeek R. The winds of (evolutionary) change: breathing new life into microbiology. J Bacteriol. 1994;176:1–6. doi: 10.1128/jb.176.1.1-6.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber TM, Eisen JA, Gish K, Bryant DA. The phylogenetic relationships of Chlorobium tepidum and Chloroflexus auranticus based upon their RecA sequences. FEMS Microbiol Lett. 1998;162:53–60. doi: 10.1111/j.1574-6968.1998.tb12978.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RS, Mukhtar T, Singh B. Evolutionary relationships among photosynthetic prokaryotes (Heliobacterium chlorum, Chloroflexus aurantiacus, cyanobacteria, Chlorobium tepidum and proteobacteria): implications regarding the origin of photosynthesis. Mol Microbiol. 1999;32:893–906. doi: 10.1046/j.1365-2958.1999.01417.x. [DOI] [PubMed] [Google Scholar]
- Gupta RS. The Phylogeny and Signature Sequences characteristics of Fibrobacters, Chlorobi and Bacteroidetes. Crit Rev Microbiol. 2004;30:123–143. doi: 10.1080/10408410490435133. [DOI] [PubMed] [Google Scholar]
- Blankenship RE. Origin and early evolution of photosynthesis. Photosynthesis Research. 1992;33:91–111. [PubMed] [Google Scholar]
- Frigaard NU, Chew AG, Li H, Maresca JA, Bryant DA. Chlorobium tepidum: insights into the structure, physiology, and metabolism of a green sulfur bacterium derived from the complete genome sequence. Photosynth Res. 2003;78:93–117. doi: 10.1023/B:PRES.0000004310.96189.b4. [DOI] [PubMed] [Google Scholar]
- Fenna RE, Matthews BW, Olson JM, Shaw EK. Structure of a bacteriochlorophyll-protein from the green photosynthetic bacterium Chlorobium limicola: crystallographic evidence for a trimer. J Mol Biol. 1974;84:231–240. doi: 10.1016/0022-2836(74)90581-6. [DOI] [PubMed] [Google Scholar]
- Blankenship RE, Olson JM, Miller M. Antenna complexes from green photosynthetic bacateria. In: Blankenship RE, Madigan MT and Bauer CE, editor. Anoxygenic photosynthetic bacteria. Dordrecht, Kluwer; 1995. pp. 399–435. [Google Scholar]
- Kuwahara T, Yamashita A, Hirakawa H, Nakayama H, Toh H, Okada N, Kuhara S, Hattori M, Hayashi T, Ohnishi Y. Genomic analysis of Bacteroides fragilis reveals extensive DNA inversions regulating cell surface adaptation. Proc Natl Acad Sci U S A. 2004;101:14919–14924. doi: 10.1073/pnas.0404172101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongodin EF, Nelson KE, Daugherty S, DeBoy RT, Wister J, Khouri H, Weidman J, Walsh DA, Papke RT, Sanchez PG, Sharma AK, Nesbo CL, MacLeod D, Bapteste E, Doolittle WF, Charlebois RL, Legault B, Rodriguez-Valera F. The genome of Salinibacter ruber: Convergence and gene exchange among hyperhalophilic bacteria and archaea. Proc Natl Acad Sci U S A. 2005;102:18147–18152. doi: 10.1073/pnas.0509073102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kainth P, Gupta RS. Signature Proteins that are Distinctive of Alpha Proteobacteria. BMC Genomics. 2005;6:94. doi: 10.1186/1471-2164-6-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths E, Ventresca MS, Gupta RS. BLAST screening of chlamydial genomes to identify signature proteins that are unique for the Chlamydiales, Chlamydiaceae, Chlamydophila and Chlamydia groups of species. BMC Genomics. 2006;7:14. doi: 10.1186/1471-2164-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao B, Parmanathan R, Gupta RS. Signature proteins that are distinctive characteristics of Actinobacteria and their subgroups. Antonie van Leeuwenhoek. 2006;(In press) doi: 10.1007/s10482-006-9061-2. [DOI] [PubMed] [Google Scholar]
- Gupta RS. Molecular signatures (unique proteins and conserved Indels) that are specific for the epsilon proteobacteria (Campylobacterales) BMC Genomics. 2006;7:167. doi: 10.1186/1471-2164-7-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao B, Gupta RS. Signature Proteins for Archaea and Its Main Subgroups and the Origin of Methanogenesis. BMC Genomics. 2007;8:86. doi: 10.1186/1471-2164-8-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren A. Prokaryote diversity and taxonomy: current status and future challenges. Philos Trans R Soc Lond B Biol Sci. 2004;359:623–638. doi: 10.1098/rstb.2003.1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RS, Griffiths E. Critical Issues in Bacterial Phylogenies. Theor Popul Biol. 2002;61:423–434. doi: 10.1006/tpbi.2002.1589. [DOI] [PubMed] [Google Scholar]
- Ragan MA, Charlebois RL. Distributional profiles of homologous open reading frames among bacterial phyla: implications for vertical and lateral transmission. Int J Syst Evol Microbiol. 2002;52:777–787. doi: 10.1099/00207713-52-3-777. [DOI] [PubMed] [Google Scholar]
