Abstract
The absence of Tsa1, a key peroxiredoxin that functions to scavenge H2O2 in Saccharomyces cerevisiae, causes the accumulation of a broad spectrum of mutations including gross chromosomal rearrangements (GCRs). Deletion of TSA1 also causes synthetic lethality in combination with mutations in RAD6 and several key genes involved in DNA double-strand break repair. In the present study we investigated the causes of GCRs and cell death in these mutants. tsa1-associated GCRs were independent of the activity of the translesion DNA polymerases ζ, η, and Rev1. Anaerobic growth reduced substantially GCR rates of WT and tsa1 mutants and restored the viability of tsa1 rad6, tsa1 rad51, and tsa1 mre11 double mutants. Anaerobic growth also reduced the GCR rate of rad27, pif1, and rad52 mutants, indicating a role of reactive oxygen species in GCR formation in these mutants. In addition, deletion of TSA1 or H2O2 treatment of WT cells resulted in increased formation of Rad52 foci, sites of repair of multiple DNA lesions. H2O2 treatment also induced the GCRs. Our results provide in vivo evidence that oxygen metabolism and reactive oxygen species are important sources of DNA damages that can lead to GCRs and lethal effects in S. cerevisiae.
Keywords: dsDNA break, gross chromosomal rearrangement, translesion DNA synthesis, peroxiredoxin
Maintaining genome stability is crucial for cell growth and cell survival. Different genetic disorders, including most human cancers, are associated with different forms of genome instability (1–3). One type of genomic instability observed frequently in many cancers is gross chromosomal rearrangements (GCRs), such as translocations, deletions of chromosome arms, interstitial deletions, inversions, amplifications, chromosome fusions, and aneuploidy. Multiple genes and pathways that suppress GCRs have been characterized by using the yeast Saccharomyces cerevisiae as a model system (4–6). These include genes involved in DNA replication, recombination, and repair, cell-cycle checkpoints that function during DNA replication and repair, and pathways involved in telomere maintenance, chromatin assembly, and detoxification of reactive oxygen species (ROS) (refs. 7 and 8 and references therein). The importance of these pathways for suppressing genome instability is further emphasized by their roles in preventing the development of human cancer (7, 9, 10).
Although many pathways that function in the suppression of GCRs have been discovered, little is known about the causes and origin of GCRs. It is generally thought that a major cause of DNA damage that leads to mutations is ROS, which are generated as a normal part of oxygen metabolism but are also produced by ionizing radiation, metabolism of exogenous compounds, and pathological processes such as infection and inflammation (11, 12). ROS, such as the superoxide radical, H2O2, and the hydroxyl radical, can attack almost all cell components and can induce many types of DNA damage, including single-stranded DNA breaks and dsDNA breaks (DSB), base and sugar modifications, and DNA–protein cross-links (13). These lesions can exert a number of deleterious effects including the induction of mutations, aging, and cell death. Given the broad range of ROS sources and the highly reactive nature of these species, it is not surprising that ROS have been implicated in a number of disease states (11, 14).
Aerobic organisms have a wide array of mechanisms to prevent the negative effects of ROS. Peroxiredoxins, previously termed the thioredoxin peroxidases, have received considerable attention in recent years as a new and expanding family of thiol-specific antioxidant proteins (15, 16). Peroxiredoxins are abundant, ubiquitously distributed peroxidases that use cysteine as the primary site of oxidation during the reduction of peroxides. Many organisms have more than one peroxiredoxin, including five peroxiredoxins identified in S. cerevisiae and six in human cells. Among the five peroxiredoxins in S. cerevisiae, Tsa1 possesses the most potent ability to scavenge H2O2 (17). Tsa1 is also the most significant contributor to genome stability and prevents a broad spectrum of mutations including GCRs (18, 19). Tsa1 is essential for cell survival in rad6 or recombination-defective mutants and results in slow growth when combined with mutations that cause defects in the DNA damage and replication checkpoints, sister chromatid cohesion, and postreplication repair (20). Interestingly, mice lacking PrdxI, a homologue of Tsa1, have increased erythrocyte ROS and increased risk of malignant cancers (21).
In the present study we investigated the causes of GCRs and cell death in tsa1 and other mutants with genome instability phenotypes. Our results provide in vivo evidence that the oxygen metabolism and ROS are important sources of endogenous DNA damages that lead to spontaneous GCR and lethal effects in S. cerevisiae.
Results
tsa1-Associated GCRs Are Independent of the Translesion DNA Polymerases.
Translesion DNA synthesis (TLS) involves DNA polymerase ζ, whose two subunits are encoded by REV3 and REV7, DNA polymerase η, encoded by RAD30, and the Rev1 deoxycytidyl transferase, all of which function to bypass different DNA replication blocking lesions and in some cases contribute to mutagenesis (22). To determine whether these TLS polymerases play a role in tsa1-associated mutator phenotypes, we analyzed the dependence of canavanine-resistant mutations (Canr) and GCRs [canavanine- and 5-fluoroorotic acid-resistant (Canr-5FOAr) mutations] in a tsa1 mutant on Rev3, Rev1, and Rad30 activity (Table 1). As expected, deletion of REV3 or REV1 reduced the Canr rate of a WT, whereas the Canr rate of the rad30 mutant was similar to that of the WT strain (23, 24). The GCR rates of these single mutants were similar to that of a WT strain (Table 1). The Canr rate of a tsa1 rev3 mutant was reduced compared with the tsa1 mutant but was higher than that of WT, whereas the GCR rate was similar to that of a tsa1 mutant. These observations indicate that the Canr mutator phenotype of the tsa1 mutant is partially Rev3-dependent, whereas the GCR phenotype appears to be Rev3-independent. The role of Rev1 in tsa1-associated phenotypes was similar to that found for Rev3. The tsa1 rad30 mutant displayed a small, but significant, decrease in the Canr rate compared with the tsa1 mutant, suggesting that Rad30 plays a minor role in the mutations that arise in the tsa1 mutant. Again, the GCR rate of the tsa1 rad30 mutant was similar to that of the tsa1 mutant. Altogether, these results indicate that TLS mediated by DNA polymerases ζ, η, and Rev1 contributes to the Canr mutator phenotype observed in tsa1 mutants but does not contribute substantially to tsa1-associated GCRs.
