Abstract
Oxygenic photosynthesis is an important bioenergetic process that maintains the Earth's atmosphere and allows carbon fixation. A critical enzyme in this process, the cytochrome b6f complex, differs from other protein complexes of the same family by an unusual covalently attached cofactor chemically defined as a c′ heme. We have identified a set of pioneer proteins that carry the biogenesis of this c′ heme and started their characterization. They are encoded by the genomes of all organisms performing oxygenic photosynthesis, whatever their phylogenetic distances. These proteins are thus among the few that distinguish photosynthetic cells evolving oxygen from other types of living cells.
Keywords: biogenesis, heme, system IV, chloroplast, Chlamydomonas
Two major enzymes of the energy-transducing membrane, ubihydroquinone:cytochrome c oxidoreductases (bc1 complexes) and plastohydroquinone:plastocyanin oxidoreductase (b6f complexes), are closely related both in their function and structural organization. They transfer electrons from a lipid-soluble transporter to a water-soluble protein transporter while building a transmembrane proton gradient. The protein moiety of these enzymes is rather well conserved, from bacteria to mitochondria and chloroplasts, particularly for the cytochrome b domain. Major progress of our understanding of the structural organization of the hemes and Fe–S center within these energy-transducing enzymes was made a few years ago with the resolution of the crystal structure of bc1 complexes from mitochondrial respiratory chains (1, 2) and the homologous b6f complexes, active in oxygenic photosynthesis (3, 4). A striking feature of the b6f complex that differentiates it from the bc1 complex is the presence of an extra c heme bound to the protein backbone of cytochrome b6.
The c-type hemes are usually covalently attached to the protein via two thioether bonds between the heme vinyl groups and the thiols of two cysteine residues in a conserved CXXCH motif. Usually the histidine residue from this motif provides one of the axial ligands to heme iron. The extra c heme found in b6f complexes shows unusual binding and iron-coordination features. It is a high-spin c′ heme with a pentacoordinated central iron that lacks amino acid axial ligands but has instead a unique axial ligand, either hydroxyl or water (3–5). Furthermore, it is attached by a unique thioether bond to cytochrome b6 near the quinone-binding site Qi on the negative side (n side) of the membrane. It is hereafter referred to as the ci′ heme.
Covalent binding of a c heme to the apocytochrome backbone comprises several critical steps that include reduction of the cysteine ligands, a ferrous heme supply, and heme binding. Heme (iron protoporphyrin IX), being a hydrophobic and cytotoxic macrocycle, requires specific pathways to be safely delivered to its subcellular destinations; up to now three distinct multifactorial cytochrome c biogenesis pathways have been described (6). The common feature of these three pathways is that they operate on the positive side (p side) of membranes, opposite to the side where membrane insertion of the protein backbone occurs.
Here we report the identification of four gene products that functionally complement the ccb mutants from the unicellular green alga Chlamydomonas reinhardtii that were previously identified as specifically altered in cytochrome b6 maturation (7). We first show that the CCB loci control the apocytochrome to holocytochrome b6 conversion and then describe the major protein components of this unique biogenesis pathway for c-type heme binding on the n side of the membranes. The four components correspond to proteins whose function was unknown up to now. Together, they represent one of the genuine genetic signatures of living organisms that perform oxygenic photosynthesis.
Results
Identification of Four CCB Factors.
We identified the four nuclear CCB loci by complementation of the ccb mutants with an indexed library and selection for restoration of phototrophy (see Materials and Methods).
