Abstract
Type I (IFN-α/β) and type III (IFN-λs) IFNs are important components of the host antiviral response. Although type III IFNs possess intrinsic antiviral activity similar to that of type I IFNs, they signal through a specific unique receptor complex, and their functional importance for antiviral resistance is largely uncharacterized. Here, we report the first virus defense mechanism that directly targets type III IFNs. Y136 from Yaba-like disease virus, a yatapoxvirus, is a secreted glycoprotein related to protein B18 from Vaccinia virus, a known type I IFN-binding protein and a member of the Ig superfamily. Surprisingly, whereas B18 inhibits only type I IFNs, Y136 inhibits both type I and type III IFNs. Y136 inhibits IFN-induced signaling and suppresses IFN-mediated biological activities including up-regulation of MHC class I antigen expression and induction of the antiviral state. These data demonstrate that poxviruses have developed unique strategies to counteract IFN-mediated antiviral protection and highlight the importance of type III IFNs in antiviral defense. These results suggest that type III IFNs may be an effective treatment for some poxviral infections.
Keywords: antiviral response, interferon antagonists, interferon receptors, poxviruses, virus evasion
Interferons (IFNs) are defined by their ability to induce resistance to viral infection. Three types of IFNs have been described that signal through unique receptor heterodimers.
Human type I IFNs include the well characterized 13 IFN-α proteins, IFN-β, IFN-ω and the more recently identified IFN-κ (1) and IFN-ε (2). Type I IFNs signal through a common cellular IFN-α/β receptor complex, although the receptor subunits used and the precise signaling of IFN-κ and IFN-ε are less well characterized. The IFN-α/β receptor complex is composed of two unique subunits, IFN-αR1 and IFN-αR2 (3, 4). Both subunits are required to assemble the functional receptor complex for IFN-α, IFN-β and IFN-ω. Antibody-mediated neutralization of IFN-αR2 blocked IFN-κ signaling, demonstrating the requirement for IFN-αR2 (1). However, participation of IFN-αR1 subunit in the IFN-κ receptor complex has not been demonstrated. The signaling and the receptor components for IFN-ε are unknown.
IFN-γ, the sole type II IFN, binds to an IFN-γ receptor complex and induces cellular (Th1) immune responses directed toward destruction of virus-infected cells (5). The IFN-γ receptor complex consists of unique IFN-γR1 and IFN-γR2 chains.
Type III IFNs were discovered recently and are IFN-λ1, IFN-λ2 and IFN-λ3 (6), also known as IL-29, IL-28A, and IL-28B, respectively (7). They signal through an IFN-λ receptor complex composed of a unique IFN-λR1 chain and a shared IL-10R2 chain that is also the second subunit of the IL-10, IL-22, and IL-26 receptor complexes (3).
Type I and type III IFNs are produced in response to viral infections (3, 4, 8, 9). Binding of IFNs to their corresponding cellular receptor complexes, despite their differences, induces similar signaling events of the Janus kinase (JAK)-signal transducers and activators of transcription (STAT) signal transduction pathway, including phosphorylation of kinases Jak1 and Tyk2 and activation of latent transcriptional factors STAT1 and STAT2 as well as STAT3, STAT4, and STAT5 to a lesser extent (3, 4, 6, 10). Activated STATs regulate gene expression, and both types of IFNs induce very similar sets of genes, including many genes that encode important mediators of antiviral response (11, 12). Consequently, type III and type I IFNs are both able to induce an antiviral state in cells (3, 8). However, whereas type I IFN receptors are expressed in most cell types, IFN-λR1 demonstrates a more restricted pattern of expression, limiting the response to type III IFNs to primarily epithelium-like tissues (13). Therefore, although both type I and type III IFNs share similar expression pattern and biological activities, they may play distinct roles in the establishment of multifaceted antiviral response.
Experiments in vivo demonstrated that type III IFNs are important mediators of antiviral response in mucosal/epithelial tissues. IFN-λs induced potent antiviral activity against Herpes simplex virus (HSV)-2 in the vaginal infection model, whereas they were inefficient in systemic infections caused by Encephalomyocarditis virus and Lymphocytic choriomeningitis virus (14).
Importantly, Vaccinia virus (VACV) expressing murine type III IFN was highly attenuated in vivo in the intranasal infection model (15), demonstrating that type III IFNs are biologically relevant against poxviruses. Thus, neutralization of type III IFNs would provide an additional advantage to viruses replicating in mucosal/epithelial tissues. Nevertheless, the functional importance and uniqueness of type III IFNs for antiviral resistance needs further characterization and there were no reported virus defense mechanisms against these IFNs.
Because the IFN system is one of the most important defense mechanisms against viral infections, viruses have developed numerous strategies to circumvent IFN-induced antiviral protection, generally interfering with IFN expression and signaling (16, 17). The Poxviridae is a family of large dsDNA viruses (18) that encode numerous immunomodulatory proteins. VACV, the smallpox vaccine, encodes two secreted proteins that function as IFN antagonists. The B8 protein is the soluble receptor for IFN-γ (19, 20), whereas the B18 protein of VACV strain Western Reserve binds IFN-α, IFN-β and IFN-ω and suppresses interaction of IFNs with their membrane-bound receptor complexes (21–24). Many orthopoxviruses encode orthologues of B18 that are predicted to, or have been shown to, neutralize IFN-α/β. For example, Yaba-like disease virus (YLDV), a strain of Tanapoxvirus, which causes vesicular skin lesions in primates and can be transmitted to humans, encodes protein Y136 that shares 27% amino acid identity with B18 (25). However, the ability of Y136 to inhibit biological activities of IFNs is unknown. Similarly, the ability of poxvirus IFN-binding proteins to neutralize type III IFNs and novel members of the type I IFN family, IFN-κ and IFN-ε has not been investigated.
