Abstract
Nucleophosmin/B23 is a major multifunctional nucleolar phosphoprotein that plays a critical role in ribosome biogenesis and cell proliferation. Arf tumor suppressor binds B23 and enhances its sumoylation. However, the biological effects of this event remain unknown. Here we show that B23 is sumoylated on both Lysine 230 and 263 residues, but the latter is the major one. Mutation of K263, but not K230, into R abolishes its centrosomal and nucleolar residency. Moreover, Rb binds to wild-type B23, but fails to interact with K263R. Sumoylation enhances B23 binding to Rb. Consequently, B23 potently stimulates E2F1-mediated transcriptional activity, which is abolished in B23 K263R. Further, K263R mutation makes B23 vulnerable to caspase-3 cleavage and sensitizes cells to apoptosis. Surprisingly, K230R mutant strongly binds to phosphatidylinositol-3,4,5-trisphosphate and suppresses DNA fragmentation. Thus, B23 sumoylation regulates its subcellular localization, cell proliferation, and survival activities.
Keywords: centrosome, nucleolus, sumoylation, K263
Nucleophosmin (NPM)/B23 is a multifunctional protein involved in many cellular activities, and it has been attributed with both oncogenic and tumor-suppressive functions. B23 is more abundant in tumor and growing cells than in normal resting cells (1, 2). In addition, B23 is frequently found in the chromosomal translocation associated with several hematopoietic malignancies, such as acute promyelocytic leukemia (3), anaplastic large-cell lymphomas (4), and myelodysplasia/acute myeloid leukemia (5). Furthermore, certain mutations in the NPM gene incur the cytoplasmic localization of mutated B23, which has been proposed as a biomarker for certain acute myelogenous leukemia (6, 7). Overexpression of B23 induces cell cycle arrest in normal fibroblasts, whereas it promotes S phase entry in cells lacking p53. Conversely, knocking down B23 inhibits the processing of preribosomal RNA and induces cell death (8). In agreement with these observations, overexpression of B23 decreases the susceptibility of human leukemia HL-60 cells to retinoic acid-induced differentiation and apoptosis as well as UV-induced apoptosis in NIH 3T3 cells (9–11). Nonetheless, B23 can be cleaved by active caspase-3, which may influence its antiapoptotic action (12). It has been shown that B23 suppresses hypoxia or UV-induced cell death by repressing p53 phosphorylation and transcription activity (13, 14). Recently, we presented biochemical evidence revealing that B23 is a nuclear phosphatidylinositol-3,4,5-trisphosphate [PI(3,4,5)P3] receptor, and this complex directly interacts with caspase-activated DNase and inhibits its DNA fragmentation activity (15). Thus, these findings demonstrate that B23 acts as a physiologically important antiapoptotic protein. However, emerging evidence demonstrates that B23 can also act as a tumor suppressor. Npm germ-line deletion reveals that it is essential for embryonic development and the maintenance of genomic stability. Npm1 heterozygosity accelerates oncogenesis both in vitro and in vivo, supporting the fact that it has tumor-suppressive activity (16). Moreover, accumulating evidence indicates that B23 contributes to the growth-suppressing pathways through its interaction with Arf (8, 17).
Normally, the nucleolar proteins, including B23, ARF, and Mdm2, are caged in the nucleolus. They tightly bind to each other and neutralize their oncogenic and tumor-suppressive activities. In response to an elevated expression of oncogenes, ARF is up-regulated and induces apoptosis through stabilization of p53. The ability of ARF to block cell cycle progression through the MDM2–p53 pathway and suppress ribosomal biogenesis through B23 supports a role for ARF in coordinating inhibitions of growth and proliferation (18, 19). Arf can also block phosphorylation of the retinoblastoma protein by inhibiting CDK4 and CDK6, preventing exit from the G1 phase of the cell cycle (20). B23 binds Arf and stabilizes it by retarding its turnover. ARF binds B23 through the same domains that mediate nucleolar localization and Mdm2 binding, suggesting that B23 could control ARF localization and compete with Mdm2 for ARF association (21). Indeed, B23 knockdown markedly enhanced ARF–Mdm2 association and diminished ARF nucleolar localization. B23 overexpression antagonizes ARF function while increasing its nucleolar localization, suggesting that B23 inhibits ARF's p53-dependent activity by targeting it to nucleoli and impairing ARF–Mdm2 association (19). However, increasing levels of ARF after oncogenic stress promotes B23 degradation through ubiquitination and proteasome pathways and interferes with B23 nucleocytoplasmic shuttling (8, 22). Recently, it has been shown that B23 is sumoylated at only one site in Arf-null NIH 3T3 cells. Its sumoylation is increased on Arf induction, and multisumoylated species are detected (17). However, the sumoylation residues are unknown, and the physiological functions of this event remain elusive as well.
In the present study, we show that B23 is sumoylated on both K230 and K263 sites, but the latter is a predominant site. Mutation of K263 abrogates its subcellular distribution, and Arf appears to contribute to this effect. In addition, K263R mutation makes B23 susceptible to caspase-3 cleavage and decreases cell proliferation. Therefore, sumoylation of B23 mediates its physiological functions by regulating its subcellular residency and protein stability.
Results
Lysine 263 Is the Major Sumoylation Site on B23.
