Abstract
NMR spectroscopy plays a major role in the determination of the structures and dynamics of proteins and other biological macromolecules. Chemical shifts are the most readily and accurately measurable NMR parameters, and they reflect with great specificity the conformations of native and nonnative states of proteins. We show, using 11 examples of proteins representative of the major structural classes and containing up to 123 residues, that it is possible to use chemical shifts as structural restraints in combination with a conventional molecular mechanics force field to determine the conformations of proteins at a resolution of 2 Å or better. This strategy should be widely applicable and, subject to further development, will enable quantitative structural analysis to be carried out to address a range of complex biological problems not accessible to current structural techniques.
Keywords: NMR spectroscopy, structural biology
Chemical shifts are exquisitely sensitive probes of molecular structure (1–4). Indeed, it is this characteristic that is the origin of their unique value in probing in atomic detail the properties of systems ranging from simple organic and inorganic compounds to complex biological macromolecules, because it enables the resolution of distinct signals from even chemically identical groups when located in different local or global environments. In structural biology, chemical shifts are most often used to predict regions of secondary structure in native and nonnative states of proteins (2, 5), to aid in the refinement of complex structures (6), and for the characterization of conformational changes associated with partial unfolding (7) or binding (8, 9). It has also been recognized that chemical shifts can aid in the determination of the tertiary structure of proteins when used in combination with other NMR probes that report on interproton distances (NOEs) and the relative orientations of the different nuclei in a protein structure [residual dipolar couplings (RDC)] (3, 6, 10). In many important cases, however, chemical shifts are the only NMR parameters that can be obtained on a given state of a protein with any degree of completeness (7, 8, 11–15), prompting us to explore the extent to which these quantities alone can be used to determine high-resolution structures.
The unique fingerprints of proteins provided by their NMR spectra suggest that chemical shifts inherently carry sufficient information to determine their structures at high resolution, as indeed is often the case for molecules of low molecular weight (4). The structural information contained in the chemical shifts is, however, very different in nature from that provided by NOEs, because the latter report on pairwise distances between specific protons and can thus provide unequivocal information about the relative spatial locations of different residues in a protein sequence (1). The chemical shift associated with a specific atom, by contrast, is a summation of many contributing factors (16–18) so that the reliable identification of interaction partners is very difficult, even though they may be substantially influenced by contacts between residues, such as hydrogen bonding and proximity to aromatic rings, that are at very different locations in the protein sequence. If such effects could be interpreted in depth, therefore, they would enable the characterization of the detailed environment of virtually every atom in the structure and, in turn, the determination of a unique overall conformation compatible with all such environments.
It has been demonstrated recently that strategies in which experimental data are used as restraints in molecular dynamics simulations can lead to a description of the structures of proteins, at least in outline, even in highly heterogeneous states such as those adopted by natively unfolded polypeptide molecules (19, 20). Such techniques have also been shown to be able to describe, simultaneously and with high accuracy, the structures and dynamics of native states of globular proteins by using both distance information (NOEs) and NMR order parameters (21) or RDC (22). In approaches of this type, the experimental information is used essentially to complement standard force fields and to guide the sampling of conformational space toward regions consistent with experimental observations related to the specific state under investigation (19). In a similar spirit, we describe here how NMR chemical shifts can be used to define the structures of the native states of proteins at high resolution without the requirement of any additional experimental measurements.
Results
Recent advances in the analysis of chemical shifts have enabled their values to be used increasingly successfully to obtain information about a number of specific features of protein conformations, notably dihedral angles (2), in some cases with high accuracy. Such measurements do not, however, enable the high-resolution structures of proteins to be defined without the use of extensive additional information, such as that provided, for example, by NOEs or RDC (2, 23), because even small errors in dihedral angles give rise to progressive error accumulation.
Computational Strategy.
The procedure that we present here (termed CHESHIRE, protein structure determination with CHEmical SHIft REstraints) exploits the availability of fast empirical methods that have recently been developed to enable the chemical shifts to be calculated approximately but very rapidly for a given structure (16–18). Our computational strategy is based on the molecular fragment replacement approach used in ab initio structure prediction (Rosetta) (24–26) and in the analysis of RDC (23, 27) and sparse NMR data (28) including unassigned chemical shifts (29).
3PRED.