- Imhoff JF. Phylogenetic taxonomy of the family Chlorobiaceae on the basis of 16S rRNA and fmo (Fenna-Matthews-Olson protein) gene sequences. Int J Syst Evol Microbiol. 2003;53:941–951. doi: 10.1099/ijs.0.02403-0. [DOI] [PubMed] [Google Scholar]
- Overmann J, Tuschak C. Phylogeny and molecular fingerprinting of green sulfur bacteria. Arch Microbiol. 1997;167:302–309. doi: 10.1007/s002030050448. [DOI] [PubMed] [Google Scholar]
- Alexander B, Andersen JH, Cox RP, Imhoff JF. Phylogeny of green sulfur bacteria on the basis of gene sequences of 16S rRNA and of the Fenna-Matthews-Olson protein. Arch Microbiol. 2002;178:131–140. doi: 10.1007/s00203-002-0432-4. [DOI] [PubMed] [Google Scholar]
- Gruber TM, Bryant DA. Molecular systematic studies of eubacteria, using s70- type sigma factors of group 1 and group 2. J Bacteriol. 1997;179:1734–1747. doi: 10.1128/jb.179.5.1734-1747.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Nakagawa Y, Harayama S, Yamamoto S. Phylogenetic analysis and taxonomic study of marine Cytophaga-like bacteria: proposal for Tenacibaculum gen. nov with Tenacibaculum maritimum comb. nov and Tenacibaculum ovolyticum comb. nov., and description of Tenacibaculum mesophilum sp nov and Tenacibaculum amylolyticum sp nov. Int J Syst Evol Microbiol. 2001;51:1639–1652. doi: 10.1099/00207713-51-5-1639. [DOI] [PubMed] [Google Scholar]
- Rokas A, Williams BL, King N, Carroll SB. Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature. 2003;425:798–804. doi: 10.1038/nature02053. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Inferring phylogenies from protein sequences by parsimony, distance, and likelihood methods. Methods in Enzymology. 1996;266:418-27:418–427. doi: 10.1016/s0076-6879(96)66026-1. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Inferring Phylogenies. Sunderland, Mass., Sinauer Associates, Inc.; 2004. [Google Scholar]
- Le Quesne WJ. The uniquely evolved character concept and its cladistic application. Systematic Zoology. 1975;23:513–517. [Google Scholar]
- Sneath PHA, Sackin MJ, Ambler RP. Detecting evolutionary incompatibilities from protein sequences. Systematic Zoology. 1975;24:311–332. [Google Scholar]
- Estabrook GF, Johnson CSJ, McMorris FR. A mathematical foundation for the analysis of cladistic character compatibility. Mathematical Biosciences. 1976;29:181–187. [Google Scholar]
- Meacham CA, Estabrook GF. Comaptibility methods in Systematics. Ann Rev Ecol Syst. 1985;16:431–446. [Google Scholar]
- Gupta RS, Sneath PHA. Application of the character compatibility approach to generalized molecular sequence data: Branching order of the proteobacterial subdivisions. J Mol Evol. 2006;64:90–100. doi: 10.1007/s00239-006-0082-2. [DOI] [PubMed] [Google Scholar]
- Nelson KE, Fleischmann RD, DeBoy RT, Paulsen IT, Fouts DE, Eisen JA, Daugherty SC, Dodson RJ, Durkin AS, Gwinn M, Haft DH, Kolonay JF, Nelson WC, Mason T, Tallon L, Gray J, Granger D, Tettelin H, Dong H, Galvin JL, Duncan MJ, Dewhirst FE, Fraser CM. Complete genome sequence of the oral pathogenic bacterium Porphyromonas gingivalis strain W83. J Bacteriol. 2003;185:5591–5601. doi: 10.1128/JB.185.18.5591-5601.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer M, Kube M, Teeling H, Richter M, Lombardot T, Allers E, Wurdemann CA, Quast C, Kuhl H, Knaust F, Woebken D, Bischof K, Mussmann M, Choudhuri JV, Meyer F, Reinhardt R, Amann RI, Glockner FO. Whole genome analysis of the marine Bacteroidetes'Gramella forsetii' reveals adaptations to degradation of polymeric organic matter. Environ Microbiol. 2006;8:2201–2213. doi: 10.1111/j.1462-2920.2006.01152.x. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Bryant SH. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2004;32:W327–W331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY, Koonin EV. Who's your neighbor? New computational approaches for functional genomics. Nat Biotechnol. 2000;18:609–613. doi: 10.1038/76443. [DOI] [PubMed] [Google Scholar]
- Danchin A. From protein sequence to function. Curr Opin Struct Biol. 1999;9:363–367. doi: 10.1016/S0959-440X(99)80049-9. [DOI] [PubMed] [Google Scholar]
- Nishikawa K, Yoshimura F, Duncan MJ. A regulation cascade controls expression of Porphyromonas gingivalis fimbriae via the FimR response regulator. Mol Microbiol. 2004;54:546–560. doi: 10.1111/j.1365-2958.2004.04291.x. [DOI] [PubMed] [Google Scholar]
- Gogarten JP, Doolittle WF, Lawrence JG. Prokaryotic evolution in light of gene transfer. Mol Biol Evol. 2002;19:2226–2238. doi: 10.1093/oxfordjournals.molbev.a004046. [DOI] [PubMed] [Google Scholar]