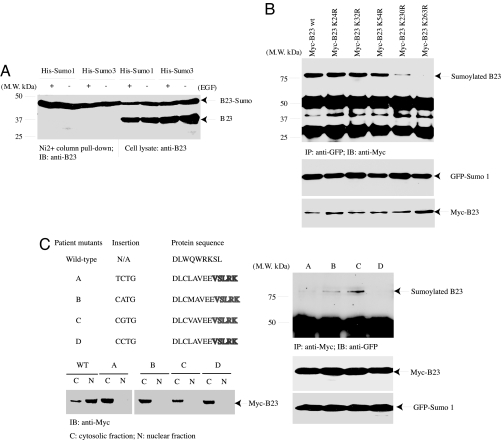
B23 is sumoylated in Arf-null NIH 3T3 cells, and its sumoylation is increased after Arf induction (17). To explore whether B23 can be sumoylated by both Sumo1 and Sumo3 isoforms, we transfected Myc-B23 into F293 cells, stably transfected with inducible His-Sumo1 and His-Sumo3, and induced His-tagged sumo expression with tetracycline. Nickel column pull-down assay reveals that B23 was robustly sumoylated by both isoforms regardless of EGF stimulation (Fig. 1A), suggesting that it can also be sumoylated in HEK293 cells. B23 can also be sumoylated by Sumo2 isoform as well (data not shown). Sumoylated lysine residues are often found within ΨKXE consensus sequences, where Ψ is an aliphatic residue and X is any amino acid. To determine which residues on B23 are sumoylated, we prepared a variety of mutants with K into R and cotransfected them into HEK293 cells with GFP-Sumo1. Immunoblotting analysis reveals that K263R mutation completely eliminates B23 sumoylation, whereas K230R partially decreases its sumoylation. K24, 32, or 54 mutation alone has no effect. Interestingly, they all tightly bound to GFP-Sumo1 regardless of the sumo modification (Fig. 1B).
Fig. 1.
K263 is the major sumoylation site on B23. (A) B23 is strongly sumoylated by Sumo1 and Sumo3. F293 cells were transfected with Myc-B23 and induced with tetracycline. The transfected cells were treated with EGF and pulled down with Nickel column. B23 was strongly sumoylated regardless of EGF stimulation. (B) K263 is the major sumoylation site. Myc-tagged B23 constructs were cotransfected into GFP-Sumo1, and the transfected proteins were immunoprecipitated with anti-GFP antibody. K263R mutation completely abolished B23 sumoylation, and K230R partially decreased its sumoylation. (C) Patient-derived B23 mutants reveal different sumoylation activities. B23 mutants with C-terminal 4 nucleotides insertion result in frame shift and encode the last 11 amino acids different from the wild-type seven residues (Upper Left). These mutants were exclusively located in the cytoplasmic but not nuclear fractions, whereas wild-type B23 mainly occurred in the nuclear fraction (Lower Left). Mutants B and C, but not A or D, were sumoylated (Right).
B23 is frequently found in the chromosomal translocation associated with several hematopoietic malignancies. Certain mutations in the NPM gene incur the cytoplasmic localization of mutated B23, which has been proposed as a biomarker for certain acute myelogenous leukemia (AML) (6, 7). For example, nucleotide insertion in patient-derived mutant A (956 to 959) in exon 12 of the C terminus of B23 elicits a frame shift by replacing the last seven amino acids (WQWRKSL) with 11 different residues (CLAVEEVSLRK). All these mutants share the same last five residues (VSLRK) and reside in the cytoplasm (Fig. 1C Left). To explore whether these patient-derived mutants can also be sumoylated, we performed a coimmunoprecipitation assay with cotransfected GFP-Sumo1 and myc-tagged B23 mutants. Immunoblotting reveals that mutants B and C were able to be sumoylated; by contrast, mutants A and D failed (Fig. 1C Right). Although mutants A and D possess different inserted nucleotides, they encode the proteins with identical sequence. Thus, B23 can be sumoylated regardless of its nuclear or cytoplasmic localization, and K263 is the major sumoylation residue. Presumably, K230 also contributes to sumoylation when B23 is multisumoylated.
K263 Sumoylation Is Essential for B23 Nucleolar Residency.
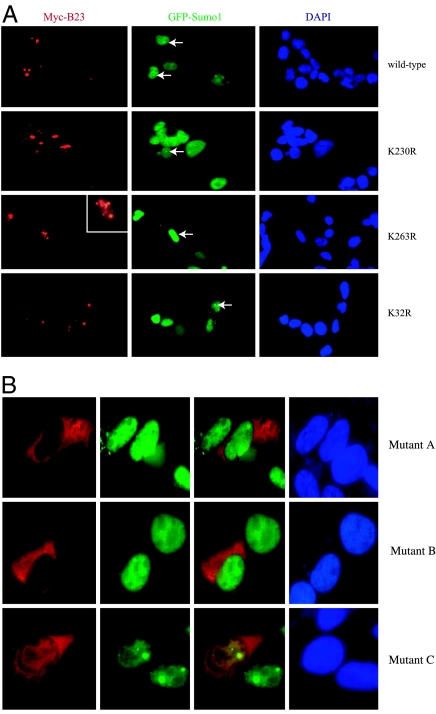
B23 mainly resides in the centrosome and the nucleolus. To determine whether sumoylation is required for its nucleolar distribution, we cotransfected Myc-B23 wild-type and mutants into HEK293 cells with GFP-Sumo1 and conducted immunofluorescent staining with anti-myc antibody and DAPI. As expected, wild-type B23, K24R, and K32R all occurred in the nucleolus (normally with three to four in number), and transfected GFP-Sumo1 was also concentrated in the nucleolus (white arrow), whereas K263R mutant was dispersed both inside and outside the nucleolus with >8 to 10 aggregates, and GFP-Sumo1 was distributed in the whole nucleoplasm (white arrow). Notably, the nuclear distribution of K230R mutant was somehow between wild-type B23 and K263R mutant, fitting with its crippled sumoylation effect (Fig. 2A). Therefore, B23 sumoylation is required for its nucleolar residency. Although single-transfected, patient-derived B23 mutants mainly reside in the cytoplasm (Figs. 1C and 2B Top), B23 mutant C distributed in both the cytoplasm and nucleus when cotransfected with GFP-Sumo1. However, mutants A and B remained in the cytoplasm regardless of GFP-Sumo1 (Fig. 2B Middle and Bottom).