In the first phase of the procedure (called 3PRED; see Methods), we use the experimental chemical shifts to predict the secondary structure of protein fragments of three and of nine residues (2). For ≈85% of the residues, we predict correctly whether they are in an α,β or coil conformation (Table 1).
Table 1.
Quality of the structures determined in the present study using chemical shift restraints compared with conventional methods
| Protein name | %α* | %β† | Nres‡ | NCS§ | %SS¶ | %da‖ | RMSDbb** | RMSDaa†† | Npp‡‡ | QRDC§§ |
|---|---|---|---|---|---|---|---|---|---|---|
| Ubiquitin | 25 | 32 | 76 | 281 | 74 | 93 | 1.33 | 2.13 | 3 | 0.55 |
| FF domain | 77 | 0 | 54 | 214 | 90 | 86 | 1.46 | 2.30 | 9 | 0.48 |
| Calbindin | 60 | 0 | 74 | 286 | 85 | 95 | 1.47 | 2.16 | 5 | 0.54 |
| HPr | 37 | 29 | 85 | 331 | 87 | 86 | 1.83 | 2.59 | 10 | 0.60 |
| Sda | 60 | 0 | 46 | 181 | 89 | 86 | 1.37 | 2.19 | 0 | 0.43 |
| MrR5 | 0 | 51 | 70 | 264 | 80 | 75 | 1.58 | 2.61 | 5 | 0.89 |
| PhS018 | 21 | 50 | 92 | 350 | 83 | 91 | 1.21 | 2.17 | 4 | 0.47 |
| Bet v 4 | 64 | 4 | 84 | 325 | 92 | 96 | 1.64 | 2.35 | 4 | 0.57 |
| Δ27-GG | 0 | 65 | 106 | 402 | 83 | 77 | 1.46 | 2.59 | 8 | 0.60 |
| TM1442 | 44 | 20 | 110 | 428 | 80 | 90 | 1.32 | 2.26 | 12 | 0.63 |
| Sen15 | 32 | 29 | 123 | 478 | 83 | 91 | 1.72 | 2.47 | 3 | 0.62 |
We used the following reference PDB structures: 1UBQ (ubiquitin), 1UCZ (FF domain), 1ICB (calbindin), 1POH (HPr), 1PV0 (Sda), 1YVC (MrR5), 2GLW (PhS018), 1H4B (Bet v 4), 1SA8 (Δ27-GG), 1SBO (TM1442), and 2GW6 (Sen15).
*Percentage of α-helical structure in the native state.
†Percentage of β-sheet structure in the native state.
‡Number of residues in the protein.
§Number of chemical shifts used as restraints.
¶Percentage of residues with the same predicted secondary structures (α, β, coil) as in the reference conformations, as determined in the 3PRED phase.
‖Percentage of predicted backbone dihedral angles within 60° from those in the reference conformations, as determined in the TOPOS phase.
**RMSD for all backbone and Cβ atoms. Residues before the first secondary structure element and after the last one are excluded from the calculations.
††RMSD on all atoms.
‡‡Number of interproton distances <5.5 Å in the reference structures but >6.5 Å in the structures determined here.
§§Estimated Q factors (see text) for the HN-N, CA-HA, CA-C, CA-CB residual dipolar couplings.
TOPOS.
In the second phase (called TOPOS; see Methods), we generate a library of trial conformations for each of these protein fragments by screening a database to search for those of similar sequence, secondary structure, and chemical-shift patterns (see Methods). This procedure provides predicted values of backbone dihedral angles that are in ≈95% of the cases within 60° from those in the high-resolution structures determined by conventional methods (Table 1).
Molecular fragment replacement.
In the third phase, these fragments are assembled (Fig. 1), and the structures in the resulting ensembles are refined with the use of a scoring function defined by a combination of chemical shifts and a force field similar to the standard ones used in classical molecular dynamics simulations (see Methods). In this phase of the procedure, we effectively exploit the tertiary information contained in the experimental chemical shifts, including orientations of aromatic rings and hydrogen bonds (17). For example, in the case of the 106-residue protein Δ27-GG, if we do not use the chemical-shift information in the refinement stage, we obtain a structure with a backbone root mean square distance (RMSD) of 3.1 Å from the reference structure [Protein Data Bank (PDB) entry 1SA8], rather than 1.46 Å as in the case in which the chemical shifts are used (Table 1). For the same protein, if we do not use the chemical-shift information at any stage of the procedure, we obtain a structure at 6.2 Å from the reference structure.