Table 1.
Translesion DNA polymerases and mutation formation
| Strain | Canr rate (× 10−7) | GCR rate (× 10−10) |
|---|---|---|
| WT | 2.6 (2.2–3.0) | 2.6 (2.1–9.5) |
| tsa1 | 25.6 (24.1–28.8) | 55.6 (36.9–80.5) |
| rev3 | 0.9 (0.5–1.6) | 1.8 (0.3–8.9) |
| tsa1 rev3 | 11.5 (8.8–14.2) | 42.7 (35.2–71.5) |
| rev1 | 0.9 (0.7–1.4) | 2.9 (0.8–5.4) |
| tsa1 rev1 | 8.7 (6.8–11.8) | 36.4 (18.7–50.8) |
| rad30 | 2.9 (2.0–3.5) | 3.5 (2.0–11.9) |
| tsa1 rad30 | 17.8 (14.2–19.2) | 81.8 (39.2–96.3) |
The numbers in parentheses indicate the low and high values for the 95% confidence interval for each rate.
Anaerobic Growth Conditions Decrease Canr and GCR Rates of WT and tsa1 Cells.
To further investigate the role of ROS in mutagenesis, WT and tsa1 strains were grown under aerobic or anaerobic conditions and the rates of Canr and GCRs were determined (Table 2). Overall, the rates of Canr and GCRs in the anaerobically grown strains were significantly reduced relative to the same strain grown aerobically (Table 2). For the WT strain, the Canr rate and the GCR rate were reduced 3.2-fold and at least 8-fold, respectively. We were not able to precisely determine the GCR rate of the WT strain under anaerobic conditions because GCRs were not detected in a large enough proportion of the cultures to allow accurate calculation of mutation rates (Table 2). A large reduction of the Canr rate and the GCR rate was also observed in the tsa1 mutant under anaerobic conditions, but again we were not able to precisely determine the GCR rate of the tsa1 mutant under anaerobic conditions. These results provide evidence that a significant proportion of the Canr mutations and GCRs in WT and tsa1 strains occurs due to oxygen metabolism under aerobic growth conditions, suggesting that ROS is a major source of endogenous DNA damage leading to Canr mutations and GCRs.
Table 2.
Effect of reduced oxygen metabolism on Canr and GCR mutation rates
| Strain | Aerobic Canr rate (× 10−7) | Anaerobic Canr rate (× 10−7) | Anaerobic suppression | Aerobic GCR rate (× 10−10) | Anaerobic GCR rate (× 10−10) | Anaerobic suppression |
|---|---|---|---|---|---|---|
| WT | 2.6 (2.2–3.0) | 0.8 (0.6–1.2) | 3.2 | 2.6 (2.1–9.5) | <0.3 | >8.7 |
| tsa1 | 25.6 (24.1–28.8) | 4.8 (3.6–10.0) | 5.3 | 55.6 (36.9–80.5) | <1.1 | >50.5 |
The numbers in parentheses indicate the low and high values for the 95% confidence interval for each rate. Anaerobic suppression represents the fold reduction of mutation rate for a given strain in anaerobic conditions relative to the same strain grown aerobically.
Anaerobic Growth Conditions Differentially Affect Spontaneous Mutation Rates in DNA Repair-Deficient Cells.
The observation described above prompted us to determine the effect of oxygen tension on the phenotypes of several representative GCR mutator mutants such as mre11, rad27, pif1, and rad52 mutants. Inactivation of MRE11, RAD27, or PIF1 increased the GCR rate 500- to 1,500-fold above WT, whereas deletion of RAD52 resulted in a more modest increase of the GCR rate (Table 3) (5, 25). Anaerobic conditions appeared to have no significant effect on the GCR rate of the mre11 mutant (95% confidence limits), suggesting that oxygen metabolism is not a major contributor to the elevated genomic instability found in the mre11 mutant. A similar result was found when a deletion of RAD50, which encodes a component of Mre11 complex, was analyzed (data not shown). In the case of the rad27 mutant, the GCR rate was reduced 4-fold under anaerobic conditions compared with normal aerobic conditions. The PIF1 gene encodes a helicase that functions in both mitochondria and the nucleus. The GCR rate of the pif1 mutant was reduced 6-fold under anaerobic conditions. Finally, the GCR rate of the rad52 mutant was also reduced 6-fold under anaerobic conditions. We then analyzed mutants with defects in DNA mismatch repair because Msh2–Msh6-dependent mismatch repair has been implicated in the repair of mispairs that result from oxidative damage to DNA and DNA precursors (26, 27). To our surprise, anaerobic conditions did not reduce the elevated Canr and hom3–10 rates of the msh2 and msh6 mutants (95% confidence limits) (Table 4). This is in contrast to the report that the reversion of base substitution mutations in the Cys-22 codon of the CYC1 gene in mismatch repair-deficient strains was significantly decreased under anaerobic conditions (27). However, our analysis used more general mutator assays than the specific base substitution reversion assays used in the previous study. Collectively, these results suggest that reduced oxygen tension differentially effects the accumulation of mutations and GCRs in different DNA repair-deficient cells.
Table 3.
GCR rates in DNA repair mutants under aerobic and anaerobic conditions
| Strains | Aerobic GCR rate (× 10−10) | Anaerobic GCR rate (× 10−10) | Anaerobic suppression |
|---|---|---|---|
| WT | 2.6 (2.1–9.5) | <0.3 | >8.7 |
| mre11 | 2,345.5 (2,104.6–3,085.0) | 1,853.1 (1,325.0–4,025.3) | 1.3 |
| rad27 | 3,469 (2,153.9–3,786.2) | 818.7 (328.2–1,828.8) | 4.2 |
| pif1 | 4,000.2 (2,712.4–5,083.2) | 652.6 (419.1–941.2) | 6.1 |
| rad52 | 63.3 (45.0–115.2) | 9.5 (2.5–23.7) | 6.6 |
The numbers in parentheses indicate the low and high values for the 95% confidence interval for each rate. Anaerobic suppression represents the fold change of GCR rate for a given strain in anaerobic conditions relative to the same strain grown aerobically.