In this report, we have determined whether a type I IFN antagonist from VACV (B18) and a related but unstudied protein from YLDV (Y136) can neutralize type III IFNs. In addition, these proteins were tested against all type I IFNs, including IFN-κ and IFN-ε that were discovered recently. Surprisingly, although these proteins both inhibit all type I IFNs, they differ in their ability to neutralize type III IFNs.
Results
Type I and Type III IFNs and Their Cellular and Viral Receptors.
Subunits of IFN receptor complexes and receptors for IL-10-related cytokines share limited sequence similarity in their extracellular domains and comprise the class II cytokine receptor family (CRF2) (3, 4, 26). In contrast, VACV protein B18 belongs to the Ig family and reveals highest similarity to other poxvirus-encoded B18 orthologues, such as Variola virus (VARV) D9 and YLDV Y136 proteins and limited similarity to cellular receptors from the IL-1 receptor family (members of the Ig superfamily), such as IL-1 receptor type II (IL-1R2) and IL-1 receptor-like 1 (IL-1RL1), and do not share significant similarity with the ligand-binding subunits of the type I and type III IFN receptor complexes, IFN-αR2 and IFN-λR1, respectively [supporting information (SI) Fig. 5 A and B]. Nevertheless, the B18 protein binds and neutralizes IFN-α, IFN-ω, and IFN-β (21–23).
Although the 13 human IFN-αs are very similar, other members of the type I IFN family demonstrate only limited similarity. For instance, the latest additions to the family, IFN-κ and IFN-ε, share <30% of aa identity with IFN-αs (SI Fig. 5 C and D). The similarity between type I and type III IFNs is even lower and ranges from 15% to 20% amino acid identity. Therefore, based on simple sequence comparison of receptors and ligands, it is not possible to predict whether Y136 (or B18) would neutralize type I and type III IFNs.
Y136 Protein Inhibits Type I IFNs from Primates but Not Rodents.
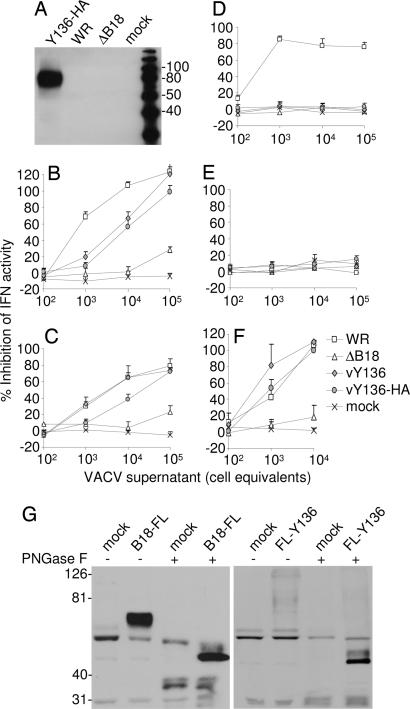
Initially, we observed that the supernatant from owl monkey kidney cells infected with YLDV contained an inhibitor of human IFN-α2 that was not present in the supernatant of mock-infected cells (data not shown). To determine whether the Y136R gene encoded this activity, the gene was expressed from recombinant VACV vAA6 (21), a VACV strain lacking the B18R gene (ΔB18). The Y136 protein was expressed with or without a C-terminal HA tag, and the recombinant viruses were called vY136 and vY136-HA. Immunoblotting showed that the supernatants of vY136-HA-infected cells contained a secreted protein of ≈80 kDa that was absent from controls (Fig. 1A). The size of the protein was greater than that of B18 (60–65 kDa) (21) because it contains 12 sites (N-X(except P)-T/S) for attachment of N-linked carbohydrate compared with five sites in B18. Consistent with Y136 being glycosylated, its secretion from infected cells was blocked by the glycosylation inhibitor tunicamycin (data not shown).
Fig. 1.
Y136 is a secreted glycoprotein that inhibits primate type I IFNs. (A) BS-C-1 cells were infected with the indicated VACVs, and the proteins in conditioned medium were analyzed by immunoblotting with HA mAb. (B–F) Different amounts of conditioned medium from cells infected with the indicated viruses were mixed with human IFN-α (B), human IFN-β (C), mouse IFN-α (D), mouse IFN-β (E), or rhesus monkey IFN-α (F), and the ability of the mixture to inhibit plaque formation by Cocal virus was determined as described in Methods. Data are expressed as the percentage inhibition of IFN antiviral activity from duplicate experiments. (G) The proteins in conditioned medium from COS cells (mock) and COS cells transfected with plasmids pEF-B18R-FL (B18-FL), and pEF-SPFL-Y136 (FL-Y136) were immunoprecipitated with FLAG antibody and analyzed by immunoblotting with FLAG mAb. Before immunoblotting, some samples were treated with PNGase F (+) to remove N-linked carbohydrates. The molecular mass markers are shown in kiloDaltons (A and G).
To determine whether Y136 would inhibit type I IFNs, different amounts of conditioned supernatant from VACV-infected cells were mixed with human IFN-α2 (Fig. 1B) or IFN-β (Fig. 1C), and the ability of the mixture to block plaque formation by Cocal virus was determined on HeLa cells. Y136, with or without a C-terminal HA tag, inhibited the antiviral activity of both human IFNs. As expected, the parental virus vAA6 and mock-infected cells did not express a secreted type I IFN inhibitor. Next, we tested the activity of Y136 against rodent IFNs and found that Y136 was unable to inhibit mouse IFN-α (Fig. 1D), mouse IFN-β (Fig. 1E), or rat IFN-α (data not shown). In contrast, the VACV B18 protein inhibited mouse IFN-α but not IFN-β (21, 27), indicating its broader species specificity. Y136 also inhibited rhesus monkey IFN-α (Fig. 1F) at least as well as did B18. Thus, Y136 is a soluble inhibitor of primate type I IFNs but did not inhibit rodent type I IFNs, and this specificity is consistent with the fact that YLDV was derived from primates (28).