Fig. 1.
Schematic illustration of the molecular fragment replacement procedure implemented for chemical shifts in the CHESHIRE procedure. The protein shown is ubiquitin, and fragments are generated with main-chain dihedral angles compatible with the information contained in the chemical shifts. The fragments are then assembled in a combinatorial manner to produce an ensemble of trial structures that are subsequently refined by exploiting the information about tertiary structure contained in the chemical shifts.
Analysis of the Quality of the Structures.
We have applied the CHESHIRE procedure to 11 proteins chosen from the literature to be representative of different structural classes and to have a well defined set (1H, 13Cα, 13Cβ, and 15N) of chemical shifts for the main-chain atoms [Table 1, Fig. 2, and supporting information (SI) Fig. 4].
Fig. 2.
Comparison of the structures, also showing side chains in the hydrophobic cores, determined from chemical-shift information using the CHESHIRE procedure and those determined by standard x-ray or NMR methods. (A) Ubiquitin (blue) and PDB entry 1UBQ (pink). (B) FF domain (blue) and PDB entry 1UZC (pink). (C) Calbindin (blue) and PDB entry 4ICB (pink). (D) HPr (blue) and PDB entry 1POH (pink).
RMSDs from the reference structures.
In all 11 cases, the overall RMSD between the structures obtained by using the CHESHIRE procedure and the corresponding previously determined high-resolution x-ray or NMR structures, which are used as reference conformations, was between 1.21 and 1.83 Å for the backbone atoms and between 2.13 and 2.61 Å for all atoms (Table 1). The average pairwise backbone RMSDs between the 10 lowest-energy structures determined for each protein range from 1.0 to 1.5 Å.
WHATIF scores.
The overall quality of the structures was assessed by the WHATIF procedure (30), which in all cases resulted in scores that are regarded as good in conventional structures (a summary of the results is reported in SI Text).
Interproton distances.
We also considered all of the interproton distances below 5.5 Å in the reference structures, i.e., distances corresponding to the upper limits defined in conventional NOE-based structure determination. In almost all cases, the corresponding distances in the structures that we determined here are within this bound, and in the few exceptions (Table 1), they exceed it by <1 Å.
Residual dipolar coupling Q factors.
As a further analysis of the quality of the structures, in one case (ubiquitin), for which 344 (HN-N, CA-HA, CA-C, CA-CB) experimental backbone RDC (31) are available, we calculated a Q factor (32) of 0.49, which is comparable to typical Q factors of structures determined from NOE information. In addition, we predicted the same types of RDC for all of the 11 reference structures by using the PALES program (33) and used them to estimate the Q factors of the structures determined here (Table 1). In the case of ubiquitin, this procedure results in an estimated Q factor of 0.55, which is close to one calculated above by using experimental RDC. For the 11 structures, these estimated Q factors range from 0.43 to 0.89.
A Self-Consistent Criterion for Convergence.
To establish, without any knowledge of previously determined structures or additional experimental information, whether the structure of a particular protein has been correctly identified, we use a two-step self-consistent criterion based only on the analysis of the structures generated by the CHESHIRE procedure.
In the first step, the CHESHIRE E score (see Methods) of the best structure generated by this procedure is compared with the expected native E score (Epred score). The Epred value for a particular sequence is found by the linear formula Epred = aNres + b, where Nres is the number of residues, and a and b are constants determined by computing the E score on 3,003 randomly selected native structures from the ASTRAL SCOP database, assuming a correlation between experimental and back-calculated chemical shifts close to the SHIFTX accuracy. As shown in SI Fig. 5, there is a linear correlation between the E score and Nres for this set of 3,003 proteins (correlation coefficient of 0.98). Therefore, we can use the Epred score to estimate the E score for the native state of a protein, even if we do not actually know the structure in advance. We can thus use the Epred score to exclude cases in which the CHESHIRE procedure fails to produce any reliable structural model.