Table 4.
Mutation rates of msh2 and msh6 mutants under aerobic and anaerobic conditions
| Strain | Aerobic Canr rate (× 10−7) | Anaerobic Canr rate (× 10−7) | Anaerobic suppression | Aerobic Hom+ rate (× 10−8) | Anaerobic Hom+ rate (× 10−8) | Anaerobic suppression |
|---|---|---|---|---|---|---|
| WT | 2.6 (2.2–3.0) | 0.8 (0.6–1.2) | 3.2 | 0.7 (0.3–1.0) | 0.7 (0.5–1.2) | 1.0 |
| msh6 | 16.1 (13.0–25.8) | 14.1 (12.1–21.0) | 1.1 | 2.8 (2.4–4.4) | 3.7 (2.3–6.3) | 0.8 |
| msh2 | 54.8 (50.2–63.1) | 44.5 (37.0–49.6) | 1.2 | 460.0 (360.1–595.3) | 332.4 (274.9–450.4) | 1.4 |
The numbers in parentheses indicate the low and high values for the 95% confidence interval for each rate. Anaerobic suppression represents the fold reduction of mutation rate for a given strain in anaerobic conditions relative to the same strain grown aerobically.
Anaerobic Growth Conditions Restore Cellular Viability of tsa1 rad6, tsa1 rad51, and tsa1 mre11 Mutants.
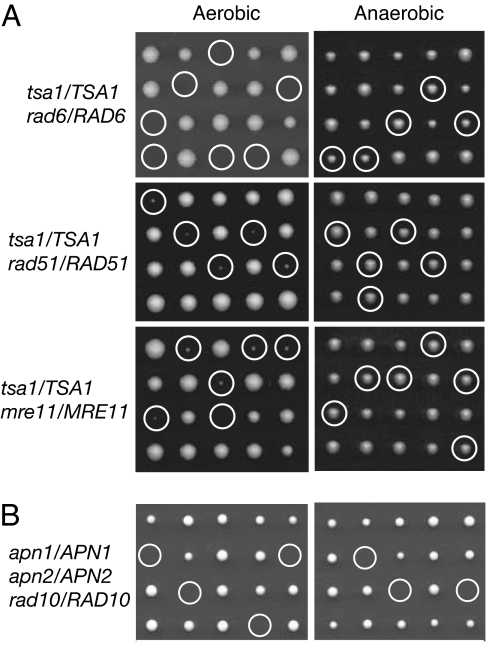
We have previously shown that combining a tsa1 mutation with rad6, rad51, or mre11 mutations, among others, results in cell death (20). To evaluate the contribution of oxygen metabolism to the inviability of these double mutants, we tested whether anaerobic conditions restore the viability of tsa1 rad6, tsa1 rad51, or tsa1 mre11 mutants. Three diploid strains (tsa1/TSA1 rad6/RAD6, tsa1/TSA1 rad51/RAD51, and tsa1/TSA1 mre11/MRE11) were sporulated and dissected, and the spore dissection plates were incubated under either anaerobic or aerobic conditions. As reported (20), a rad6 mutation was found to be lethal in combination with tsa1 under normal aerobic conditions. In contrast, all spore products grew when the dissection plates were incubated under anaerobic conditions (Fig. 1A). Genotyping of the spore clones confirmed that tsa1 rad6 mutants were viable under anaerobic conditions and appeared to have a growth rate similar to that of rad6 mutants. Similarly, lethality of tsa1 rad51 and tsa1 mre11 mutants under aerobic conditions was entirely suppressed under anaerobic conditions. Replating the tsa1 rad6, tsa1 rad51, or tsa1 me11 cells from anaerobic culture under aerobic conditions again resulted in cell death. Thus, aerobic oxygen metabolism was responsible for the death of the tsa1 rad6, tsa1 rad51, and tsa1 mre11 mutants. Additional elimination of Srs2 helicase, which probably enhances homologous recombination, did not result in the recovery of tsa1 rad6 mutants (data not shown).
Fig. 1.
Reduction of oxygen metabolism suppresses the lethality of tsa1 rad6, tsa1 rad51, and tsa1 mre11 double mutation combinations but does not rescue the inviability of apn1 apn2 rad10 triple mutants. (A) Tetrad dissection plates of diploids heterozygous for tsa1 and rad6, diploids heterozygous for tsa1 and rad51, and diploids heterozygous for tsa1 and mre11 were grown at 30°C under aerobic or anaerobic conditions for 5 days. Circles indicate inferred double mutants. (B) Tetrad dissection plates of diploids heterozygous for apn1, apn2, and rad10 mutations were incubated under aerobic or anaerobic conditions for 5 days. Circles indicate the apn1 apn2 rad10 triple mutants.
The observation that anaerobic growth restores the viability of tsa1 rad6, tsa1 rad51, and tsa1 mre11 mutants prompted us to evaluate the effect of anaerobic condition on the inviability of the apn1 apn2 rad10 triple mutant (28, 29), thought to be due to the inability to repair apurinic/apyrimidinic (AP) sites or deficient removal of 3′-blocking groups formed by the action of ROS produced during cellular metabolism (28). An apn1 apn2 mutant was crossed with a rad10 mutant, and the resulting apn1/APN1 apn2/APN2 rad10/RAD10 diploid strain was sporulated and dissected. We observed a similar proportion of spore lethality on plates grown aerobically or anaerobically, and under both conditions no apn1 apn2 rad10 spore clones were identified by genotyping (Fig. 1B). Thus, reduced oxygen metabolism does not suppress the inviability of the apn1 apn2 rad10 mutant. In contrast, deletion of UNG1 gene encoding the uracil DNA glycosylase partially suppressed the inviability of the apn1 apn2 rad10 (30). This suggests that uracil in DNA, rather than the ROS-induced DNA damage, is a critical source of the formation of spontaneous AP sites in DNA and that the lethality of the apn1 apn2 rad10 mutation combination is likely due to the inability to repair such AP sites.