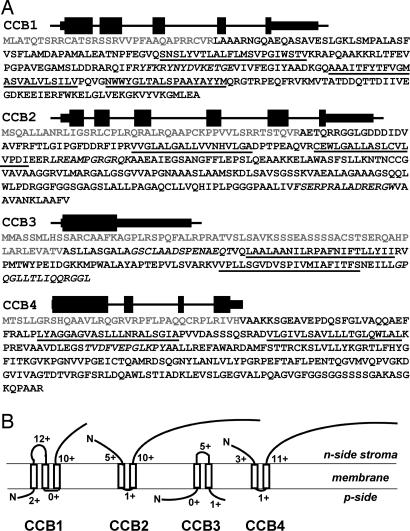
The CCB1 gene belongs to linkage group XVIII (http://genome.jgi-psf.org/Chlre3), comprises five exons, and encodes a protein of 269 residues (Fig. 1A). Taking into account the prediction of transmembrane domains made by using TMAP (8) and the prediction of the localization of positively charged segments by the “positive inside rule” (9), we conclude that CCB1 has three transmembrane domains, with its C terminus in the stroma (Fig. 1B). The ccb1-1 substitution mutant has a stop codon in the second exon [see supporting information (SI) Fig. 5].
Fig. 1.
Molecular identification of CCB factors. (A) CCB gene models and CCB protein sequences. Exons are shown by thick lines, chloroplast transit peptides as predicted by ChloroP are shown in gray, predicted transmembrane domains from sequences aligned by TMAP are underlined, and sequences of anti-peptides are shown in italic. (B) Topology of the CCB chloroplast membrane protein apparatus as predicted by TMAP and the positive inside rule. The numbers of positively charged residues are indicated for predicted mature proteins.
The CCB2 gene belongs to the XII/XIII linkage group, comprises six exons, and encodes a protein of 310 residues (Fig. 1A) that has two transmembrane domains at the N terminus and a long C-terminal part in the stroma (Fig. 1B). The ccb2-1 substitution mutant has a stop codon in the fourth exon (SI Fig. 5).
The CCB3 gene belongs to linkage group I, has no introns, and encodes a protein of 193 residues (Fig. 1A). CCB3 has a predicted horseshoe transmembrane configuration with N and C termini in the lumen (Fig. 1B), similar to some polypeptides shown to insert “spontaneously” in the thylakoid membrane (10, 11). The ccb3-1 substitution mutant has a stop codon in the middle of the coding sequence (SI Fig. 5).
The CCB4 gene belongs to linkage group VIII, comprises four exons, and encodes a protein of 306 residues (Fig. 1A). CCB4 is predicted to have the same topology as CCB2 (Fig. 1B). As discussed below, CCB4 is a paralogue of CCB2. The ccb4-1 mutant bears three substitutions, the third one modifying the second exon exit site and generating a stop codon in the third exon (SI Fig. 5).
The CCB System Is Required for Covalent Binding of Heme ci′ to Cytochrome b6.
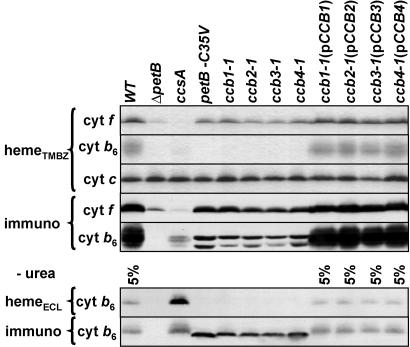
The signature of apocytochrome b6 impaired in heme ci′ binding has already been described for the petB-C35V mutant (5) where the unique covalent cysteine ligand to heme ci′ was substituted. It consists of a double band on urea SDS/polyacrylamide gels or a single band of lower apparent molecular mass than that in the WT on SDS/polyacrylamide gels concomitant with the lack of peroxidase activity (see Fig. 2). The same signature is observed in all ccb mutants, indicating that the binding of heme ci′ is prevented in these mutants. WT behavior is restored upon complementation with the appropriate cDNAs in the ccb(pCCB) transformants.
Fig. 2.