Fig. 2.
K263 is essential for B23 nucleolar residency. (A) A variety of Myc-B23 constructs were cotransfected into HEK293 cells with GFP-Sumo1. The transfected cells were stained with anti-myc antibody (red) and DAPI (blue). GFP-Sumo1 specifically distributes in the nucleolus, colocalized with wild-type B23 and B23 K32R (Top and Bottom). B23 K230R mutant mainly distributed in the nucleolus, but it also aggregated in some of the transfected cells (Upper Middle). B23 K263R coagulated in the nucleus, whereas the cotransfected GFP-Sumo1 evenly dispersed in the whole nucleus (Lower Middle). (B) Patient-derived B23 mutants A and B predominantly occurred in the cytoplasm, whereas mutant C distributed in both the nucleus and cytoplasm.
Sumoylation on K263 Is Required for B23 to Localize to the Centrosomes.
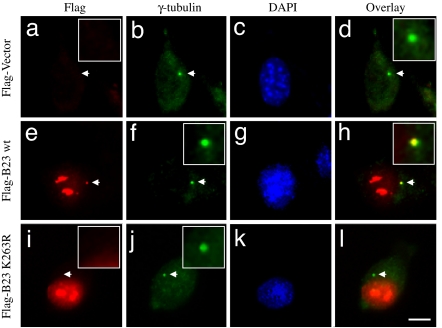
The centrosome consists of paired centrioles and many different proteins surrounding the paired centrioles. It has previously been shown that B23 localizes to centrosomes (23), in particular between the paired centrioles in unduplicated centrosomes (24), likely being involved in the pairing of centrosomes. Because the initial event of centrosome duplication is physical separation of the paired centrioles, centrosomally localized B23 suppresses centrosome duplication. Indeed, partial depletion of B23 (i.e., B23 hypomorphic mice) results in abnormal amplification of centrosomes (16). To test whether sumoylation affects B23 to localize to centrosomes, FLAG-tagged wild-type B23, K230R, and K263R were transiently transfected into NIH 3T3 cells. A vector plasmid was transfected as a control. After 24 h posttransfection, cells were coimmunostained with anti-γ-tubulin (centrosome marker) and anti-FLAG antibodies (Fig. 3). As described previously (25), FLAG-B23 wild type was readily detected at the unduplicated centrosomes (Fig. 3 e–h). Similarly, FLAG-K230R mutant was also found at centrosomes (data not shown). However, FLAG-K263R failed to localize to centrosomes (Fig. 3 i–l), suggesting that sumoylation on K263 may play a critical role in either targeting B23 to centrosomes or stable localization of B23 at centrosomes.
Fig. 3.
K263 is a critical residue for centrosomal localization of NPM/B23. NIH 3T3 cells were transfected with either FLAG-wild-type NPM/B23 (e–h) or FLAG-K263R mutant (i–l). The vector plasmid was transfected as a control (a–d). At 24 h posttransfection, cells were fixed with methanol and coimmunostained with anti-FLAG and anti-γ-tubulin antibodies. Cells were also counterstained with DAPI. The arrows point to the positions of centrosomes. (Insets) Enlarged images of the areas indicated by arrows. (Scale bar: 10 μm.)
K263R Mutation Abolishes the Centrosome Duplication-Suppressing Activity of B23.
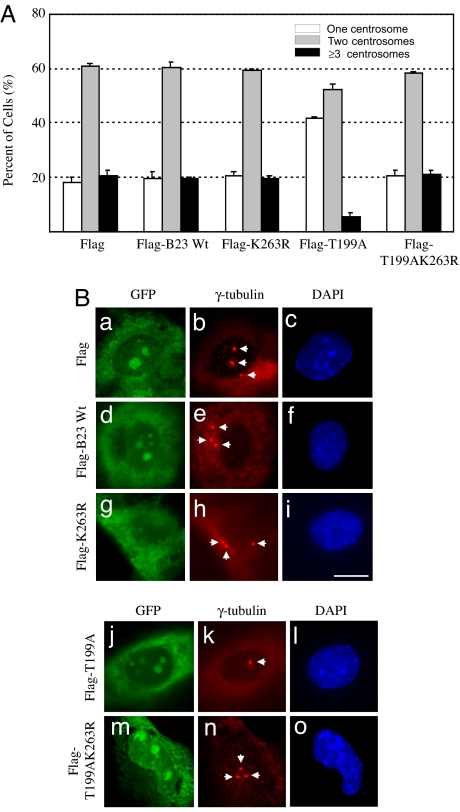
It has previously been shown that CDK2/cyclin E-mediated phosphorylation of B23 on Thr-199 promotes dissociation of B23 from centrosomes, allowing the initiation of centrosome duplication (23). Thus, when the T199A unphosphorylatable mutant is ectopically expressed, T199A continuously binds to centrosomes, resulting in suppression of centrosome duplication (25). The centrosome duplication-suppressive activity of T199A can be readily assessed experimentally by suppression of centrosome amplification in cells containing abnormally amplified centrosomes such as p53−/− mouse embryonic fibroblasts (MEFs) (26). Because T199A mutant suppresses centrosome duplication through centrosome-binding, if sumoylation on K263 is indeed critical for B23 to localize to centrosomes, introduction of K263R mutation into the T199A mutant should abolish the activity to suppress centrosome amplification. We next tested this hypothesis. We transfected FLAG-tagged B23 wild-type, T199A, K263R, and T199A/K263R double mutant into p53−/− MEFs. The vector was transfected as a control. The plasmid-encoding GFP was cotransfected (15:1 molar ratio) as a transfection marker. At 72 h posttransfection, GFP-positive cells were examined for their centrosome profiles by γ-tubulin immunostaining (Fig. 4A). Representative immunostaining images are shown in Fig. 4B. Expression of FLAG-wild-type B23 did not affect the frequency of centrosome amplification (≈20%), whereas expression of T199A mutant resulted in suppression of centrosome amplification (≈5%) as expected. However, introduction of K263R mutation into the T199A mutant completely abolished the activity to suppress centrosome amplification. From these findings, we concluded that sumoylation on K263 is essential for (stable) localization of B23 at centrosomes.