The Absence of TSA1 Results in Increased Rad52-YFP Focus Formation.
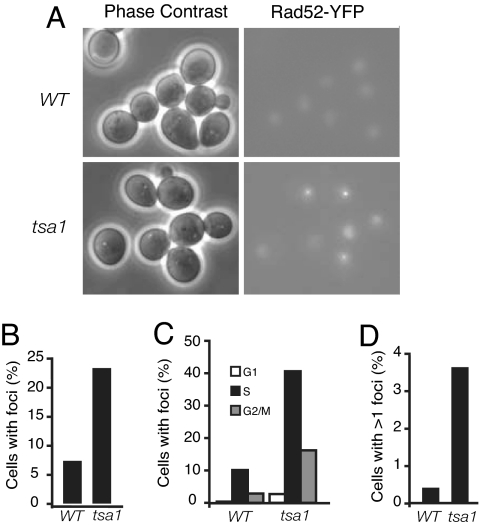
To further investigate the nature of tsa1-associated DNA lesions and the involvement of Rad52-dependent DNA repair and recombination in response to ROS-induced DNA damage, the formation of Rad52-YFP foci was monitored in tsa1 mutants. These foci are sites of repair of multiple DNA lesions, such as DSBs (31). Unsynchronized exponential growing WT cells showed diffuse nuclear localization of Rad52-YFP with few cells (6.6%) containing spontaneous Rad52-YFP foci (Fig. 2 A and B). Quantification of the appearance of Rad52-YFP foci for each phase of the cell cycle revealed that these foci occurred in 0.4% of G1, 10.1% of S, and 2.9% of G2/M phase cells (Fig. 2C). Spontaneous repair foci were observed primarily in S phase, implying that DNA lesions were being recognized and repaired during replication. Deletion of TSA1 caused an increased incidence of Rad52 foci (22.5%) (Fig. 2 A and B). These foci occurred predominantly in S and G2/M phase cells with 2.7% of G1, 40.6% of S, and 16.2% of G2/M phase cells showing foci (Fig. 2C). Furthermore, the frequency of cells with more than one Rad52-YFP focus increased from 0.38% in WT cells to 3.6% in tsa1 cells (Fig. 2D). These observations suggest that the absence of Tsa1 results in DNA damages that require Rad52-dependent recombinational repair during DNA replication, consistent with the synthetic lethality of tsa1 and rad52 mutations.
Fig. 2.
Analysis of spontaneous Rad52-YFP foci in exponentially growing WT and tsa1 cells. (A) A representative image shows that the tsa1 strain exhibits an increase in Rad52-YFP foci compared with the WT strain. The fluorescence images are maximum projections of optical sections (0.2-μm intervals) spanning the entire volume of the cell. (B) Graph shows the percentage of cells that contain Rad52-YFP foci. (C) Graph shows the percentage of cells in G1, S, or G2/M phase that contain Rad52-YFP foci. (D) Graph shows the percentage of cells that contain more than one Rad52-YFP focus. Values reported are the mean of three independent measurements.
H2O2 Treatment Increases Rad52-YFP Focus Formation.
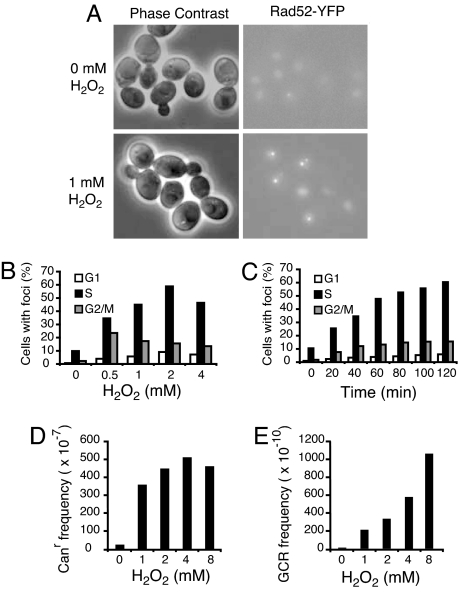
We then investigated the involvement of RAD52-dependent DNA repair in response to exogenous H2O2 treatment. DNA damage was induced in the Rad52-YFP strain by exposure to 0.5, 1, 2, and 4 mM H2O2 for 1 h, resulting in 74%, 50%, 41%, and 23% cell survival, respectively. A significant increase in the proportion of cells containing Rad52 foci was observed (Fig. 3A), with the greatest increase in Rad52-YFP foci in S phase cells (Fig. 3 A and B). The proportion of cells with foci increased with increasing H2O2 concentration up to 2 mM H2O2 and with increasing time of exposure to H2O2 (Fig. 3 B and C). The proportion of cells with Rad52 foci and H2O2 dose were not directly proportional; this may be because of the fact that one focus may be the site of multiple repair events (31) and also that the higher concentrations of H2O2 (>2 mM) induced higher levels of cell death. Altogether, exogenous H2O2-induced DNA damage, like endogenous ROS in the absence of Tsa1, resulted in a significant increase of Rad52 foci in S-phase cells.
Fig. 3.