We then generated recombinant VACV B18 and YLDV Y136 proteins from uninfected mammalian cells to investigate comprehensively whether these viral proteins can block the activity of various IFNs. The B18R and Y136R genes were cloned into mammalian expression vectors, which enabled a FLAG epitope to be fused to the viral protein at either the C terminus (B18-FL and Y136-FL) or the N terminus (FL-B18 and FL-Y136). COS-1 cells were transfected with the plasmids, and 3 days later, conditioned media were collected and analyzed by immunoblotting (Fig. 1G and data not shown). Plasmids producing the highest amounts of secreted proteins, pEF-B18-FL and pEF-SPFL-Y136, were selected for further analyses and biological assays. Immunoblotting revealed that B18-FL and FL-Y136 were secreted from COS cells with sizes of ≈60–65 kDa and 70 kDa, respectively (Fig. 1G). These results are in accord with previously published data for B18 protein (21) and with results observed for Y136 secreted from vY136-HA-infected cells (Fig. 1A). Treatment with peptide N-glycosidase F (PNGase F) reduced the apparent molecular masses of these proteins to ≈50 kDa for B18 and 45 kDa for Y136, confirming that they are glycosylated.
IFN-κ and IFN-ε Signal Through Canonical Type I IFN Receptor Complex.
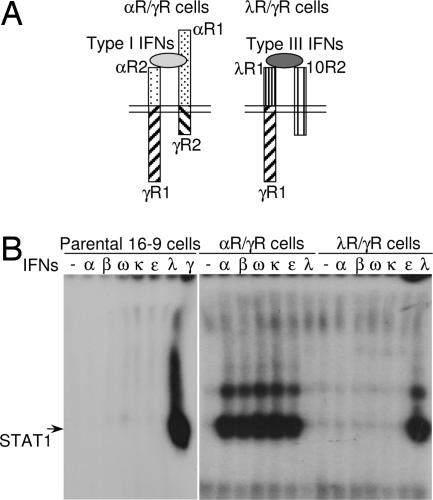
To characterize IL-10, IL-22, and IFN-λ receptor complexes, we had created a series of reporter hamster cell lines that respond to these human cytokines specifically (6, 29, 30). Cytokines demonstrate various degrees of species specificity. Hamster cells are not responsive to human IFNs and IL-10-related cytokines (Fig. 2 and refs. 6, 29, and 30). Therefore, appropriate human receptor subunits must be expressed in hamster cells to render them responsive to a given human cytokine. One receptor subunit in each receptor complex determines signal transduction specificity (26). When the natural intracellular domain of a signaling receptor subunit is replaced by the IFN-γR1 intracellular domain in a reconstituted functional receptor complex for a particular cytokine, this cytokine induces IFN-γ-like signaling and biological activities that can be uniformly measured. This approach allowed us to generate hamster cell lines that signal specifically in response to a single human cytokine and to more easily monitor signaling of cytokines, such as IFN-λs, which induce weak signaling in intact cells because of the low level of receptor expression.
Fig. 2.
IFN-κ and IFN-ε signal through canonical type I IFN receptor complex. (A) Two hamster cell lines expressing modified human type I and type III IFN receptor complexes are shown schematically. The αR/γR cells (Right) express chimeric IFN-αR2/IFN-γR1 (αR2/γR1) and IFN-αR1/IFN-γR2 (αR1/γR2) receptor chains; and λR/γR cells (Left) express chimeric IFN-λR1/IFN-γR1 (λR1/γR1) and intact IL-10R2 chains. (B) The response of the parental, αR/γR and λR/γR hamster cells to various type I and type III IFNs was evaluated by measuring IFN-induced STAT1 activation in EMSA. The cells were left untreated or treated with various stimuli: recombinant E. coli-produced human IFN-α2 (α; 1,000 units/ml = 4 ng/ml) and IFN-λ1 (λ; 4 ng/ml), or conditioned medium from COS cells transfected with plasmids encoding either human IFN-β (β), IFN-ω (ω), IFN-κ (κ), IFN-ε (ε) or hamster IFN-γ (γ). Each type I IFN was used at a concentration of 1,000 IFN-α-equivalent units/ml, as determined by their antiviral potency on human cells and the ability to induce STAT activation in comparison with standard IFN-α2.
Therefore, to detect signaling in response to either type I or type III human IFNs, we used hamster cells expressing human IFN-λR1/IFN-γR1 and IL-10R2 chains (6) (Fig. 2A) and created hamster cells responsive to human type I IFNs. These hamster cells express human chimeric IFN-αR2/IFN-γR1 and IFN-αR1/IFN-γR2 chains that were generated by replacing the intracellular and transmembrane domains of IFN-αR2 and IFN-αR1 by the corresponding domains of IFN-γR1 and IFN-γR2, respectively (Fig. 2A). Expression of receptors was confirmed by flow cytometry (SI Fig. 6A). Hamster cells expressing modified human type I and type III IFN receptor complexes were designated αR/γR and λR/γR cells, respectively. The ability of these cells to respond to various type I and type III IFNs was tested by measuring cytokine-induced STAT1 activation in electrophoretic mobility shift assay (EMSA).