Fig. 4.
K263R mutation abolishes the centrosome duplication-suppressing activity of NPM/B23. (A) p53−/− MEFs were transfected with FLAG-wild-type NPM/B23, FLAG-K263R, FLAG-T199A, or FLAG-T199A/K263R. The plasmid vector was transfected as a negative control. The GFP plasmid was also cotransfected as a transfection marker at 1:15 molar ratio. At 72 h posttransfection, cells were fixed with 10% formalin, immunostained with anti-γ-tubulin antibody, and counterstained with DAPI. The centrosome profiles of the GFP-positive transfectants were determined and are shown here. The results are shown as average ± SE from three experiments. In each experiment, >200 cells were examined. (B) Representative immunostaining images are shown. The arrows point to the positions of centrosomes. (Scale bar: 10 μm.)
Sumoylation of B23 Is Required for Its Binding to Retinoblastoma Protein.
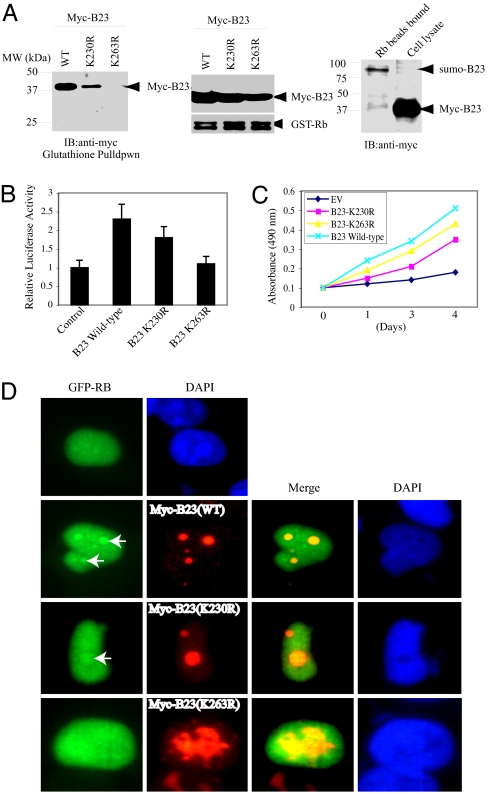
Hyperphosphorylated retinoblastoma (Rb) protein is imported into nucleoli late in S or G2 phase through binding to B23 (27). To explore whether sumoylation plays any role in mediating the interaction between B23 and Rb, we conducted an in vitro binding assay with recombinant GST-Rb protein. Wild-type B23 strongly bound to Rb, and B23(K230R) displayed a crippled binding affinity. In contrast, B23(K263R) did not associate with Rb at all (Fig. 5A Left and Middle). GST pull-down assay reveals that sumoylated B23 possesses stronger affinity to Rb than unsumoylated B23 (Fig. 5A Right). Hypophosphorylated Rb associates with E2F1 and inhibits its transcription activity. To assess the effect of B23 sumoylation on Rb's suppressive activity, we conducted luciferase assay with E2F1 report plasmid. Compared with control vector, transfection of wild-type B23 construct substantially increased E2F1 promoter activity, which was partially decreased in B23 (K230R). The stimulatory effect was completely abolished in K263R mutant (Fig. 5B). Cell proliferation assay with B23 stably transfected cells showed that overexpression of wild-type B23 and unsumoylated mutants evidently enhanced cell growth compared to control empty vector cells. Interestingly, K263R mutant cells displayed a higher growth rate than K230R cells (Fig. 5C), suggesting that E2F1 transcriptional activity elevation by B23 might not be the sole effect contributing to cell proliferation. Immunofluorescent staining reveals that transfected Rb colocalized with B23 wild-type and K230R mutant in the nucleolus; however, Rb evenly occurred in the nucleoplasm when it was cotransfected with B23 K263R mutant (Fig. 5D). Therefore, these results demonstrate that B23 interaction with Rb or its nucleolar localization relieves its transcriptional repression activity on E2F1 promoter.
Fig. 5.