Induction of Rad52-YFP foci and genome instability by H2O2 treatment in the WT strain. (A) A representative image shows Rad52-YFP foci in untreated cells and after exposure to 1 mM H2O2 for 1 h. The fluorescence images are maximum projections of optical sections (0.2-μm intervals) spanning the entire volume of the cell. Note that one of the H2O2-treated cells contains two Rad52-YFP foci. (B) Graph shows the percentage of cells in G1, S, or G2/M phase that contain Rad52-YFP foci after exposure to the indicated dose of H2O2 for 1 h. (C) Graph shows the percentage of cells in G1, S, or G2/M phase that contain Rad52-YFP foci after exposure to 1 mM H2O2 for up to 2 h. Values reported are the mean of three independent measurements. (D) Induction of the Canr mutation frequency by different doses of H2O2. The median H2O2-induced Canr mutation frequency from five independent trials is presented. (E) Induction of the Canr-5FOAr mutation frequency by different doses of H2O2. The median H2O2-induced Canr-5FOAr mutation frequency from five independent trials is presented.
H2O2 Treatment Increases Genome Rearrangements.
We further investigated whether exogenous H2O2 treatment would increase the frequency of GCRs in WT cells. Exponential growing cells were treated with 1, 2, 4, and 8 mM H2O2 for 2 h, resulting in 37%, 27%, 19%, and 12% cell survival, respectively. A dose-dependent increase in the frequency of Canr and Canr-5FOAr mutations was consistently observed. A representative experiment showing a median H2O2-induced mutation frequency is presented (Fig. 3 D and E). The maximum frequency of Canr mutations was induced 18-fold above that of untreated cells (Fig. 3D). The induced Canr mutation frequency did not increase further after exposure to higher doses of H2O2, suggesting that the maximum mutation frequency level may represent saturation of the DNA damage-handling capacity of the WT strain. Treatment with H2O2 also increased the frequency of GCRs (Canr-5FOAr mutants) in a dose-dependent manner (Fig. 3E). Treatment with 8 mM H2O2, which resulted in ≈12% survival, increased the frequency of cells containing GCRs ≈100-fold. We observed that WT cells in stationary phase were more resistant to H2O2-induced killing and H2O2-induced mutagenesis (data not shown), paralleling the observation that H2O2-induced Rad52 foci were seen mostly in S-phase cells. To gain further insight into the types of GCRs induced by H2O2, the structure of the GCRs from eight independent Canr-5FOAr clones was determined as described (8). Treatment with H2O2 resulted in recovery of translocation, telomere addition, and interstitial deletion classes of GCRs. None of the eight Canr-5FOAr isolates resulted from independent point mutations in the CAN1 and URA3 genes. These results indicate that H2O2-induced DNA damages induce the formation of GCRs.
Discussion
In previous studies we demonstrated that the peroxiredoxin Tsa1 plays a key role in preventing genome instability, presumably by detoxifying H2O2, and that both recombination and postreplication repair are key pathways for maintaining the viability of tsa1 mutants (20). The results from the present study extend these observations and support a number of critical findings. First, the TLS polymerases ζ, η, and Rev1 do not contribute significantly to tsa1-associated GCR formation, although they do contribute to the formation of Canr mutations, which are primarily base substitution mutations (18). Second, decreased oxygen tension substantially reduced GCR rates of WT and tsa1 mutants. The same conditions also reduced the GCR rates of some mutants that have elevated GCR rates such as rad27, pif1, and rad52 mutants but not of other mutants such as mre11 and rad50. Third, decreased oxygen tension restored the viability of tsa1 rad6, tsa1 rad51, and tsa1 mre11 double mutants but not that of the apn1 apn2 rad10 triple mutant, supporting the view that recombination and postreplication repair are key pathways in preventing the deleterious effects of oxidative damage to DNA. Fourth, the absence of Tsa1 or treatment with H2O2 resulted in an increased frequency of Rad52-YFP foci, indicative of increased DNA repair. And fifth, treatment of WT cells with H2O2 significantly increased the frequency of GCRs. These results demonstrate that oxygen metabolism and ROS are a major source of DNA lesions leading to mutations, GCRs, and cell death and that the Tsa1, along with recombination and postreplication repair, plays a key role in suppressing genome instability and cell death induced by endogenous oxidative stress.
ROS-related DNA lesions, including those induced by exogenous H2O2 treatment, are mutagenic, and anaerobic growth conditions reduce their mutagenic effects, such as point mutations and poly(GT) tract instability (13, 27, 32–35). The work presented here demonstrates that reduced oxygen metabolism significantly reduces the rate of spontaneous GCRs in WT strains and tsa1 as well as other mutants and that treatment with H2O2 substantially induces GCRs, indicating that the ROS-induced DNA lesions are an important source of GCRs. Consistent with this, it has been observed in mammalian cells that oxygen metabolism is partially responsible for chromosome breaks and neuronal apoptosis in cells deficient for nonhomologous DNA end joining (36).
The effect of oxygen metabolism on the GCR rate varied depending on the genetic defect underlying increased GCR rates. Among the mutants tested, in addition to tsa1, anaerobic conditions significantly reduced the GCR rates observed in a pif1 mutant lacking a DNA helicase that functions in both mitochondria and the nucleus and in a rad52 mutant lacking a key recombination protein. This suggests that Pif1- and Rad52-dependent recombination play important roles in either repairing or preventing aberrant repair of ROS-induced DNA damage that has the potential to result in GCRs. Anaerobic conditions also reduced the GCR rate of a rad27 mutant, although to a lesser extent. It is generally thought that Rad27 functions in processing flapped DNA molecules that occur during sealing of Okazaki fragments and during base excision repair (37). Our results suggest that Rad27 functions to prevent GCRs both by preventing DNA replication errors during Okazaki fragment synthesis and repairing mutagenic DNA damage induced by endogenous ROS. In contrast, anaerobic growth conditions had little or no effect on the GCR rate of mre11 and rad50 mutants. This suggests that the GCRs that occur in Mre11 complex-defective mutants may not originate from oxidative DNA damage but more likely reflect S-phase defects due to an inability of Mre11 complex-defective mutants to maintain the structure of replication forks, repair spontaneously damaged replication forks, or mount an appropriate S-phase checkpoint response to spontaneously damaged replication forks (38, 39). However, the Mre11 complex must play some role in response to oxidative DNA damage because it is essential for the repair of tsa1-associated lethal DNA damage, as is indicated by the synthetic lethality of a tsa1 mutation in combination with mre11, rad50, or xrs2 mutations as well as by the observation that anaerobic conditions suppress this lethality; possibly these latter effects reflect the role of the Mre11 complex in recombination similar to that of Rad52 (40). Taken together, the difference in the effect of ROS on increased GCRs in different mutants probably reflects the diversity of DNA lesions that can lead to GCRs.