Biochemical study of heme-binding mutants. Detection of heme peroxidase activity by using tetramethylbenzidine (hemeTMBZ) or chemiluminescence (hemeECL) and immunodetection (immuno) of cytochrome (cyt) f and cytochrome b6 in cells of the following strains: WT, cytochrome b6 deletion mutant ΔpetB, system II maturation mutant ccsA, cytochrome b6 substitution mutant petB-C35V, ccb mutants (ccb1-1, ccb2-1, ccb3-1, and ccb4-1), and complemented transformants [ccb1-1(pCCB1), ccb2-1(pCCB2), ccb3-1(pCCB3), ccb4-1(pCCB4)]. Polypeptides were separated on SDS/polyacrylamide gels with urea except when indicated (−urea). Loading was 100% (see cyt c) except when indicated to be 5% (in which case samples were mixed with 95% ΔpetB).
Because the biogenesis of chloroplast c-type cytochromes such as cytochrome f is known to involve maturation system II, the peroxidase activity detected for cytochrome f in the ccb mutants allows us to reject the hypothesis that this pathway was altered in these mutants. Moreover, maturation system II mutants such as the ccsA mutant (12) shown in Fig. 2 are not affected in heme ci′ binding.
Thus, the four CCB factors are specifically required for covalent binding of heme ci′ to cytochrome b6 and define an additional maturation system for c-type cytochromes.
CCBs Subcellular Localization.
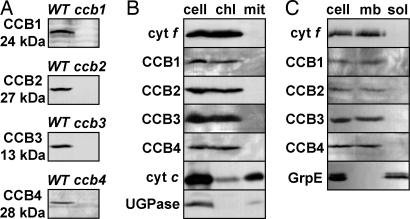
Using polyclonal antibodies raised against the four CCB factors, we determined their subcellular localization. The mature CCB1 (24 kDa), CCB2 (27 kDa), CCB3 (13 kDa), and CCB4 (28 kDa) proteins are found in the WT strain and are absent in the corresponding ccb mutant (Fig. 3A). Their apparent molecular masses are consistent with those expected from their protein sequences. Chloroplast preparations show some contamination by mitochondria (see the mitochondrial protein cytochrome c shown as control in Fig. 3B). Pure mitochondrial fractions can be obtained with no chloroplast contamination (see the chloroplast protein cytochrome f shown as control in Fig. 3B). The mature CCBs are present in chloroplasts and absent in mitochondria (Fig. 3B). The good separation of the membrane and the soluble fraction is illustrated by two protein controls, the transmembrane thylakoid protein cytochrome f and the soluble chloroplast protein GrpE (Fig. 3C). The four mature CCBs are associated with the chloroplast membrane fraction (Fig. 3C).
Fig. 3.
Biochemical study of CCB factors. (A) CCB immunodetection with CCB anti-peptide antibodies in cells of WT and ccb (as given in Fig. 2) strains. (B) Presence of CCBs in chloroplasts (chl) and absence of CCBs in mitochondria (mit). (C) CCBs are membrane proteins (mb). Markers were as follows: cytochrome f (cyt f), chloroplast and membrane; holocytochrome c (cyt c), mitochondria; UDP-glucose pyrophosphorylase (UGPase), cytoplasm; and GrpE, soluble (sol). Polypeptides were separated on SDS/polyacrylamide gels with urea.
Discussion
A Specific and Conserved Pathway for Covalent Heme Binding in Thylakoid Membranes.