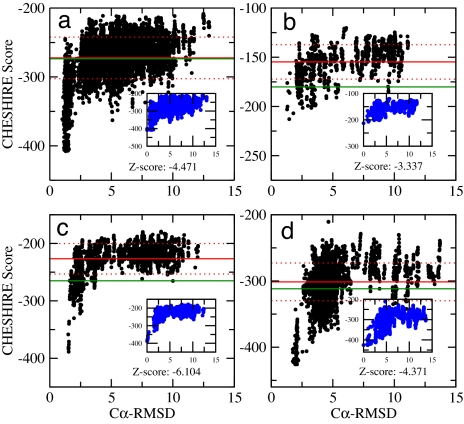
In the second step of the self-consistent criterion, the landscapes of the E score are calculated as a function of the RMSD value from the structure having the best E score (see Fig. 3). Funneled landscapes indicate that the CHESHIRE procedure has generated good structural models, because structures with good E scores but significantly different conformations are absent. For any structure, we define its Z score as Z = (E − μ)/σ, where μ and σ are the average and the standard deviation, respectively, of the distribution of the E values of the structures that we generated. Given this definition, a conformation whose E score is more than three standard deviations better than the average is characterized by Z < −3. Our results show that it is very unlikely that a structure will simultaneously have a very low value of the molecular mechanics energy and a very close agreement with the experimental chemical shifts; we can therefore conclude that the CHESHIRE structure determination is robust in these cases.
Fig. 3.
Landscapes of the CHESHIRE scores (E) for four of the proteins analyzed in this work. The landscapes report the E scores as a function of the RMSD from the reference structures (see Table 1) or the structures of minimal CHESHIRE scores (Insets). The proteins are those shown in Fig. 2. In all cases, the Z scores of the best structures are below −3, indicating that the landscapes are funneled toward the native structure. The averages and standard deviations are shown by red horizontal lines. The Epred energies are also shown as horizontal green lines. (a) Ubiquitin. (b) FF domain. (c) Calbindin. (d) HPr. See also Fig. 2.
In addition to the four cases shown in Fig. 3, we illustrate this two-step criterion for the 150-residue low-molecular-weight protein tyrosine phosphatase (YwIE) from Bacillus subtilis (SI Fig. 6), for which this criterion is not satisfied and therefore the structure is not described. In this case, the Epred value (−589) is lower than the lowest E score produced in the fragment assembly procedure (−568); in addition, the landscape of the E score is clearly not funneled.
Thus, our results suggest that the current implementation of the CHESHIRE procedure is able to define structures of proteins up to 120 residues in length. We anticipate that this limit will be increased as our ability to predict the chemical shifts corresponding to given structures improves.
Conclusions
We have shown with 11 representative examples that it is possible to determine high-resolution structures of protein molecules by using NMR chemical shifts as the only source of experimental information. This approach should enable structures to be determined by NMR spectroscopy much more rapidly than is possible at present, thereby enhancing the value of this technique in applications such as high-throughput structural genomics (34). The development of progressively accurate methods of calculating chemical shifts, particularly for side chains, which were not used in the present study, will progressively enable higher-resolution structures of proteins of increasing sizes to be determined. Indeed, because chemical shifts can already be measured experimentally with high accuracy, repositories of such data could be used to update regularly structures determined in this way at increasing resolution.
Because chemical shifts are sensitive to the dynamics on the microsecond time scale, the chemical-shift restraints can be treated as ensemble averages as described for other NMR observables (19, 21, 22). This approach should enable a description of the structural and dynamical properties of specific proteins under a variety of conditions to be obtained. Furthermore, recent studies are showing that new approaches can permit NMR spectra to be obtained and assigned for systems that have previously appeared inaccessible to this spectroscopic technique, including large or transient multimolecular assemblies (12, 14, 15), low-populated states involved in enzymatic catalysis, allosteric communication, and protein folding (7, 8), and proteins associated with membranes (13). The ability to define detailed structures from chemical shifts by using the type of approach described in the present study could be crucial in addressing the structural challenges associated with such systems and hence play an increasingly important and unique role in structural and molecular biology.
Methods
Chemical-Shift-Based Prediction of Secondary Structure Propensities.
In the first step of the CHESHIRE procedure, chemical shifts are used to predict the secondary structure of the protein. The method that we developed, termed 3PRED, uses Bayesian inference to predict the secondary structure of amino acids from the known chemical shifts in combination with the intrinsic secondary structure propensity of amino acids triplets
 |
The probability distributions Pδ measure the likelihood for individual amino acids of forming specific secondary structures S given a set of experimentally measured chemical shifts (δHα, …, δCβ). The second set of probability distributions P3 take into account the intrinsic propensities of fragments of three consecutive amino acids (Q1, Q2, Q3) to form given secondary structures (S1, S2, S3). The P3 distributions act as smoothing potentials to increase the accuracy of the assignments derived from chemical shifts alone through the Pδ distributions.