ROS-related DNA lesions that occur in the absence of TSA1 are lethal in the absence of appropriate repair, probably because the lesions result in replication blocks or replication-associated DSBs. A number of results are consistent with this view. The absence of TSA1 results in increased Rad52-YFP foci, predominantly during S phase; these are thought to be sites of Rad52-dependent recombination and DSB repair (31). Mutations in TSA1 or RAD52 result in increased GCR rates, and these increases depend on oxygen metabolism. tsa1 mutations cause synthetic lethality when combined with rad51, rad52, mre11, rad50, xrs2, or rad6 mutations with the tsa1 rad6 combination causing the most severe phenotype (20); reduced oxygen tension restores the viability and growth of these double mutants. Rad51, Rad52, and the Mre11 complex all are involved in recombinational DSB repair (40), consistent with the view that recombination is required for repairing the lesions that occur in tas1 mutants. Interestingly, in Escherichia coli, mutation fur, ubiH, or ubiE, most likely leading to an increased level of ROS, causes cell death in the absence of RecA-dependent recombinational repair (41). The fur recA synthetic lethality is ascribed to elevated chromosomal fragmentation. The role of the Rad6 protein in dealing with tsa1-associated lesions is less clear. Rad6 regulates two pathways of postreplication repair, one of which is error-prone and involves the TLS polymerases and Rad18; the other is error-free and involves recombination or template switching and Rad5 (42–44). In addition, Rad6 plays a role in checkpoint activation through yet a different pathway involving Bre1 (45). Our experiments indicate that defects in TLS only partially account for tsa1-associated accumulation of Canr mutations and are unlikely to account for GCRs, making it unlikely that RAD6-mediated TLS alone promotes survival of a tsa1 mutant. In contrast, both rad5 and rad18 mutations result in a slow growth phenotype and increased GCR rates when combined with a tsa1 mutation (19, 20), suggesting that both error-prone and error-free postreplication repair play roles in the survival of a tsa1 mutant. In addition, checkpoint defects result in slow growth and increased GCR rates when combined with a tsa1 mutation (20). These results raise the possibility that all three pathways regulated by Rad6 may cooperate in promoting the survival of tsa1 mutants.
Based on the data presented here, we conclude that spontaneous GCRs and cell death observed in many mutants are substantially due to endogenous ROS produced by oxygen metabolism. It should be noted that intracellular generation of ROS is an inevitable and physiologically important process (12). The true source of oxidative stress is not ROS generation per se but rather an imbalance in ROS production, appropriate detoxification, and repair. Thus, normal oxygen metabolism could have deleterious biological consequences in cells with compromised ROS defenses and DNA repair. Our result linking intracellular ROS production and GCR or cell death may provide insights into the understanding of the pathological scenarios associated with chronic oxidative stress such as chronic infection, inflammation, neurodegeneration, and hereditary genome instability syndromes.
Materials and Methods
Media and Strains.
S. cerevisiae strains were grown in standard media including yeast extract/peptone/dextrose (YPD) medium or synthetic complete (SC) medium lacking appropriate amino acids. Canr mutants caused by inactivation of the CAN1 gene by base substitutions, frameshifts, small deletions, or insertions were selected on SC-arginine dropout plates containing 60 mg/liter canavanine. Hom+ revertants caused by reversion of a +1 insertion in the HOM3 gene (hom3–10) were selected for on SC-threonine dropout plates. Canr-5FOAr mutants resulting from the loss of the region including CAN1 and URA3 on chromosome V were selected on SC-arginine and uracil dropout plates containing 60 mg/liter canavanine and 1 g/liter 5FOA (5, 8).
The strains used for mutation rate assays and genetic interaction analysis were all isogenic to the S288c strain RDKY3615 MATa, ura3–52, leu2Δ1, trp1Δ63, his3Δ200, lys2ΔBgl, hom3–10, ade2Δ1, ade8, and hxt13::URA3 (5). Gene disruptions were made by standard PCR-based methods. Double or triple mutants were usually constructed by intercrossing and tetrad dissection. The relevant genotypes of these strains are as follows: RDKY3615 WT; RDKY5502 tsa1::kanMX4; RDKY5521 rev3::HIS3; RDKY5678 tsa1::kanMX4 rev3::HIS3; MEHY1530 rev1::HIS3; MEHY1531 tsa1::kanMX4 rev1::HIS3; RDKY5523 rad30::HIS3; RDKY5679 tsa1::kanMX4 rad30::HIS3; RDKY4718 rad27::HIS3; RDKY4421 rad52::HIS3; RDKY3633 mre11::HIS3; RDKY4399 pif1::kanMX4; RDKY3812 msh2::kanMX4; and RDKY5507 msh6::TRP1. The parental strain used for the analysis of Rad52-YFP foci, UM74–3B MATa, his3-11,15, leu2-3,112, trp1-1, ura33-1, bar1::LEU2, and RAD52-YFP, was provided by A. Nicolas (Institut Curie, Paris, France) (46), and then the tsa1::kanMX4 mutation was introduced into this strain.
Fluctuation Analysis.
The rate of accumulation of Canr mutations and GCRs in cell populations was determined by fluctuation analysis as described (18) except that the cell cultures were incubated under aerobic or anaerobic conditions without agitation. The number of mutant cells per culture among 15–35 parallel cultures was calculated, and the median value was used to determine the mutation rate of a given strain by the method of the median (47). The 95% confidence intervals for a median rate were calculated based on order statistics with the formula available on request and were used to compare mutation rates.
Anaerobic Growth Conditions.