Parental hamster cells were unresponsive to either type I or type III human IFNs (Fig. 2B). All type III IFNs were able to activate STAT1 only in λR/γR cells and not in αR/γR cells (Fig. 2B and SI Fig. 6B and data not shown), confirming that type III IFNs signal through a unique receptor complex composed of IFN-λR1 and IL-10R2 and do not cross-react with the type I IFN receptor complex. To obtain human IFN-β, IFN-ω, IFN-κ, and IFN-ε, their genes were cloned and expressed in COS-1 cells. The relative amounts of IFNs in COS cell-conditioned media were determined in IFN-α-equivalent units per milliliter based on antiviral assays (Fig. 3B and data not shown) in comparison with antiviral potency of recombinant Escherichia coli-produced IFN-α2 in similar assays. IFN-α-equivalent units, determined in antiviral assays, correlated very well with STAT1-inducing ability of various IFNs in EMSA (Fig. 2B, SI Fig. 6B, and data not shown). Recombinant IFN-α2 and COS cell-produced IFN-β, IFN-ω, IFN-κ and IFN-ε were used to demonstrate that all type I IFNs, including IFN-κ and IFN-ε, signal through the canonical type I IFN receptor complex composed of IFN-αR1 and IFN-αR2. None of the type I IFNs was able to induce signaling through the type III IFN receptor complex (Fig. 2B). That the recently identified IFN-κ and IFN-ε signal through the same receptor complex as all of the other type I IFNs has not been demonstrated previously.
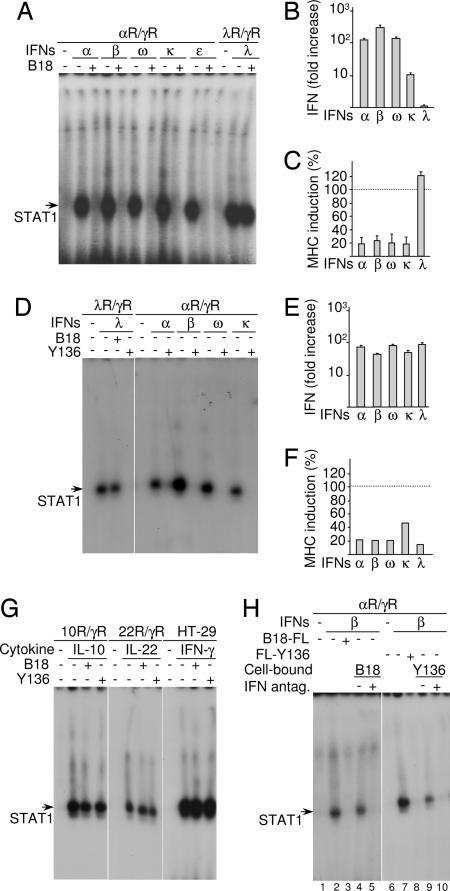
Fig. 3.
Effects of B18 and Y136 proteins on IFN signaling and biological activities. (A and D) αR/γR and λR/γR cells were left untreated or treated with 1,000 units/ml of various type I IFNs [recombinant IFN-α2 (α) and COS cell produced IFN-β (β), IFN-ω (ω), IFN-κ (κ) and IFN-ε (ε)] or recombinant IFN-λ1 (λ; 4 ng/ml) with or without COS cell-conditioned medium containing B18-FL or FL-136 protein. STAT1 activation in cells was then evaluated by EMSA. (B and E) Antiviral protection in response to type I and type III IFNs with or without B18 or Y136 was evaluated on HT-29 cells infected with VSV with a cytopathic effect (CPE) reduction assay. The ordinate represents the fold increase of the amount of IFNs required to achieve 50% protection against virus-induced CPE in the presence of B18 versus in its absence. (C and F) The inhibition by B18 or Y136 of IFN-induced MHC class I antigen expression in HT-29 cells was evaluated by flow cytometry. The level of IFN-induced MHC class I antigen expression in the presence of B18 or Y136 is shown as a percentage of the IFN-induced (without B18 and Y136; type I IFNs, 1,000 units/ml; IFN-λ1, 4 ng/ml) over the basal level of MHC class I antigen expression. (G) HT-29, 10R/γR and 22R/γR cells were left untreated or treated with 4 ng/ml of recombinant IFN-γ, IL-10 and IL-22, respectively, with or without COS cell-conditioned medium containing either B18-FL or FL-Y136 protein, and STAT1 activation in cells was evaluated by EMSA. (H) COS cells were transfected with expression plasmids for either B18 (cell-bound B18, +, lane 5) or Y136 (cell-bound Y136, +, lane 10) or were mock transfected (cell-bound IFN antagonists, −, lanes 4 and 9) and were incubated with IFN-β (1,000 units/ml, β, lanes 4, 5, 9, and 10) in 100 μl of medium for 1 h. COS cells were removed by centrifugation, and cell-free supernatants were used to treat αR/γR cells. In control assays, αR/γR cells were left untreated (lanes 1 and 6) or treated with IFN-β (1,000 units/ml, β, lanes 2, 3, 7, and 8) with (lanes 3 and 8) or without (lanes 2 and 7) soluble COS cell produced B18-FL or FL-Y136 and STAT1 activation in cells was evaluated by EMSA.
VACV B18 Is a Specific Antagonist of all Human Type I IFNs and Not Type III IFNs.
We used λR/γR and αR/γR reporter cell lines to evaluate whether B18 protein can inhibit signaling induced by either all type I IFNs, including IFN-κ and IFN-ε, or type III IFNs. The λR/γR and αR/γR cells were treated by IFN-λs, and IFN-α2, IFN-β, IFN-ω, IFN-κ, and IFN-ε, respectively, with or without B18 protein (Fig. 3A). We found that B18 blocked the ability of all type I IFNs to induce STAT1 activation in αR/γR cells. In contrast, type III IFN signaling was not affected by B18 protein. Therefore, B18 inhibited signaling induced by all human type I IFNs, but not by type III IFNs (Fig. 3A).