B23 binds Rb and stimulates E2F1 transcriptional activity. (A) In vitro binding assay. Purified GST-Rb protein was incubated with lysate of HEK293 cells transfected with wild-type B23, B23 K230R, or K263R, respectively. Wild-type B23 robustly bound to Rb, and K230R displayed a decreased affinity. By contrast, K263R mutant did not bind to Rb at all (Left and Middle). Sumoylated B23 binds GST-Rb stronger than unsumoylated B23 (Right). (B) E2F1 luciferase activity assay. Wild-type B23 strongly enhanced E2F1 luciferase activity, and K230R revealed a reduced effect. Compared with control plasmid, K263R failed to affect E2F1 luciferase activity. (C) Cell growth curve. Stably transfected PC12 cells were cultured in tetracycline-free medium. Four thousand cells were seeded in a 48-well plate, and MTT assay was conducted at different time intervals. (D) Immmuofluorescent staining of GFP-Rb and B23 cotransfected HEK293 cells. Single-transfected Rb localized in the whole nucleus. It concentrated in the nucleolus when cotransfected with wild-type B23 and B23 (K230R) (Top, Upper Middle, and Lower Middle). Rb uniformly distributed in the nucleus of B23 K263R-transfected HEK293 cells (Bottom).
K263R Mutation Destabilizes B23 and Increases DNA Fragmentation During Apoptosis.
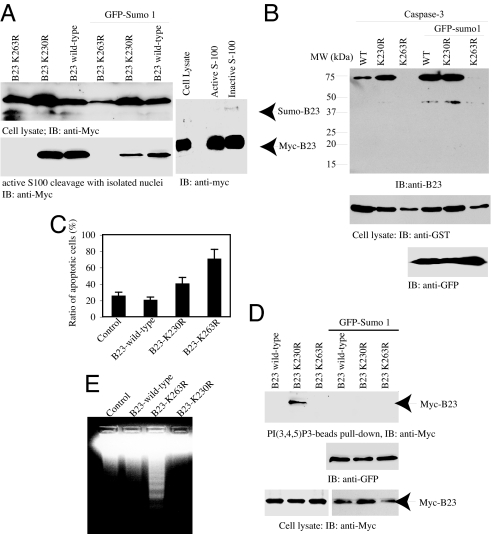
Overexpression of B23 protects cells from apoptosis, and it prevents DNA fragmentation on binding to PI(3,4,5)P3 (15). Nonetheless, B23 can be cleaved by active caspase-3. To explore whether sumoylation plays any role in mediating its proteolytic degradation, we transfected Myc-B23 wild-type and mutants into HeLa cells in the presence or absence of GFP-Sumo1 and treated the transfected cells with staurosporine. Compared with wild-type B23 and K230R mutant, B23(K263R) was completely degraded after drug treatment regardless of GFP-Sumo1. The isolated transfected nuclei incubated in the active cell-free apoptotic solution revealed similar results (Fig. 6A Left). Nevertheless, the sumoylated B23 was still unstable in the active apoptotic solution (Fig. 6A Right). In vitro apoptotic cleavage with active caspase-3 and purified GST-B23 recombinant proteins revealed the same result (Fig. 6B). To further investigate its role in regulating cell survival, we prepared stably transfected PC12 cells with wild-type and mutants and treated the cells with staurosporine. Quantitative analysis of apoptotic cells with MR-(DEVD)2, a cell-permeable fluorescent dye activated by active caspase-3 or −7, showed that 70% of K263R cells and 40% of K230R cells were apoptotic. By contrast, the ratios were dropped to 25% and 20% for empty vector and wild-type B23 cells, respectively (Fig. 6C).
Fig. 6.
K263R mutation destabilizes B23 and increases DNA fragmentation during apoptosis. (A) K263R mutation makes B23 sensitive to apoptotic degradation. HeLa cells were transfected with Myc-B23 wild-type and mutant constructs and treated with sturosporine, or the isolated nuclei were incubated with active S-100. Wild-type B23 and its K230R mutant in the absence of GFP-Sumo1 were more resistant against apoptotic proteolytic cleavage than the cells cotransfected with GFP-Sumo1 (Left). Both sumoylated and unsumoylated B23 can be degraded by active S-100 (Right). (B) Caspase-3 cleavage assay of purified B23 recombinant protein. GST-tagged B23 wild-type and mutant constructs were cotransfected into HEK293 cells with or without GFP-Sumo1, and the GST recombinant proteins were purified and subjected to active caspase-3 cleavage (Upper). The expression of transfected constructs was verified (Lower). (C) Quantitative analysis of apoptosis in the stably transfected cells. Data are mean ± SD values of triplicate determinations taken from a single experiment. The experiments were replicated three times. (D) B23 K230R strongly binds to PI(3,4,5)P3. In vitro binding assay with PIP3 beads and cell lysate from HEK293 cells cotransfected with various B23 constructs in the presence or absence of GFP-Sumo1 (Top). The expression of transfected constructs was verified (Middle and Bottom). (E) DNA fragmentation assay. PC12 cells were stably transfected with myc-B23 wild-type and various mutants and treated with 1 μM staurosporine overnight. The extracted DNA samples were analyzed on 2% agarose gel.