YPD medium with or without agar was supplemented with Tween 80 and ergosterol to a final concentration of 1.32 g/liter and 12 mg/liter, respectively. For determining mutation rates, liquid cultures in plates were placed in an airtight jar containing a disposable hydrogen- and carbon dioxide-generating envelope (BD BBL GasPak Plus; Becton Dickinson, Sparks, MD) and grown anaerobically at 30°C for 5–7 days to yield enough cells for analysis. For anaerobic growth of dissected spores, dissection plates were placed immediately after dissection in the anaerobic jar and grown at 30°C for 5 days. Anaerobic conditions were monitored with the redox indicator Anaerotest (Merck, Darmstadt, Germany) placed inside the airtight jar.
Fluorescence Microscopy.
Strains of the desired genotype carrying the RAD52-YFP fusion were grown at 23°C and harvested for microscopy during midlog phase. In the case of monitoring Rad52-YFP foci after DNA damage, H2O2 was added to the concentrations indicated and the cultures were further incubated at 23°C for the indicated times before harvesting. Cells were washed twice in SC and then immobilized on a glass slide precoated by a solution of 1% (wt/vol) low-melting agarose. Fluorescent and phase-contrast images were captured with a Leica microscope (DMRXA) equipped with a cooled CCD camera MicroMAX (Princeton Instruments, Trenton, NJ) under control of MetaMorph software (Molecular Devices, Downingtown, PA). Images were obtained at 0.2-μm intervals along the z axis of each cell using a piezo-controlled stepper. Foci were inspected and counted by examining all of the focal planes intersecting each cell and by overlaying the Rad52-YFP signal with the phase-contrast image processed with software ImageJ (http://rsb.info.nih.gov/ij), and cells were also classed as G1 (unbudded), S (small-budded), or G2/M (large-budded). Three independent experiments each counting at least 300 cells were performed for each strain or condition.
Induction of Mutations by H2O2 Treatment.
Log-phase cells in YPD were washed twice with distilled water and resuspended in a volume of distilled water that was equal to the starting culture volume, and then the cells were incubated with the indicated concentration of H2O2 for 2 h at 30°C. The treated cells were washed twice with distilled water and resuspended in the original volume of YPD. Aliquots were removed to determine the percentage of cell survival. Cells were also cultured overnight and plated onto YPD and selection plates to determine the total number of viable cells and the number of mutants, respectively. Mutation frequencies were calculated as the number of mutants divided by the total number of viable cells. Potential jackpot cultures were eliminated from the frequency calculations. The median frequency from at least five independent trials is reported. The structure of the GCRs from independent Canr-5FOAr clones was determined as described (8).
Acknowledgments
We thank Christopher Putnam, Jorrit Enserink, Martine Heude, Laurence Vernis, and Roland Chanet for comments on the manuscript, members of the R.D.K. and Giuseppe Baldacci laboratories for helpful discussions, Giuseppe Baldacci for his support and encouragement, Alain Nicolas for the RAD52-YFP strain, and Amandine Martin for technical help. This work was supported by the Centre National de la Recherche Scientifique Action Thématique et Incitative sur Programme, Institut Curie, Fondation Recherche Medicale (M.-E.H.), and National Institutes of Health Grant GM26017 (to R.D.K.). S.R. is supported by a Ph.D. training fellowship from the Institut Curie.
Abbreviations
- GCR
gross chromosomal rearrangement
- ROS
reactive oxygen species
- DSB
dsDNA break
- TLS
translesion DNA synthesis
- Canr
canavanine-resistant
- Canr-5FOAr
canavanine- and 5-fluoroorotic acid-resistant
- YPD
yeast extract/peptone/dextrose
- SC
synthetic complete.
Footnotes
The authors declare no conflict of interest.
References
- 1.Kolodner RD, Marsischky GT. Curr Opin Genet Dev. 1999;9:89–96. doi: 10.1016/s0959-437x(99)80013-6. [DOI] [PubMed] [Google Scholar]
- 2.Kops GJ, Weaver BA, Cleveland DW. Nat Rev Cancer. 2005;5:773–785. doi: 10.1038/nrc1714. [DOI] [PubMed] [Google Scholar]
- 3.De Lange T. Cold Spring Harbor Symp Quant Biol. 2005;70:197–204. doi: 10.1101/sqb.2005.70.032. [DOI] [PubMed] [Google Scholar]
- 4.Huang D, Koshland D. Genes Dev. 2003;17:1741–1754. doi: 10.1101/gad.1089203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen C, Kolodner RD. Nat Genet. 1999;23:81–85. doi: 10.1038/12687. [DOI] [PubMed] [Google Scholar]
- 6.Tennyson RB, Ebran N, Herrera AE, Lindsley JE. Genetics. 2002;160:1363–1373. doi: 10.1093/genetics/160.4.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kolodner RD, Putnam CD, Myung K. Science. 2002;297:552–557. doi: 10.1126/science.1075277. [DOI] [PubMed] [Google Scholar]
- 8.Schmidt KH, Pennaneach V, Putnam CD, Kolodner RD. Methods Enzymol. 2006;409:462–476. doi: 10.1016/S0076-6879(05)09027-0. [DOI] [PubMed] [Google Scholar]
- 9.Nagy R, Sweet K, Eng C. Oncogene. 2004;23:6445–6470. doi: 10.1038/sj.onc.1207714. [DOI] [PubMed] [Google Scholar]
- 10.Fearson ER. Science. 1997;278:1043–1050. doi: 10.1126/science.278.5340.1043. [DOI] [PubMed] [Google Scholar]