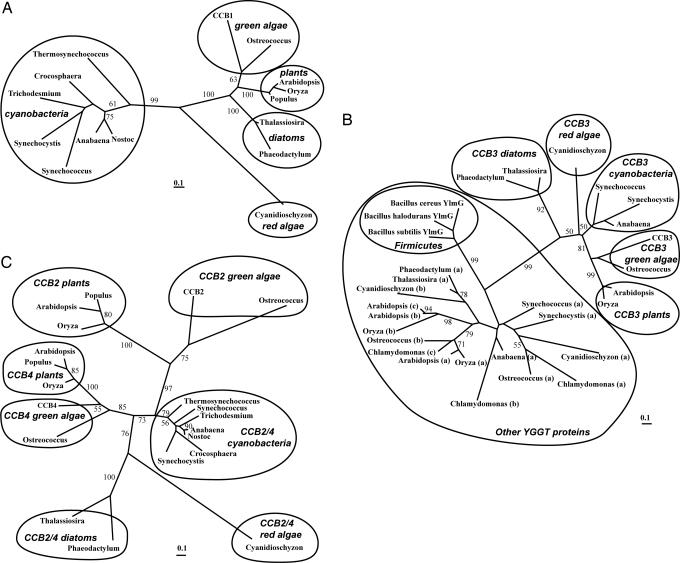
The four proteins identified here are conserved among all organisms performing oxygenic photosynthesis whose genome sequences are currently available. (See phylogenetic trees in Fig. 4 and alignments in SI Fig. 6. Accession numbers of the sequences are provided in SI Table 1.) None of these proteins had been previously assigned any physiological function. CCB1 is ubiquitous among organisms performing oxygenic photosynthesis, where it is present as a unique orthologue (Fig. 4A). CCB3 belongs to the large YGGT protein family (European Molecular Biology Laboratory InterPro accession no. IPR003425), which is found in both plastids and bacteria. This family of integral membrane proteins of unknown function was named after the Escherichia coli yggT gene, and its members are characterized by a conserved YGGT motif. Except for one YGGT member suggested by one study in Streptococcus to be involved in some cell division processes (13), CCB3 is the first protein of this family that has a clearly assigned function. Whereas bacteria contain one YGGT member (two in cyanobacteria), photosynthetic eukaryotes contain up to four, all predicted to be chloroplast-localized. Sequence alignments of the whole family allowed us to generate a phylogenetic tree (Fig. 4B) in which single members of each photosynthetic organism segregate in a cluster with C. reinhardtii CCB3, clearly defining its functional orthologues.
Fig. 4.
Unrooted maximum likelihood phylogenetic trees. The scale bar represents the number of changes per sites. Bootstrap values of ≥50% are indicated. Accession numbers of the sequences are given in SI Table 1. (A) CCB1 phylogeny. (B) CCB3 and other YGGT phylogeny from cyanobacteria, firmicutes, and photosynthetic eukaryotes. Letters in parentheses used to name non-CCB3 YGGT proteins are arbitrary. (C) CCB2 and CCB4 phylogeny.
CCB2 and CCB4 are paralogues with a low similarity level (SI Fig. 6C). They derive from a unique cyanobacterial ancestor, as shown in Fig. 4C, which suggests that they probably act as a heterodimer in the chloroplast (14). CCB4 is closer to the cyanobacterial protein than CCB2. The branch distances of the CCB2 cluster indicate that this gene is less constrained than CCB4. The duplication probably occurred after endosymbiosis between a cyanobacterium and a phagotrophic eukaryote. Red algae chloroplasts derived from the same endosymbiosis event, as in the green lineage, whereas diatoms arose by a secondary endosymbiosis when a heterotrophic eukaryote engulfed a red alga. The analysis of the scarce genomic data from a red alga and two diatoms identifies unique proteins with low similarities to cyanobacterial CCB2/CCB4 (SI Fig. 6F). The position of these orthologues in the tree shown in Fig. 4C, between the CCB2 and CCB4 branches, suggests that gene duplication occurred before the separation of the green and red lineages. The single orthologue found in red alga and diatoms suggests that one of the duplicated copies of the ancestral CCB2/CCB4 gene was lost or became too divergent to be identified in these lineages.
Diversity of Cytochrome c Maturation Systems.
Three distinct systems, system I (Ccm), system II (CCS/Res), and system III (heme lyase), have evolved to attach heme c by two thioether bonds on a CXXCH motif in different organisms and compartments on the p side of membranes (6). Mitochondrial cytochromes c and c1 of euglenids and trypanosomatids contain a single thioether bond on the heme-binding motif XXXCH. Their maturation probably occurs on the p side of membranes by a different biogenesis pathway, because none of the three previously known c-type maturation systems (15) or system CCB (this work) is encoded within the genome of trypanosomatids.