In vitro PI(3,4,5)P3 lipid-binding assay demonstrated that B23 (K230R) robustly bound to the lipid. Surprisingly, this interaction was disrupted when cotransfected with GFP-Sumo1. Interestingly, wild-type B23 and B23 (K263R) failed to associate with PI(3,4,5)P3 lipid regardless of GFP-Sumo1 (Fig. 6D), fitting with the previous finding that B23 interacts with PI(3,4,5)P3 lipid and requires growth factor stimulation (15). Compared with control cells, DNA fragmentation assay reveals that overexpression of wild-type B23 slightly diminished DNA degradation. By contrast, robust DNA fragmentation was detected in K263R cells, which was completely blocked in K230R cells (Fig. 6E). These results are consistent with our previous observations that the PI(3,4,5)P3/B23 complex potently binds to caspase-activated DNase and inhibits its DNA fragmentation activity. Collectively, these data demonstrate that B23 sumoylation plays an essential role in protecting it from apoptotic degradation, promoting cell survival.
Discussion
In this report, we first provide evidence that sumoylation of B23 regulates its subcellular residence, including the centrosomal and nucleolar distribution. Arf triggers B23 multiple sumoylation, but K263 residue is the major site. B23 robustly binds Rb and stimulates E2F1-mediated transcriptional activity, whereas B23 K263R fails. Further, we show that B23 sumoylation protects B23 from apoptotic degradation.
Rb protein is phosphorylated at multiple sites by CDK4 and CDK6 to drive cells to exit the G1 phase and enter the S phase (28, 29). Hypophosphorylated Rb inhibits cell cycle progression by tightly associating with E2F1 and preventing its transcriptional activity, and Rb phosphorylation releases its inhibitory sequestration on E2F1 (30). Hyperphosphorylated Rb protein is imported into nucleoli late in the S or G2 phase by binding to B23 (27). Here we also found that B23 robustly binds to Rb, which is regulated by its sumoylation on K263 (Fig. 6). When cotransfected with wild-type B23 or K230R, Rb is concentrated in the nucleolus, colocalizing with B23. Nevertheless, it does not reside in the nucleolus when cotransfected with B23 K263R, which is consistent with its failure to interact with this unsumoylated mutant. Consequently, overexpression of wild-type B23 enhanced E2F1 transcriptional activity, whereas B23 K263R failed. In agreement with this observation, B23 evidently provokes cell proliferation. B23 up-regulates E2F1 transcriptional activity by sequestrating Rb from E2F1 and inhibits its repressive activity on E2F1, resulting in elevation of the transcriptional activity and a higher growth rate. However, both K230R and K263R mutants displayed decreased cell proliferative effects compared to wild-type B23 cells, suggesting that sumoylation on B23 contributes to its accelerating activity (Fig. 6). It remains unknown why K263R cells exhibited an even faster proliferation rate than K230R cells. Presumably, K263R loss of its suppressive activity on the centrosomal duplication might somehow enhance cell division, leading to an escalation of cell cycle progression (Figs. 3 and 4).
B23 acts as a physiological substrate of caspase-3 (12). Here we show that sumoylation plays a protective role in preventing B23 from apoptotic degradation (Fig. 6 A and B). Our previous finding suggests that the phosphatidylinositol-4,5-biphosphate-3 (PIP3)/B23 complex associates with caspase-activated DNase and suppresses its DNA fragmentation activity (15). Consistent with its susceptibility to caspase cleavage, B23 K263R cells displayed demonstrable DNA fragmentation compared to wild-type B23 or control cells in response to staurosporine treatment. Quantitative analysis of apoptotic cells further confirmed this observation (Fig. 6 C–E). Surprisingly, K230R exhibited a robust PIP3-binding affinity even in the absence of growth factor stimulation. Correlatively, K230R completely blocked DNA fragmentation. However, the activity was lost in the presence of GFP-Sumo1, indicating that K263 sumoylation interferes with K230R's PIP3 lipid-binding affinity. A large body of evidence demonstrates that sumoylation promotes cell survival. For example, sumoylation augments cell survival against hypoxia- or desferroxamine-induced injury by modulating adaptive responses in salivary epithelial cells (31). Although caspase-8 can be sumoylated at lysine156 and triggers its nuclear localization, it did not impair caspase-8 activation (32). Conceivably, sumoylation of antiapoptotic proteins can selectively protect them from proteolytic degradation, leading to an increment of cell survival.
It remains unknown exactly how B23 is sumoylated. ARF plays an essential role in triggering B23 sumoylation (17). It has been proposed before that ARF might be a component of SUMO E3 ligase. Nevertheless, ARF can only selectively provoke its binding targets, but not nonbinding proteins' sumoylation (17), suggesting that it is not a SUMO E3 ligase. However, ARF cannot directly block the deconjugating SUMO protease SENP1 either (33), indicating that ARF-induced sumoylation is not mediated through blocking the deconjugating reaction. ARF preferentially triggers its binding substrate sumoylation, which occurs in the nucleoli. Conceivably, the E3 ligase for B23 might be an ARF-binding nucleolar protein.
Materials and Methods
Cells and Transfection.
His-Sumo stably transfected HEK293 cells were grown in complete medium containing 1× DMEM supplemented with 10% FBS containing 10 μg/ml Blasticidin, 100 μg/ml Hygromycin in a humidified incubator at 37°C, and 5% CO2. Expression of the His-S-SUMO proteins was induced by adding tetracycline to the culture medium at a final concentration of 1 μg/ml (a generous gift from Van G. Wilson, Texas A&M University, College Station, TX). NIH 3T3 and p53−/− MEF cells were maintained in complete medium [DMEM supplemented with 10% FBS, penicillin (100 units/ml), and streptomycin (100 μg/ml)] in an atmosphere containing 10% CO2. Anti-FLAG monoclonal antibody and anti-γ-tubulin monoclonal antibody were from Sigma–Aldrich (St. Louis, MO), anti-GFP monoclonal antibody was from Roche Diagnostics (Indianapolis, IN), and rabbit anti-γ-tubulin polyclonal antibody was generated in K.J.'s laboratory.