- 11.Hussain SP, Hofseth LJ, Harris CC. Nat Rev Cancer. 2003;3:276–285. doi: 10.1038/nrc1046. [DOI] [PubMed] [Google Scholar]
- 12.Finkel T. Curr Opin Cell Biol. 2003;15:247–254. doi: 10.1016/s0955-0674(03)00002-4. [DOI] [PubMed] [Google Scholar]
- 13.Newcomb TG, Loeb LA. In: DNA Repair in Prokaryotes and Lower Eukaryotes. Nickoloff JA, Hoekstra MF, editors. Totawa, NJ: Humana; 1998. pp. 65–84. [Google Scholar]
- 14.Bladier C, de Haan JB, Kola I. In: Antioxidant and Redox Regulation of Genes. Sen CK, Sies H, Baeuerle PA, editors. San Diego, CA: Academic; 2000. pp. 425–449. [Google Scholar]
- 15.Rhee SG, Chae HZ, Kim K. Free Radical Biol Med. 2005;38:1543–1552. doi: 10.1016/j.freeradbiomed.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 16.Wood ZA, Schroder E, Robin Harris J, Poole LB. Trends Biochem Sci. 2003;28:32–40. doi: 10.1016/s0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]
- 17.Park SG, Cha MK, Jeong W, Kim IH. J Biol Chem. 2000;275:5723–5732. doi: 10.1074/jbc.275.8.5723. [DOI] [PubMed] [Google Scholar]
- 18.Huang ME, Rio AG, Nicolas A, Kolodner RD. Proc Natl Acad Sci USA. 2003;100:11529–11534. doi: 10.1073/pnas.2035018100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith S, Hwang JY, Banerjee S, Majeed A, Gupta A, Myung K. Proc Natl Acad Sci USA. 2004;101:9039–9044. doi: 10.1073/pnas.0403093101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang ME, Kolodner RD. Mol Cell. 2005;17:709–720. doi: 10.1016/j.molcel.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 21.Neumann CA, Krause DS, Carman CV, Das S, Dubey DP, Abraham JL, Bronson RT, Fujiwara Y, Orkin SH, Van Etten RA. Nature. 2003;424:561–565. doi: 10.1038/nature01819. [DOI] [PubMed] [Google Scholar]
- 22.Friedberg EC, Lehmann AR, Fuchs RP. Mol Cell. 2005;18:499–505. doi: 10.1016/j.molcel.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 23.Huang ME, de Calignon A, Nicolas A, Galibert F. Curr Genet. 2000;38:178–187. doi: 10.1007/s002940000149. [DOI] [PubMed] [Google Scholar]
- 24.McDonald JP, Levine AS, Woodgate R. Genetics. 1997;147:1557–1568. doi: 10.1093/genetics/147.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Myung K, Chen C, Kolodner RD. Nature. 2001;411:1073–1076. doi: 10.1038/35082608. [DOI] [PubMed] [Google Scholar]
- 26.Ni TT, Marsischky GT, Kolodner RD. Mol Cell. 1999;4:439–444. doi: 10.1016/s1097-2765(00)80346-9. [DOI] [PubMed] [Google Scholar]
- 27.Earley MC, Crouse GF. Proc Natl Acad Sci USA. 1998;95:15487–15491. doi: 10.1073/pnas.95.26.15487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guzder SN, Torres-Ramos C, Johnson RE, Haracska L, Prakash L, Prakash S. Genes Dev. 2004;18:2283–2291. doi: 10.1101/gad.1232804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guillet M, Boiteux S. EMBO J. 2002;21:2833–2841. doi: 10.1093/emboj/21.11.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guillet M, Boiteux S. Mol Cell Biol. 2003;23:8386–8394. doi: 10.1128/MCB.23.22.8386-8394.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lisby M, Mortensen UH, Rothstein R. Nat Cell Biol. 2003;5:572–577. doi: 10.1038/ncb997. [DOI] [PubMed] [Google Scholar]
- 32.Gralla EB, Valentine JS. J Bacteriol. 1991;173:5918–5920. doi: 10.1128/jb.173.18.5918-5920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salmon TB, Evert BA, Song B, Doetsch PW. Nucleic Acids Res. 2004;32:3712–3723. doi: 10.1093/nar/gkh696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bjelland S, Seeberg E. Mutat Res. 2003;531:37–80. doi: 10.1016/j.mrfmmm.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 35.Vongsamphanh R, Wagner JR, Ramotar D. DNA Repair (Amsterdam) 2006;5:235–242. doi: 10.1016/j.dnarep.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 36.Karanjawala ZE, Murphy N, Hinton DR, Hsieh CL, Lieber MR. Curr Biol. 2002;12:397–402. doi: 10.1016/s0960-9822(02)00684-x. [DOI] [PubMed] [Google Scholar]
- 37.Liu Y, Kao HI, Bambara RA. Annu Rev Biochem. 2004;73:589–615. doi: 10.1146/annurev.biochem.73.012803.092453. [DOI] [PubMed] [Google Scholar]
- 38.Mirzoeva OK, Petrini JH. Mol Cancer Res. 2003;1:207–218. [PubMed] [Google Scholar]
- 39.Trenz K, Smith E, Smith S, Costanzo V. EMBO J. 2006;25:1764–1774. doi: 10.1038/sj.emboj.7601045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krogh BO, Symington LS. Annu Rev Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- 41.Kouzminova EA, Rotman E, Macomber L, Zhang J, Kuzminov A. Proc Natl Acad Sci USA. 2004;101:16262–16267. doi: 10.1073/pnas.0405943101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Broomfield S, Hryciw T, Xiao W. Mutat Res. 2001;486:167–184. doi: 10.1016/s0921-8777(01)00091-x. [DOI] [PubMed] [Google Scholar]
- 43.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 44.Zhang H, Lawrence CW. Proc Natl Acad Sci USA. 2005;102:15954–15959. doi: 10.1073/pnas.0504586102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Giannattasio M, Lazzaro F, Plevani P, Muzi-Falconi M. J Biol Chem. 2005;280:9879–9886. doi: 10.1074/jbc.M414453200. [DOI] [PubMed] [Google Scholar]
- 46.Loeillet S, Palancade B, Cartron M, Thierry A, Richard GF, Dujon B, Doye V, Nicolas A. DNA Repair (Amsterdam) 2005;4:459–468. doi: 10.1016/j.dnarep.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 47.Lea DE, Coulson CA. J Genet. 1948;49:264–285. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]