- Gogarten JP, Townsend JP. Horizontal gene transfer, genome innovation and evolution. Nat Rev Microbiol. 2005;3:679–687. doi: 10.1038/nrmicro1204. [DOI] [PubMed] [Google Scholar]
- Bruck I, Goodman MF, O'Donnell M. The essential C family DnaE polymerase is error-prone and efficient at lesion bypass. J Biol Chem. 2003;278:44361–44368. doi: 10.1074/jbc.M308307200. [DOI] [PubMed] [Google Scholar]
- Daubin V, Ochman H. Bacterial genomes as new gene homes: the genealogy of ORFans in E. coli. Genome Res. 2004;14:1036–1042. doi: 10.1101/gr.2231904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beiko RG, Harlow TJ, Ragan MA. Highways of gene sharing in prokaryotes. Proc Natl Acad Sci U S A. 2005;102:14332–14337. doi: 10.1073/pnas.0504068102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths E, Gupta RS. The use of signature sequences in different proteins to determine the relative branching order of bacterial divisions: evidence that Fibrobacter diverged at a similar time to Chlamydia and the Cytophaga- Flavobacterium-Bacteroides division. Microbiology. 2001;147:2611–2622. doi: 10.1099/00221287-147-9-2611. [DOI] [PubMed] [Google Scholar]
- Gupta RS. Evolutionary Relationships Among Photosynthetic Bacteria. Photosynth Res. 2003;76:173–183. doi: 10.1023/A:1024999314839. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein databases search programs. Nucleic Acids Research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TIGR TIGR Unfinished Microbial Genome Database. . 2007. http://www.tigr.org/tdb/ufmg/index.shtml
- Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ. Multiple sequence alignment with Clustal x. Trends Biochem Sci. 1998;23:403–405. doi: 10.1016/s0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Kimura M. The Neutral Theory of Molecular Evolution. Cambridge, Cambridge University Press; 1983. [Google Scholar]
- Van de PY, De Wachter R. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Appl Biosci. 1994;10:569–570. doi: 10.1093/bioinformatics/10.5.569. [DOI] [PubMed] [Google Scholar]
- Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- Schmidt HA, Strimmer K, Vingron M, von Haeseler A. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18:502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits in phylogenies: an approach using the bootstap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP, version 3.5c. Seattle, WA, University of Washington; 1993. [Google Scholar]
- Gupta RS. Protein Phylogenies and Signature Sequences: A Reappraisal of Evolutionary Relationships Among Archaebacteria, Eubacteria, and Eukaryotes. Microbiol Mol Biol Rev. 1998;62:1435–1491. doi: 10.1128/mmbr.62.4.1435-1491.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RS. The phylogeny of Proteobacteria: relationships to other eubacterial phyla and eukaryotes. FEMS Microbiol Rev. 2000;24:367–402. doi: 10.1111/j.1574-6976.2000.tb00547.x. [DOI] [PubMed] [Google Scholar]
- Gupta RS, Pereira M, Chandrasekera C, Johari V. Molecular signatures in protein sequences that are characteristic of Cyanobacteria and plastid homologues. Int J Syst Evol Microbiol. 2003;53:1833–1842. doi: 10.1099/ijs.0.02720-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A conserved indel (3 aa deletion) in ClpB protein that is mainly specific for the Bacteroidales, Flavobacteriales and Flexibacteraceae species. Partial sequence alignment of the ClpB protein containing this indel region is shown. The boxed region is missing in the Bacteroidales, Flavobacteriales and Flexibacteraceae species. Dashes in the alignment show identity with the amino acid on the top line. The ClpB homologs from C. phaebacteroidetes and Methanospirillum hungatei also lack the boxed region, which could be due to LGT. The beta and gamma proteobacteria contain a larger insert in this region, which has likely occurred independently.
Proteins that are specific for the Bacteroides Genus. All significant hits for these proteins are from the following sequenced Bacteroides species unless otherwise indicated: B. thetaiotaomicron VPI-5482, B. fragilis NCTC 9343 and YCH46.
Proteins specific for Flavobacteria that are missing in several species. All significant hits for these proteins are also from Flavobacteriales species. However, unlike the proteins listed in Table 5, these proteins are either present in only a small number of Flavobacteria or are missing from many species.
Chlorobi-specific proteins that are missing in some species. All significant hits for these proteins are also from Chlorobi species. However, unlike the proteins listed in Table 6, these proteins are not present in all Chlorobi species.