Nickel Column Pull-Down.
His-Sumo stably transfected F293 cells were transfected with Myc-B23 and induced with tetracycline for 24 h. His-tagged proteins were pulled down with Nickel column. After extensive washing with the above lysis buffer without EDTA, the coprecipitated proteins were analyzed by immunoblotting with anti-myc antibody.
Immunofluorescence Microscopy.
Cells were fixed with either methanol at −20°C for 20 min or 10% formalin at room temperature for 20 min. The fixed cells were blocked by 10% normal goat serum in PBS for 1 h and incubated with primary antibodies for 1 h. Cells were then incubated with secondary antibodies (Alexa Fluor 594-tagged goat anti-mouse IgG, Alexa Fluor 488-tagged goat anti-rabbit IgG, or Alexa Fluor 594-tagged goat anti-mouse IgG antibodies; Molecular Probes, Eugene, OR) for 1 h and counterstained for DNA with DAPI. After incubation with antibodies, cells were washed extensively in PBS. Cells were examined under a fluorescence microscope (Microphot-FX; Nikon, Tokyo, Japan) using a ×60 objective lens. The images were captured with a SPOT CCD camera (Diagnostic Instruments, Sterling Heights, MI).
DNA Fragmentation Assay.
Oligonucleosomal fragmentation of genomic DNA was determined as described below. In brief, 3 × 106 cells in 10 ml of medium were incubated with 250 nM staurosporine for various times. At the end of incubation, cells were pelleted, washed twice with ice-cold PBS, and lysed on ice for 60 min in 250 μl of 1% Nonidet P-40/proteinase K (0.5 mg/ml) in PBS. Samples were centrifuged, the supernatants were removed and incubated with 5 μl of RNase A (10 mg/ml) at 37°C for 40 min, and 1 ml of anhydrous ethanol was added. Tubes were placed at −20°C for 20 min and then centrifuged to pellet DNA. After the samples were dry, the same amount of DNA (10 μg) was electrophoresed at 80 V for 3 h through a 2% agarose gel containing ethidium bromide in TAE buffer [0.04 M Tris-acetate/0.001 M EDTA (pH 8.0)].
MTT Assay.
The same number (4 × 1,000) of rat PC12 cells stably transfected with empty vector, B23 wild-type, B23 K230R, and B23 K263R were cultured in tetracycline-free medium. At different intervals, cells were incubated with 0.5 mg of MTT per ml fresh medium at 37°C for 1 h. The formazan products were dissolved in DMSO and quantified by measurement of the absorbance at 490 nm, which represents the number of proliferating cells.
Statistical Analysis.
The results were expressed as means ± SEM calculated from the specified numbers of determination. Student's t test was used to compare individual data with control value.
Acknowledgments
We thank Dr. Junying Yuan (Harvard Medical School, Boston, MA) for GFP-Sumo1 plasmid. This work was supported by National Institutes of Health Grants R01 NS045627 (to K.Y.) and R01 CA95925 (to K.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Chan WY, Liu QR, Borjigin J, Busch H, Rennert OM, Tease LA, Chan PK. Biochemistry. 1989;28:1033–1039. doi: 10.1021/bi00429a017. [DOI] [PubMed] [Google Scholar]
- 2.Feuerstein N, Spiegel S, Mond JJ. J Cell Biol. 1988;107:1629–1642. doi: 10.1083/jcb.107.5.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Redner RL. Leukemia. 2002;16:1927–1932. doi: 10.1038/sj.leu.2402720. [DOI] [PubMed] [Google Scholar]
- 4.Chiarle R, Gong JZ, Guasparri I, Pesci A, Cai J, Liu J, Simmons WJ, Dhall G, Howes J, Piva R, Inghirami G. Blood. 2003;101:1919–1927. doi: 10.1182/blood-2002-05-1343. [DOI] [PubMed] [Google Scholar]
- 5.Yoneda-Kato N, Look AT, Kirstein MN, Valentine MB, Raimondi SC, Cohen KJ, Carroll AJ, Morris SW. Oncogene. 1996;12:265–275. [PubMed] [Google Scholar]
- 6.Falini B, Mecucci C, Tiacci E, Alcalay M, Rosati R, Pasqualucci L, La Starza R, Diverio D, Colombo E, Santucci A, et al. N Engl J Med. 2005;352:254–266. doi: 10.1056/NEJMoa041974. [DOI] [PubMed] [Google Scholar]
- 7.Alcalay M, Tiacci E, Bergomas R, Bigerna B, Venturini E, Minardi SP, Meani N, Diverio D, Bernard L, Tizzoni L, et al. Blood. 2005;106:899–902. doi: 10.1182/blood-2005-02-0560. [DOI] [PubMed] [Google Scholar]