Next, we determined whether B18 can inhibit the antiviral activities of a broad range of IFNs on colorectal adenocarcinoma (HT-29) cells that respond to both type I and type III IFNs (6). The ability of various IFNs to protect HT-29 cells against infection by Vesicular stomatitis virus (VSV) was measured in the presence or absence of B18 (Fig. 3B) as the reduction of virus-mediated cytopathic effect (CPE). Type I and type III IFNs demonstrate comparable antiviral potency (107 to 108 units/mg) against VSV in HT29 cells (6). The antiviral activity of all type I IFNs was inhibited strongly by B18. In the presence of B18, much higher amounts of type I IFNs were required to overcome the neutralizing effect of B18 and achieve 50% protection of the cells from CPE (Fig. 3B). Noticeably, B18 had different neutralizing activity against different type I IFNs. It had greatest neutralizing ability toward IFN-β, which is the earliest IFN produced by cells in response to viral infection (16). We also found that type III IFNs were not affected by B18 (Fig. 3B).
The effect of B18 on IFN activity was also examined by measurement of IFN-mediated induction of MHC class I antigen expression on HT-29 cells by flow cytometry. The up-regulation of MHC class I antigen expression in response to type I but not type III IFNs was reduced by B18 (Fig. 3C).
Y136 Neutralizes Signaling and Biological Activities of both Type I and Type III IFNs.
Considering the limited similarity between B18 and Y136 (27% amino acid identity), we performed a similar series of experiments with Y136. As shown in Fig. 3D, Y136 completely neutralized the ability of both type I and type III IFNs to activate STAT1 in reporter cell lines. In agreement with its ability to block IFN signaling, Y136 also inhibited antiviral protection and up-regulation of MHC class I antigen expression in HT-29 cells in response to both type I and type III IFNs (Fig. 3 E and F).
Because type III IFNs reveal a similar degree of amino acid identity to both type I IFNs and IL-10-related cytokines, we investigated whether viral receptors cross-react with IL-10-related cytokines. With the use of 10R/γR and 22R/γR reporter cell lines that respond to human IL-10 and IL-22, respectively (29, 30), we demonstrated that Y136 and B18 did not inhibit IL-10 or IL-22 signaling (Fig. 3G). Neither viral protein suppressed IFN-γ signaling in HT-29 cells (Fig. 3G).
Because B18 is present on the cell surface and in solution (24, 31), we investigated whether COS cells expressing B18 or Y136 retain biologically active proteins on the cell surface (Fig. 3H). These cells were washed to remove soluble IFN-binding proteins, and then incubated with IFN-β. The medium was then harvested and used in EMSA with reporter cell lines to determine whether the COS cells had removed IFN-β from the supernatant. If IFN-β was still present in the medium it would induce STAT1 activation. Parental untreated COS cells were used as a control. As shown in Fig. 3H, COS cells expressing B18 or Y136 sequestered IFN-β from the medium, whereas parental COS cells did not. These experiments demonstrated that some B18 and Y136 is retained on the cell surface, whereas some is also secreted.
Type I and Type III IFNs Compete for Binding to Y136.
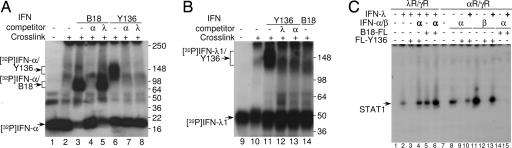
Next, we characterized the interaction of IFNs with B18 and Y136 by covalent cross-linking (Fig. 4 A and B). Radiolabeled IFN-α2-P (32) and His-Strep-IFN-λ1-P were cross-linked to either B18 or Y136 in solution with or without an excess of unlabeled (cold) IFNs as competitors, and the cross-linked complexes were analyzed. The major radiolabeled bands of ≈20 kDa (Fig. 4A) and ≈45 kDa (Fig. 4B) correspond to free IFN-α2-P and His-Strep-IFN-λ1-P, respectively. These bands did not change upon cross-linking, demonstrating that IFN-α2 and IFN-λ1 are monomers in solution. Cross-linking of radiolabeled IFN-α2-P to B18 resulted in the appearance of additional complexes of ≈70–90 kDa (Fig. 4A), whereas the presence of B18R protein did not change the pattern of cross-linking of radiolabeled His-Strep-IFN-λ1-P (Fig. 4B). Addition of excess cold type I IFN, but not type III IFN, competed with radiolabeled IFN-α2-P for binding to B18, demonstrating the specificity of the interaction (Fig. 4A).
Fig. 4.
Interaction of viral IFN antagonists with IFNs. (A and B) Untreated [32P]-IFN-α2-P and [32P]-His-Strep-IFN-λ1-P were loaded as controls (lanes 1 and 9). [32P]-labeled IFNs were cross-linked in solution in the absence of viral receptors (lanes 2 and 10) and to either B18-FL or FL-Y136 proteins with (lanes 4, 5, 7, 8, 12, and 13) or without (lanes 3, 5, 11, and 14) addition of a 100-fold excess of unlabeled competitor IFN-α2 or IFN-λ1, as indicated. The cross-linked complexes were analyzed by SDS/PAGE. Positions of molecular mass markers are shown on the right. (C) λR/γR and αR/γR cells were left untreated (lanes 1 and 7) or treated with IFN-λ1 (4 ng/ml, lanes 2–6) or IFN-α2 (α, lanes 8–10, 14 and 15) and IFN-β (β, lanes 11–13), respectively, with (+) or without (−) COS cell produced FL-Y136 or B18-FL protein (100 μl). Where indicated (bold letters) the excess of IFN-α2 (10,000 units/ml, α, lanes 4 and 6) was added to λR/γR cells, or the excess of IFN-λ1 (100 ng/ml, +, lanes 10, 13 and 15) was added to αR/γR cells. IFN-induced STAT1 activation in cells was evaluated by EMSA.