The propensities P3 were computed by considering all of the structures in the ASTRAL SCOP database (35) having <25% sequence identity according to the secondary structure classification provided by the program STRIDE (36). For the calculations of the probabilities Pδ, chemical shifts were calculated by applying SHIFTX (17) to the same set of structures to obtain an extensive database (3PRED-DB), which consisted of 939,639 calculated chemical shifts for each atom type.
Once the probabilities P3 and Pδ are known, for computational convenience they can be recast into pseudoenergies as
Thus, the pseudoenergy E of a secondary structure assignment S for a protein of sequence Q and chemical shifts Δ can be approximated as
 |
The most likely secondary structure S and the single propensities (PH, PB, PC) are then computed by averaging the assignments with the pseudoenergy function E. We used a Monte Carlo scheme in which E is minimized by a search in the space of the N-dimensional vectors S in which at each move the secondary structure assignment of a single amino acid is changed. Predictions were obtained by considering 106 such steps at a pseudotemperature T = 1.
Chemical-Shift-Based Prediction of Dihedral Restrains: TOPOS.
In the second step of the CHESHIRE procedure, the secondary structure propensities computed by 3PRED are used as input in TOPOS, an algorithm based on an approach similar to that of TALOS (2), to predict the backbone torsion angles that are most compatible with the experimental chemical shifts. In TOPOS, for each protein segment of three residues centered at position i in the sequence (the target), the similarity to a triplet centered at position j in a sequence in the ASTRAL SCOP database (the source) is evaluated by computing the similarity function σ(i, j)
 |
where Δδ is the secondary chemical shift of a given atom of the source and target protein segment; the parameters kh and ks were both set to 0.2, and the values of the remaining parameters and of the amino acid similarity matrix ΔResType were taken from Cornilescu et al. (2). The first terms in Eq. 3 are similar to the TALOS scoring function, the only substantial difference being that we do not consider HN chemical shifts. By contrast, the term ks log Pn+j(Sn+j) is the secondary structure bias present in TOPOS but not in TALOS. To avoid overfitting problems due to the use of a limited database, TOPOS uses the same extensive database of 3PRED.
The fragments with the highest σ scores, typically 200–500, are then clustered together according to the distance of the backbone torsion angles of the central amino acid. Finally, the average dihedral Φ and Ψ angles for the three best-scoring clusters are reported as prediction.
Prediction of the Structures of Fragments.
The CHESHIRE method is based on the molecular fragment replacement approach, which has been shown to be successful for the determination of protein structures with RDC (27) and in ab initio structure determination (37). In the present method, two types of fragments, of three and nine amino acids, respectively, are selected from the ASTRAL SCOP PDB database. The scoring function takes into account three contributions: (i) the score Eshifts between the experimental chemical shifts of the fragment of the protein considered and the chemical shifts of the structure in the database, (ii) the score Erestr for the compatibility with the dihedral angle restraints obtained with TOPOS, and (iii) the score Esecstr for the match between the predicted secondary structure and the secondary structure of the fragment
where the weights are set as
Chemical-shift score.
The chemical-shift score used in the fragment selection is similar to the score used by TOPOS, the only differences are that (i) the ΔResType is not included and (ii) the effect of residues i − 1 and i + 1 on residue i are not taken into account.
 |
where Eshift(i, j) is given by
 |
Dihedral angle restraint score.
The term Erestr penalizes fragments that have torsion angles that are incompatible with the predictions of TOPOS. A fragment is compatible if its distance, on the Ramachandran plot, with at least one of the predicted values is <60°.
Secondary structure score.
The secondary structure score penalizes database segments with secondary structures that differ from those predicted by 3PRED:
 |
where P(Sj, i) is the probability to have the secondary structure assignment Sj at position i.
This step of the CHESHIRE procedure provides at each position along the sequence ten fragments of length three and five fragments of length nine. These fragments are used to generate the low-resolution structures, as described below.
Generation of Low-Resolution Structures.
Molecular representation.