- 8.Itahana K, Bhat KP, Jin A, Itahana Y, Hawke D, Kobayashi R, Zhang Y. Mol Cell. 2003;12:1151–1164. doi: 10.1016/s1097-2765(03)00431-3. [DOI] [PubMed] [Google Scholar]
- 9.Hsu CY, Yung BY. Int J Cancer. 2000;88:392–400. doi: 10.1002/1097-0215(20001101)88:3<392::aid-ijc11>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 10.Wu MH, Chang JH, Yung BY. Carcinogenesis. 2002;23:93–100. doi: 10.1093/carcin/23.1.93. [DOI] [PubMed] [Google Scholar]
- 11.Wu MH, Chang JH, Chou CC, Yung BY. Int J Cancer. 2002;97:297–305. doi: 10.1002/ijc.1606. [DOI] [PubMed] [Google Scholar]
- 12.Chou CC, Yung BY. Mol Pharmacol. 2001;59:38–45. doi: 10.1124/mol.59.1.38. [DOI] [PubMed] [Google Scholar]
- 13.Maiguel DA, Jones L, Chakravarty D, Yang C, Carrier F. Mol Cell Biol. 2004;24:3703–3711. doi: 10.1128/MCB.24.9.3703-3711.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li J, Zhang X, Sejas DP, Bagby GC, Pang Q. J Biol Chem. 2004;279:41275–41279. doi: 10.1074/jbc.C400297200. [DOI] [PubMed] [Google Scholar]
- 15.Ahn JY, Liu X, Cheng D, Peng J, Chan PK, Wade PA, Ye K. Mol Cell. 2005;18:435–445. doi: 10.1016/j.molcel.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 16.Grisendi S, Bernardi R, Rossi M, Cheng K, Khandker L, Manova K, Pandolfi PP. Nature. 2005;437:147–153. doi: 10.1038/nature03915. [DOI] [PubMed] [Google Scholar]
- 17.Tago K, Chiocca S, Sherr CJ. Proc Natl Acad Sci USA. 2005;102:7689–7694. doi: 10.1073/pnas.0502978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brady SN, Yu Y, Maggi LB, Jr, Weber JD. Mol Cell Biol. 2004;24:9327–9338. doi: 10.1128/MCB.24.21.9327-9338.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Korgaonkar C, Hagen J, Tompkins V, Frazier AA, Allamargot C, Quelle FW, Quelle DE. Mol Cell Biol. 2005;25:1258–1271. doi: 10.1128/MCB.25.4.1258-1271.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao L, Samuels T, Winckler S, Korgaonkar C, Tompkins V, Horne MC, Quelle DE. Mol Cancer Res. 2003;1:195–206. [PubMed] [Google Scholar]
- 21.Enomoto T, Lindstrom MS, Jin A, Ke H, Zhang Y. J Biol Chem. 2006;281:18463–18472. doi: 10.1074/jbc.M602788200. [DOI] [PubMed] [Google Scholar]
- 22.Lindstrom MS, Zhang Y. Cell Biochem Biophys. 2006;46:79–90. doi: 10.1385/CBB:46:1:79. [DOI] [PubMed] [Google Scholar]
- 23.Okuda M, Horn HF, Tarapore P, Tokuyama Y, Smulian AG, Chan PK, Knudsen ES, Hofmann IA, Snyder JD, Bove KE, Fukasawa K. Cell. 2000;103:127–140. doi: 10.1016/s0092-8674(00)00093-3. [DOI] [PubMed] [Google Scholar]
- 24.Shinmura K, Tarapore P, Tokuyama Y, George KR, Fukasawa K. FEBS Lett. 2005;579:6621–6634. doi: 10.1016/j.febslet.2005.10.057. [DOI] [PubMed] [Google Scholar]
- 25.Tokuyama Y, Horn HF, Kawamura K, Tarapore P, Fukasawa K. J Biol Chem. 2001;276:21529–21537. doi: 10.1074/jbc.M100014200. [DOI] [PubMed] [Google Scholar]
- 26.Tarapore P, Horn HF, Tokuyama Y, Fukasawa K. Oncogene. 2001;20:3173–3184. doi: 10.1038/sj.onc.1204424. [DOI] [PubMed] [Google Scholar]
- 27.Takemura M, Ohoka F, Perpelescu M, Ogawa M, Matsushita H, Takaba T, Akiyama T, Umekawa H, Furuichi Y, Cook PR, Yoshida S. Exp Cell Res. 2002;276:233–241. doi: 10.1006/excr.2002.5523. [DOI] [PubMed] [Google Scholar]
- 28.Chen PL, Scully P, Shew JY, Wang JY, Lee WH. Cell. 1989;58:1193–1198. doi: 10.1016/0092-8674(89)90517-5. [DOI] [PubMed] [Google Scholar]
- 29.Hinds PW, Mittnacht S, Dulic V, Arnold A, Reed SI, Weinberg RA. Cell. 1992;70:993–1006. doi: 10.1016/0092-8674(92)90249-c. [DOI] [PubMed] [Google Scholar]
- 30.Lee WH, Xu Y, Hong F, Durfee T, Mancini MA, Ueng YC, Chen PL, Riley D. Cold Spring Harb Symp Quant Biol. 1994;59:97–107. doi: 10.1101/sqb.1994.059.01.013. [DOI] [PubMed] [Google Scholar]
- 31.Nguyen HV, Chen JL, Zhong J, Kim KJ, Crandall ED, Borok Z, Chen Y, Ann DK. Am J Pathol. 2006;168:1452–1463. doi: 10.2353/ajpath.2006.050782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Besnault-Mascard L, Leprince C, Auffredou MT, Meunier B, Bourgeade MF, Camonis J, Lorenzo HK, Vazquez A. Oncogene. 2005;24:3268–3273. doi: 10.1038/sj.onc.1208448. [DOI] [PubMed] [Google Scholar]
- 33.Chen L, Chen J. Oncogene. 2003;22:5348–5357. doi: 10.1038/sj.onc.1206851. [DOI] [PubMed] [Google Scholar]