In contrast, the incubation of Y136 with either [32P]-IFN-α2-P or [32P]-His-Strep-IFN-λ1-P, followed by covalent cross-linking resulted in formation of complexes of ≈100–160 kDa for [32P]-IFN-α2-P (Fig. 4A) and ≈130–190 kDa for [32P]-His-Strep-IFN-λ1-P (Fig. 4B). An excess of either unlabeled type I or type III IFN inhibited formation of these complexes, demonstrating that Y136 protein interacts with both types of IFNs in a competitive manner (Fig. 4 A and B).
Similarly, immunoprecipitation of complexes containing viral receptors and radiolabeled IFN-α with FLAG mAb demonstrated that all type I IFNs, and not type III IFNs, competed with IFN-α for binding to B18-FL (SI Fig. 7A). In contrast, both type I and type III IFNs competed with IFN-α for binding to FL-Y136 protein (SI Fig. 7B). Significantly, B18 also competed with FL-Y136 for binding with radiolabeled IFN-α (SI Fig. 7B).
Binding competition of ligands and receptors was also demonstrated by EMSA (Fig. 4C). An excess of IFN-α sequestered FL-Y136 and consequently restored signaling by type III IFNs in λR/γR cells. Similarly, an excess of IFN-λ1 bound FL-Y136 and thereby inhibited its ability to neutralize IFN-α and IFN-β signaling in αR/γR cells. However, an excess of IFN-λ1 did not prevent B18-FL inhibiting IFN-α signaling in αR/γR cells.
Discussion
Recently, a new type of IFN, designated type III IFN or IFN-λ, was discovered and demonstrated to possess intrinsic antiviral activity, similar to those of type I IFNs. Type III IFNs are effective against several viruses in epithelium-like cells expressing type III IFN receptors (3, 6, 7). Moreover, expression of mouse type III IFN by VACV caused dramatic virus attenuation in mice (15), showing that these type III IFNs can be important in vivo and might be used for the treatment of poxvirus infections. Because VACV expresses B18, a type I IFN antagonist, these experiments also demonstrated that, in the presence of type III IFNs, inhibition of only type I IFNs was inadequate for efficient virus propagation in vivo. Therefore, strategies to neutralize the activity of type III IFNs are important and biologically relevant for poxviruses and should provide survival advantage in the host. Nevertheless, the functional significance of type III IFNs against many viruses and their relative importance compared with type I IFNs remains largely uncharacterized. Hitherto, no specific virus defense mechanism targeting type III IFNs was known.
Here, we demonstrate that the poorly characterized IFN-κ and IFN-ε signal through the canonical type I IFN receptor complex (Fig. 2). Furthermore, in addition to those type I IFNs investigated previously (21–24), we demonstrate that VACV B18 also inhibits IFN-κ and IFN-ε (Fig. 3 and SI Fig. 7). B18 bound all type I IFNs and blocked their signaling and biological activities such as antiviral protection and up-regulation of MHC class I antigen expression, demonstrating that B18 is a specific antagonist of all human type I IFNs. However, B18 was unable to interact with type III IFNs and had no effect on their signaling and biological activities (Figs. 3 and 4 and SI Fig. 7), suggesting that type III IFNs may be more potent for the treatment of certain poxvirus infections.
In contrast, the Y136 protein from YLDV not only neutralized all human type I IFNs but also acted as an antagonist of type III IFNs. Y136 interacted with all type I and type III IFNs and neutralized their ability to induce signal transduction and biological activities in IFN-responsive cells (Figs. 3 and 4 and SI Fig. 7). Although type I and type III IFNs demonstrate only 15–20% amino acid identity (see uppercase letters in consensus sequence in SI Fig. 5D) and use distinct receptor complexes, they competed for binding to Y136. Y136 differs substantially from B18 (SI Fig. 5 A and B) and so how Y136 binds ligands from two very distantly related families is unclear and needs further investigation. The ability of YLDV to inhibit type III IFNs as well as type I IFNs is interesting because infections caused by Yatapoxviruses are restricted to the dermis (28), where type III IFN receptors are expressed. Orthopoxviruses, in contrast, may cause systemic infections.
Another difference between B18 and Y136 was the species specificity of the type I IFNs that these virus proteins bound and inhibited. Whereas B18 can inhibit a broad range of IFNs including mouse IFN-α, Y136 inhibited only primate and not rodent type I IFNs (Fig. 1). This specificity fits with the host range of Yatapoxviruses being restricted to primates (28) whereas several Orthopoxviruses, such as Ectromelia virus, Monkeypox virus, Cowpox virus, and probably VACV, infect rodents.
Although all three types of IFNs and IL-10-related cytokines belong to the same cytokine family (CRF2 cytokine family) and share limited primary and structural similarity (26), B18 and Y136 proteins did not inhibit the actions of other CRF2 cytokines, such as type II IFN (IFN-γ), IL-10 or IL-22 (Fig. 3G).
B18 is secreted from infected cells but is also present on the cell surface where it can protect uninfected cells from type I IFNs (22, 24, 31). Similarly, we demonstrated that cells expressing Y136 retained some of the viral protein on the cell surface (Fig. 3H), and this cell surface protein still acted as an efficient IFN antagonist. The ability of both viral proteins to exist as both soluble and cell-surface forms provides a very effective mechanism to inhibit IFN activities in a localized infected area. Cells invaded by a virus produce type I and type III IFNs that activate neighboring cells, making these resistant to subsequent virus infection. However, cells infected with VACV or YLDV produce IFN antagonists that, after release, may bind to both infected and neighboring uninfected cells to protect these from IFN. Thus, virus spread is unhindered by these IFNs. Importantly, Y136 and B18 have strong neutralizing capabilities toward IFN-β (Fig. 3), the first IFN produced by virus-infected cells (16).