In the initial low-resolution structure generation, a coarse-grained representation of the protein chain was used in which only backbone atoms are explicitly modeled (H, N, Cα, C′, O); side chains are represented by a single Cβ atom. Bond lengths and angles, and the ω backbone torsion angle are kept fixed, while the Φ and Ψ torsion are given the freedom to move.
Energy function.
The energy function used for the low-resolution structure generation is a linear combination of terms that model different features of folded proteins:
 |
In the following text, we illustrate the meaning of these energy terms.
Pairwise interactions.
Evdw, Eelec, and EEEF1 model van der Waals, electrostatic, and solvation, respectively. The first two were adapted from the CHARMM PARAM19 (38) and the third from ref. 39. The pairwise potential of mean force EPMF was implemented by using all known PDB structures in the ASTRAL SCOP database following Zhou and Zhou (40).
Secondary structure packing.
To model correctly the packing of secondary structure elements, the potential of Baker and coworkers (41) (ESS, ESH, and EHH) was implemented.
Cooperative hydrogen bonding.
This term (ECHB) was implemented according to ref. 42 to favor the formation of β-sheets by β-strands distant in sequence.
Structure generation protocol.
Low-resolution structures were generated by using a Monte Carlo algorithm carried out in an extended configuration space Γ given by the Cartesian product of the protein chain coordinates and a “virtual secondary structure” string
where N and M are, respectively, the numbers of atoms and amino acids in the protein chain. These M additional discrete degrees of freedom are used to switch on and off energy terms that depend on the secondary structure of the protein.
Starting from a fully extended chain, conformations are generated by 20,000 Monte Carlo moves using a simulated annealing protocol. Two kinds of moves are applied. In the first (fragment substitution), the torsion angles and the secondary structure string in a randomly selected three- or nine-residue window of the protein chain are replaced with those from a fragment of known structure. In the second, local backbone moves, the torsion angles, but not the secondary structure, of a window of four amino acids are randomly perturbed. The score of the new conformation is calculated, and the move is accepted according to the Metropolis criterion. For each of the proteins studied here, 10,000 trial structures were generated in this way.
Refinement.
Molecular representation.
In the third stage of the CHESHIRE procedure, all atoms, including polar hydrogen atoms, are represented explicitly from the trial structures generated from the previous low-resolution stage. In a first phase, bond lengths, angles, and the ω backbone torsion angles are kept fixed, while the Φ, Ψ, and side chain torsion angles are let free to move. Structures are then optimized by using the energy function described below. Finally, the best-scoring structures are further refined by repeated minimizations and side chain optimizations using the Dunbrack and Cohen rotamers library (43).
Initial structures were obtained by adding the missing atoms to the low-resolution structures according to the following protocol. (i) A fully extended all-atom protein chain is generated by using ideal geometries. (ii) Target Φ and Ψ angles are set to those of the source chain. (iii) An energy minimization of 10,000 steps is performed to remove steric clashes. (iv) An additional energy minimization of 10,000 steps is performed by restraining interbackbone distances to the original ones. (v) A final energy minimization of 10,000 steps is performed without any restraint.
Structure screening.
All structures containing steric clashes as well as those with a radius of gyration larger than Rmax = 2.83 × M0.34, where M is the number of amino acids in the protein (44), were discarded.
Energy function.
The CHESHIRE energy function is a combination of a physicochemical term (EFF) and of a term that describes the correlation (C) between experimental and predicted chemical shifts:
where EFF is a background force field given by
and log(1 + C)capp is given by
where
 |
Here, corrX is the correlation between the experimental and the back-calculated chemical shifts for atoms of type X, kha = 18, and kn = kca = kcb = 1. The term C is capped at 3.5 to avoid correlations between experimental and back-calculated chemical shift exceeding the error of SHIFTX. With this choice of values, the correlations are biased until they reach a threshold of ≈0.8 for Hα atoms and 0.9 for N, Cα, and Cβ atoms.
Force field.
All terms in EFF except Ehb are the same defined in Eq. 10; the Ehb term models backbone hydrogen bond following Kortemme et al. (45).
Chemical-shift correlation capping.
The chemical-shift correlation term C is capped at 3.5 to avoid correlations between experimental and back-calculated chemical shift that are better that the error of SHIFTX. With this choice of values, the correlations are biased until they reach a threshold of ≈0.8 for Hα atoms and 0.9 for N, Cα, and Cβ atoms.
Structure generation protocol.