Interestingly, the presence of CCB orthologues is restricted to organisms performing oxygenic photosynthesis. No orthologues of CCB1, CCB2, or CCB4 are found in Bacillus subtilis, although it has an atypical respiratory bc complex that also contains a heme covalently bound to its cytochrome b6 subunit on the n side of the membrane (16). YGGT family member YlmG is present, but it clusters with the bulk of YGGT proteins, away from the CCB3 branch (Fig. 4B). YlmG belongs to a gene cluster encoding four hypothetical proteins of unknown function; the three other genes (ylmD, ylmE, and ylmF) show no strong similarity to known cytochrome c maturation factors. Whether the ylmD–ylmG cluster encodes a cytochrome c maturation system remains to be demonstrated. The identification of a distinct system in B. subtilis would establish a diversity of cytochrome c maturation systems on the n side of membranes that would be reminiscent of the wide diversity of maturation systems that exists on the p side of membranes.
CCBs in Heme Delivery.
The chloroplast membrane localization of CCB factors is consistent with their role in heme binding to cytochrome b6 on the stromal side of the membranes. Interestingly, most of the highly conserved residues are found in the region of the CCB proteins predicted to be oriented toward the n side of the membrane. The opposite is observed for proteins belonging to systems I and II. In these p side, cytochrome c maturation systems, conserved tryptophan and tyrosine residues, which have been identified as critical in heme interactions (6, 17), are all located in protein domains next to the p side of the membrane. Because the four CCB proteins contain conserved tryptophan and/or tyrosine residues next to the n side of the thylakoid membranes, they could participate in heme chaperoning and delivery to apocytochrome b6. Based on the remarkably high sequence conservation of CCB3 with a typical WYP motif on its stromal side, it is tempting to suggest that this factor would play a role in the catalysis of thioether bond formation. However, any reliable functional assignment will require further site-directed mutagenesis studies to produce mutants altered in ci′ heme binding that still accumulate mutated versions of each CCB.
In contrast to the other cytochrome c maturation systems, in which cysteine residues have been identified as being critical in apocytochrome and heme reduction (6), no conserved cysteine residues were found in any of the CCBs. There are several possible explanations for this finding. It may reflect the higher reducing potential of the stroma and cyanobacterial cytoplasm versus that of the lumen or periplasm, the particular heme-binding process by a unique thioether bond, the easier access to ferrous heme produced by the neighboring ferrochelatase, or merely the existence of an additional protein factor that has escaped our genetic screen.
The requirement for this additional heme-binding machinery that we propose to call system IV indicates a central role for the ci′ heme bound to cytochrome b6 in oxygenic photosynthesis. Because this heme is located close to the reducing side of the photosynthetic membranes, its importance should most likely be found in the need for a combination of cyclic and linear electron flows to achieve the ATP/NADPH ratios required for carbon fixation at the expense of water as the ultimate source of electrons. The functional characterization of this ci′ heme will certainly shed light on this key issue in the near future.
Materials and Methods
Strains and Culture Conditions.
C. reinhardtii triple mutant strains ccb:arg7 (arginine auxotrophic):cw15 (cell-wall deficient) were isolated by crosses, and complemented transformants ccb(pCCB) were generated during this work. The other mutant strains of C. reinhardtii have been described previously: ΔpetB (18), ccsA-B6 (12), ccb mutants (7), and petB-C35V (5). Cells were grown at 25°C in Tris acetate phosphate (TAP) medium, pH 7.2, under dim light [6 microeinsteins (μE)·m−2·s−1] and collected during the exponential phase at 2 × 106 cells per ml. Arginine auxotrophic strains were grown in TAP supplemented with 100 mg/liter arginine.