In conclusion, our study provides a previously uncharacterized defense mechanism from poxviruses to circumvent the antiviral activity of host type III IFNs. We demonstrated that YLDV protein Y136 inhibits both type I IFN and type III IFNs. In contrast, VACV B18 inhibited activities of type I, and not type III, IFNs. In addition, the fact that some viruses acquired strategies to inhibit type III IFNs underscores the importance of these cytokines for antiviral protection. Further studies are required to determine the biological significance of inhibiting type III IFNs for the pathogenesis and life cycle of YLDV and whether other poxviruses possess functional Y136 orthologues. Nevertheless, data presented here and the previous demonstration that expression of type III IFNs from VACV caused a dramatic reduction in virulence (15), suggest that type III IFNs may be potent reagents for treating some poxvirus infections.
Materials and Methods
Construction of Plasmids and Recombinant VACVs.
Several mammalian expression plasmids were created to produce recombinant B18, Y136, IFN-β, IFN-ω, IFN-κ, IFN-ε and His-Strep-IFN-λ1-P proteins and to express αR1/γR2 and αR2/γR1 receptors (see SI Methods for details).
Recombinant vY136 and vY136-HA VACVs encoding Y136 and Y136-HA were generated by introducing the Y136R gene into VACV genome (SI Methods).
Transfection and Flow Cytometry.
COS-1 cells, SV40 transformed fibroblast-like simian CV-1 cells, were transfected as described (6), and conditioned media (supernatants) were collected at 72 h and used as a source of the expressed proteins. Chinese hamster ovary 16-9 cells, containing a transfected human HLA-B7 gene, were transfected as described (6).
To detect changes in MHC class I antigen expression, human colorectal adenocarcinoma HT-29 cells were treated with IFNs, and their MHC expression was analyzed by flow cytometry as described (6). COS cell supernatants containing B18 or Y136 (100 μl/2 ml) were used to inhibit IFNs.
Immunoprecipitation, Immunoblotting, and EMSA.
COS cell supernatants (1 ml) were treated with FLAG M2 mAb (1 μg; Sigma, St. Louis, MO) and protein A/G-Agarose beads (12 μl; Santa Cruz Biotechnology, Santa Cruz, CA) at 4°C for 16 h, and precipitates were separated by SDS/PAGE and analyzed by immunoblotting with FLAG mAb. N-Glycosidase (PNGase F, 1 μl; New England Biolabs, Beverly, MA) was added where indicated.
Similarly, the supernatants of BS-C-1 cells infected with the indicated VACVs at 5 plaque-forming units (pfu) per cell for 18 h were collected and analyzed by SDS/PAGE and immunoblotting with HA mAb.
To detect STAT1 activation, cells were treated with COS cell supernatants or purified recombinant proteins (IFN-α2, IFN-γ, IL-10, and IL-22; PeproTech, Rocky Hill, NJ) for 15 min and used for EMSAs with the γ-activated sequence (GAS) probe as described (6). For neutralizing experiments, IFNs were preincubated with B18 or Y136 (COS cell supernatants, 100 μl) for 1 h at 22°C.
Virus Infection, Antiviral Protection, and IFN Inhibition Assays.
Antiviral assays were performed as described (6). An equal number of HT-29 cells was plated in wells of 96-well microtiter plates and treated with 2-fold serial dilutions of IFNs for 24 h. COS cell supernatants containing B18 or Y136 proteins (50 μl in 250 μl/well) were used in selected wells. Twenty-four hours later, the cells were challenged with VSV and incubated further until controls showed full killing by virus. Cells not killed were visualized by staining with crystal violet.
BS-C-1 cells were mock-infected or infected at 5 pfu per cell with VACV strain vAA6 (ΔB18), vY136, or vY136-HA for 24 h. The supernatants were collected, and virions were removed by centrifugation and filtration of the resulting supernatants through a 0.1 μm filter. The filtrate was then tested for inhibition of various IFNs by using Cocal virus plaque formation assay as described (21). Rhesus monkey IFN-α was assayed on BS-C-1 cells. Mouse IFN-α, mouse IFN-β and rat IFN-α were assayed on mouse L929 cells.
Cross-Linking.
IFN-α2-P was created as described. (32). His-Strep-IFN-λ1-P was expressed in COS cells and purified by affinity chromatography (IBA, Göttingen, Germany). The proteins were labeled with [32P]ATP and used for cross-linking as reported (6).
Supplementary Material
Acknowledgments
We thank J. Langer, A. Zdanov, and A. Lasfar for helpful suggestions. This work was supported in part by The Wellcome Trust, the U.K. Department of Heath, U.S. Public Health Services Grants R01 AI051139 and AI057468 from the National Institute of Allergy and Infectious Disease to S.V.K. G.L.S. is a Wellcome Trust Principal Research Fellow.
Abbreviations
- YLDV
Yaba-like disease virus
- CRF2
class II cytokine receptor family.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0610352104/DC1.
References
- 1.LaFleur DW, Nardelli B, Tsareva T, Mather D, Feng P, Semenuk M, Taylor K, Buergin M, Chinchilla D, Roshke V, et al. J Biol Chem. 2001;276:39765–39771. doi: 10.1074/jbc.M102502200. [DOI] [PubMed] [Google Scholar]
- 2.Chen J, Wood WI. 6300475. Interferon PRO655 U.S. Patent. 2003:1–37.
- 3.Kotenko SV, Langer JA. Int Immunopharmacol. 2004;4:593–608. doi: 10.1016/j.intimp.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 4.Pestka S, Krause CD, Walter MR. Immunol Rev. 2004;202:8–32. doi: 10.1111/j.0105-2896.2004.00204.x. [DOI] [PubMed] [Google Scholar]
- 5.Novelli F, Casanova JL. Cytokine Growth Factor Rev. 2004;15:367–377. doi: 10.1016/j.cytogfr.2004.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, Shah NK, Langer JA, Sheikh F, Dickensheets H, Donnelly RP. Nat Immunol. 2003;4:69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- 7.Sheppard P, Kindsvogel W, Xu W, Henderson K, Schlutsmeyer S, Whitmore TE, Kuestner R, Garrigues U, Birks C, Roraback J, et al. Nat Immunol. 2003;4:63–68. doi: 10.1038/ni873. [DOI] [PubMed] [Google Scholar]
- 8.Ank N, West H, Paludan SR. J Interferon Cytokine Res. 2006;26:373–379. doi: 10.1089/jir.2006.26.373. [DOI] [PubMed] [Google Scholar]
- 9.Kotenko SV, Donnelly RP. In: The Interferons: Characterization and Application. Meager A, editor. Weinheim, Germany: Wiley-VCH; 2006. pp. 141–163. [Google Scholar]
- 10.Dumoutier L, Tounsi A, Michiels T, Sommereyns C, Kotenko SV, Renauld JC. J Biol Chem. 2004;279:32269–32274. doi: 10.1074/jbc.M404789200. [DOI] [PubMed] [Google Scholar]
- 11.Doyle SE, Schreckhise H, Khuu-Duong K, Henderson K, Rosler R, Storey H, Yao L, Liu H, Barahmand-pour F, Sivakumar P, et al. Hepatology. 2006;44:896–906. doi: 10.1002/hep.21312. [DOI] [PubMed] [Google Scholar]
- 12.Marcello T, Grakoui A, Barba-Spaeth G, Machlin ES, Kotenko SV, MacDonald MR, Rice CM. Gastroenterology. 2006;131:1887–1898. doi: 10.1053/j.gastro.2006.09.052. [DOI] [PubMed] [Google Scholar]
- 13.Lasfar A, Lewis-Antes A, Smirnov SV, Anantha S, Abushahba W, Tian B, Reuhl K, Dickensheets H, Sheikh F, Donnelly RP, et al. Cancer Res. 2006;66:4468–4477. doi: 10.1158/0008-5472.CAN-05-3653. [DOI] [PubMed] [Google Scholar]
- 14.Ank N, West H, Bartholdy C, Eriksson K, Thomsen AR, Paludan SR. J Virol. 2006;80:4501–4509. doi: 10.1128/JVI.80.9.4501-4509.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartlett NW, Buttigieg K, Kotenko SV, Smith GL. J Gen Virol. 2005;86:1589–1596. doi: 10.1099/vir.0.80904-0. [DOI] [PubMed] [Google Scholar]
- 16.Takaoka A, Yanai H. Cell Microbiol. 2006;8:907–922. doi: 10.1111/j.1462-5822.2006.00716.x. [DOI] [PubMed] [Google Scholar]
- 17.Haller O, Kochs G, Weber F. Virology. 2006;344:119–130. doi: 10.1016/j.virol.2005.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moss B. In: Fields Virology. Knipe DM, Howley PM, editors. Vol. 2. Philadelphia: Lippencott–Raven; 2001. pp. 2849–2883. [Google Scholar]
- 19.Alcami A, Smith GL. J Virol. 1995;69:4633–4639. doi: 10.1128/jvi.69.8.4633-4639.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mossman K, Upton C, Buller RM, McFadden G. Virology. 1995;208:762–769. doi: 10.1006/viro.1995.1208. [DOI] [PubMed] [Google Scholar]
- 21.Symons JA, Alcami A, Smith GL. Cell. 1995;81:551–560. doi: 10.1016/0092-8674(95)90076-4. [DOI] [PubMed] [Google Scholar]
- 22.Colamonici OR, Domanski P, Sweitzer SM, Larner A, Buller RM. J Biol Chem. 1995;270:15974–15978. doi: 10.1074/jbc.270.27.15974. [DOI] [PubMed] [Google Scholar]
- 23.Liptakova H, Kontsekova E, Alcami A, Smith GL, Kontsek P. Virology. 1997;232:86–90. doi: 10.1006/viro.1997.8527. [DOI] [PubMed] [Google Scholar]
- 24.Alcami A, Symons JA, Smith GL. J Virol. 2000;74:11230–11239. doi: 10.1128/jvi.74.23.11230-11239.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee HJ, Essani K, Smith GL. Virology. 2001;281:170–192. doi: 10.1006/viro.2000.0761. [DOI] [PubMed] [Google Scholar]
- 26.Kotenko SV. Cytokine Growth Factor Rev. 2002;13:223–240. doi: 10.1016/s1359-6101(02)00012-6. [DOI] [PubMed] [Google Scholar]
- 27.Smith VP, Alcami A. J Virol. 2002;76:1124–1134. doi: 10.1128/JVI.76.3.1124-1134.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith GL. In: Poxviruses. Mercr AA, Schmidt A, Weber O, editors. Basel: Burkhauser; 2007. pp. 113–125. [Google Scholar]
- 29.Kotenko SV, Krause CD, Izotova LS, Pollack BP, Wu W, Pestka S. EMBO J. 1997;16:5894–5903. doi: 10.1093/emboj/16.19.5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kotenko SV, Izotova LS, Mirochnitchenko OV, Esterova E, Dickensheets H, Donnelly RP, Pestka S. J Biol Chem. 2001;276:2725–2732. doi: 10.1074/jbc.M007837200. [DOI] [PubMed] [Google Scholar]
- 31.Morikawa S, Ueda Y. Virology. 1993;193:753–761. doi: 10.1006/viro.1993.1184. [DOI] [PubMed] [Google Scholar]
- 32.Li BL, Langer JA, Schwartz B, Pestka S. Proc Natl Acad Sci USA. 1989;86:558–562. doi: 10.1073/pnas.86.2.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.