Complementation of ccb Strains and Sequencing of ccb Alleles.
An indexed cosmid library with the ARG7 gene included in the cosmid vector (19) was used to transform ccb:cw15:arg7 mutants by electroporation (20) after optimization of procedures (R.K., unpublished work). Transformation efficiency was measured after a selection for arginine autotrophy. Phototrophic colonies were selected on minimal medium under light intensities of 40–100 μE·m−2·s−1 and became visible after ≈2 weeks. Pools of cosmids that yielded phototrophic transformants were analyzed to determine the complementing cosmids. Borders of cosmids were sequenced, allowing us to pinpoint ≈40 kb regions in the nuclear genome (http://genome.jgi-psf.org/Chlre3). Subclones were tested to identify the complementing genes. cDNAs were obtained to verify the splicing models. Each cDNA could complement the corresponding ccb mutant. Plasmids pCCB were generated by cloning in pBlueScript II KS(−) vector the WT CCB cDNA coding region with the corresponding 5′ promoter region and 3′ UTR. They were then used to complement ccb mutant strains. The cDNA of CCB1 was obtained from EST BP092753 (GenBank accession no. BP092753; Kazusa DNA Research Institute, Kisarazu, Japan), the cDNA of CCB2 was amplified from a cDNA library (21), the cDNA of CCB3 was obtained from EST AV626941 (GenBank accession no. AV626941; Kazusa DNA Research Institute), and the cDNA of CCB4 was obtained by RT-PCR. C. reinhardtii genomic DNA was isolated (22) from ccb strains, and amplification products covering the entire coding regions were sequenced to identify ccb allele mutations.
Protein Preparation and Analysis.
Cell preparation and fractionation of membranes and soluble proteins were performed at 4°C. Liquid culture (200 ml at 2 × 106 cells per ml) was collected by centrifugation and resuspended in a 1-ml final volume of 5 mM Hepes-KOH and 10 mM EDTA at pH 7.5 supplemented with protease inhibitors (Roche Diagnostics, Basel, Switzerland; global materials no. 11873580001; 200 μM phenylmethylsulfonyl fluoride, 1 mM benzamidine, and 5 mM ε-aminocaproic acid). DTT and Na2CO3 were added at a final concentration of 0.1 M to 200 μl of the cell suspension, and the rest of the cell suspension (800 μl) was sonicated twice for 10 sec on ice and centrifuged for 10 min in a Beckman Coulter (Fullerton, CA) TLA-100.3 rotor at 270,000 × g. DTT and Na2CO3 were added at a final concentration of 0.1 M to the supernatant containing soluble proteins. To eliminate residual soluble proteins, the membrane pellet was resuspended in 800 μl of the original buffer, sonicated, and centrifuged as described above; the resulting pellet of membrane proteins was resuspended in 800 μl of 0.1 M DTT and 0.1 M Na2CO3 in the presence of protease inhibitors. Before electrophoresis, samples were solubilized in 2% (mass/vol) SDS and 6% (mass/vol) sucrose, boiled for 50 sec, and centrifuged for 5 min at 15,000 × g. Equal volumes of cells and membrane and soluble proteins were loaded on the gel. The cell wall-deficient cw15 cells were broken with a nebulizer to isolate chloroplasts (23) and mitochondria (24). Polypeptides were separated on 12–18% SDS/polyacrylamide gels (22) in the presence or absence of 8 M urea. Proteins were electrotransferred onto Immobilon-NC membranes in a semidry blotting apparatus at 0.8 mA·cm−2 for 45 min. Heme peroxidase activity on gels was detected by using 3,3′,5,5′-tetramethylbenzidine (22) or on blots by using chemiluminescence (25). Immunodetection used antisera raised against C. reinhardtii cytochrome f, cytochrome b6, and chaperone GrpE at a 1:10,000 dilution and against barley UDP-glucose pyrophosphorylase (AgriSera, Vännäs, Sweden; catalog no. AS05 086) at a 1:1,000 dilution. Immunolabeled bands were visualized by the chemiluminescence method (Amersham Biosciences, Piscataway, NJ; product code RPN2106). Rabbit anti-CCB peptides (Fig. 1A) were produced by Eurogentec (Seraing, Belgium) and used at dilutions of 1:100 for α-CCB1, 1:400 for α-CCB2, 1:2,500 for α-CCB3, and 1:400 for α-CCB4 (Fig. 3).
Analytical Software and Phylogeny.
Predictions for chloroplast targeting and transit peptide cleavage sites were obtained by using the ChloroP 1.1 server (26). The transmembrane domains were predicted from multiple sequence alignments by using the TMAP transmembrane prediction program (8). Multiple sequence alignments were performed by using ClustalW version 1.83 (27) with default parameters and manual editing. Maximum likelihood trees were generated by using PhyML version 2.4.4 (28) with 1,000 bootstrap replicates, the Jones–Taylor–Thornton (JTT) model, estimation of proportion of invariable sites, and four substitution rate categories with estimated γ distribution parameter.
Supplementary Material
Acknowledgments
We thank S. Bujaldon and J. Girard-Bascou for assistance in the isolation of the ccb:arg7:cw15 mutants, J.-D. Rochaix (University of Geneva, Geneva, Switzerland) for the generous gift of an indexed cosmid library, Kazusa DNA Research Institute (Kisarazu, Japan) for EST clones, A. Atteia (Centre National de la Recherche Scientifique/Commissariat à l'Energie Atomique/Institut National de la Recherche Agronomique, Université Joseph Fourier, Grenoble) for expertise in mitochondria purification, S. Gribaldo (Institut Pasteur, Paris, France) for advice in phylogeny, and P. Cardol and M. Guergova-Kuras for critical reading of the manuscript. This work was supported by Unité Mixte de Recherche 7141, Centre National de la Recherche Scientifique/Université Pierre et Marie Curie (Paris 6). D.S.-M. was supported by a fellowship from the Ministère délégué de l'Enseignement Supérieur et à la Recherche.
Abbreviations
- n side
negative side
- p side
positive side.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. EF190472–EF190475).
This article contains supporting information online at www.pnas.org/cgi/content/full/0702340104/DC1.
References
- 1.Xia D, Yu CA, Kim H, Xia ZZ, Kachurin AM, Zhang L, Yu L, Deisenhofer J. Science. 1997;277:60–66. doi: 10.1126/science.277.5322.60. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Z, Huang L, Shulmeister VM, Chi YI, Kim KK, Hung LW, Crofts AR, Berry EA, Kim SH. Nature. 1998;392:677–684. doi: 10.1038/33612. [DOI] [PubMed] [Google Scholar]
- 3.Stroebel D, Choquet Y, Popot J-L, Picot D. Nature. 2003;426:413–418. doi: 10.1038/nature02155. [DOI] [PubMed] [Google Scholar]
- 4.Kurisu G, Zhang H, Smith JL, Cramer WA. Science. 2003;302:1009–1014. doi: 10.1126/science.1090165. [DOI] [PubMed] [Google Scholar]
- 5.de Vitry C, Desbois A, Redeker V, Zito F, Wollman F-A. Biochemistry. 2004;43:3956–3968. doi: 10.1021/bi036093p. [DOI] [PubMed] [Google Scholar]
- 6.Kranz R, Lill R, Goldman B, Bonnard G, Merchant S. Mol Microbiol. 1998;29:383–396. doi: 10.1046/j.1365-2958.1998.00869.x. [DOI] [PubMed] [Google Scholar]
- 7.Kuras R, de Vitry C, Choquet Y, Girard-Bascou J, Culler D, Büschlen S, Merchant S, Wollman F-A. J Biol Chem. 1997;272:32427–32435. doi: 10.1074/jbc.272.51.32427. [DOI] [PubMed] [Google Scholar]
- 8.Persson B, Argos P. J Mol Biol. 1994;237:182–192. doi: 10.1006/jmbi.1994.1220. [DOI] [PubMed] [Google Scholar]
- 9.Gavel Y, Steppuhn J, Herrmann R, von Heijne G. FEBS Lett. 1991;282:41–46. doi: 10.1016/0014-5793(91)80440-e. [DOI] [PubMed] [Google Scholar]
- 10.Mant A, Woolhead CA, Moore M, Henry R, Robinson C. J Biol Chem. 2001;276:36200–36206. doi: 10.1074/jbc.M102914200. [DOI] [PubMed] [Google Scholar]
- 11.Zygadlo A, Robinson C, Scheller HV, Mant A, Jensen PE. J Biol Chem. 2006;281:10548–10554. doi: 10.1074/jbc.M512687200. [DOI] [PubMed] [Google Scholar]
- 12.Xie Z, Merchant S. J Biol Chem. 1996;271:4632–4639. doi: 10.1074/jbc.271.9.4632. [DOI] [PubMed] [Google Scholar]
- 13.Fadda D, Pischedda C, Caldara F, Whalen MB, Anderluzzi D, Domenici E, Massida O. J Bacteriol. 2003;185:6209–6214. doi: 10.1128/JB.185.20.6209-6214.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsukatani Y, Miyamoto R, Itoh S, Oh-Oka H. J Biol Chem. 2004;279:51122–51130. doi: 10.1074/jbc.M410432200. [DOI] [PubMed] [Google Scholar]
- 15.Allen JWA, Ginger ML, Ferguson SJ. Biochem J. 2004;383:537–542. doi: 10.1042/BJ20040832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu J, Le Brun N. J Biol Chem. 1998;273:8860–8866. doi: 10.1074/jbc.273.15.8860. [DOI] [PubMed] [Google Scholar]
- 17.Uchida T, Stevens JM, Daltrop O, Harvat EM, Hong L, Ferguson SJ, Kitagawa T. J Biol Chem. 2004;279:51981–51988. doi: 10.1074/jbc.M408963200. [DOI] [PubMed] [Google Scholar]
- 18.Kuras R, Wollman F-A. EMBO J. 1994;13:1019–1027. doi: 10.1002/j.1460-2075.1994.tb06350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Depège N, Bellafiore S, Rochaix J-D. Science. 2003;299:1572–1575. doi: 10.1126/science.1081397. [DOI] [PubMed] [Google Scholar]
- 20.Shimogawara K, Fujiwara S, Grossman A, Usuda H. Genetics. 1998;148:1821–1828. doi: 10.1093/genetics/148.4.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Atteia A, Franzén L-G. Eur J Biochem. 1996;237:792–799. doi: 10.1111/j.1432-1033.1996.0792p.x. [DOI] [PubMed] [Google Scholar]
- 22.de Vitry C, Finazzi G, Baymann F, Kallas T. Plant Cell. 1999;11:2031–2044. doi: 10.1105/tpc.11.10.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schroda M, Vallon O, Whitelegge JP, Beck CF, Wollman F-A. Plant Cell. 2001;13:2823–2839. doi: 10.1105/tpc.010202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Erickson M, Gardelstöm P, Samuelsson G. Plant Physiol. 1995;107:479–483. doi: 10.1104/pp.107.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feissner R, Xiang Y, Kranz RG. Anal Biochem. 2003;315:90–94. doi: 10.1016/s0003-2697(02)00658-9. [DOI] [PubMed] [Google Scholar]
- 26.Emanuelsson O, Nielsen H, von Heijne G. Protein Sci. 1999;8:978–984. doi: 10.1110/ps.8.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thompson JD, Higgins DG, Gibson TJ. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guindon S, Gascuel O. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.