After addition of the side chain atoms, the E scores of all structures were computed, and the best 500 structures were selected for refinement. The refinement consisted of a simulated annealing Monte Carlo run of 10,000 steps. The use of a Monte Carlo strategy enables us to use a bias on the chemical shifts without requiring the derivatives of the cost function as would be necessary in a molecular dynamics scheme. After refinement, structures were ranked according to their scores, and the best-scoring one was selected as the final result.
Software.
All simulations were performed with the package almost (“all atom molecular simulation toolkit”; www.open-almost.org). The additional modules used in this project can be requested (amc82@cam.ac.uk).
Supplementary Material
Acknowledgments
This work was supported by the Swiss National Science Foundation (A.C.), the European Union (A.C., X.S., C.M.D., and M.V.), the Leverhulme Trust (X.S., C.M.D., and M.V.), the Wellcome Trust (C.M.D.), and the Royal Society (M.V.).
Abbreviations
- PDB
Protein Data Bank
- RDC
residual dipolar couplings
- RMSD
root mean square distance.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0610313104/DC1.
References
- 1.Wüthrich K. NMR of Proteins and Nucleic Acids. New York: Wiley; 1986. [Google Scholar]
- 2.Cornilescu G, Delaglio F, Bax A. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 3.Wishart DS, Case DA. Methods Enzymol. 2001;338:3–34. doi: 10.1016/s0076-6879(02)38214-4. [DOI] [PubMed] [Google Scholar]
- 4.Sanders JKM, Hunter BK. Modern NMR Spectroscopy. Oxford, UK: Oxford Univ Press; 1993. [Google Scholar]
- 5.Wishart DS, Sykes BD, Richards FM. Biochemistry. 1992;31:1647–1651. doi: 10.1021/bi00121a010. [DOI] [PubMed] [Google Scholar]
- 6.Clore GM, Gronenborn AM. Proc Natl Acad Sci USA. 1998;95:5891–5898. doi: 10.1073/pnas.95.11.5891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Korzhnev DM, Salvatella X, Vendruscolo M, Di Nardo AA, Davidson AR, Dobson CM, Kay LE. Nature. 2004;430:586–590. doi: 10.1038/nature02655. [DOI] [PubMed] [Google Scholar]
- 8.Eisenmesser EZ, Millet O, Labeikovsky W, Korzhnev DM, Wolf-Watz M, Bosco DA, Skalicky JJ, Kay LE, Kern D. Nature. 2005;438:117–121. doi: 10.1038/nature04105. [DOI] [PubMed] [Google Scholar]
- 9.Dominguez C, Boelens R, Bonvin AM. J Am Chem Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 10.Osapay K, Theriault Y, Wright PE, Case DA. J Mol Biol. 1994;244:183–197. doi: 10.1006/jmbi.1994.1718. [DOI] [PubMed] [Google Scholar]
- 11.Dyson HJ, Wright PE. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 12.Fiaux J, Bertelsen EB, Horwich AL, Wuthrich K. Nature. 2002;418:207–211. doi: 10.1038/nature00860. [DOI] [PubMed] [Google Scholar]
- 13.Chill JH, Louis JM, Miller C, Bax A. Protein Sci. 2006;15:684–698. doi: 10.1110/ps.051954706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christodoulou J, Larsson G, Fucini P, Connell SR, Pertinhez TA, Hanson CL, Redfield C, Nierhaus KH, Robinson CV, Schleucher J, Dobson CM. Proc Natl Acad Sci USA. 2005;101:10949–10954. doi: 10.1073/pnas.0400928101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sprangers R, Gribun A, Hwang PM, Houry WA, Kay LE. Proc Natl Acad Sci USA. 2005;102:16678–16683. doi: 10.1073/pnas.0507370102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu XP, Case DA. J Biomol NMR. 2001;21:321–333. doi: 10.1023/a:1013324104681. [DOI] [PubMed] [Google Scholar]
- 17.Neal S, Nip AM, Zhang H, Wishart DS. J Biomol NMR. 2003;26:215–240. doi: 10.1023/a:1023812930288. [DOI] [PubMed] [Google Scholar]
- 18.Meiler J. J Biomol NMR. 2003;26:25–37. doi: 10.1023/a:1023060720156. [DOI] [PubMed] [Google Scholar]
- 19.Vendruscolo M, Dobson CM. Philos Trans R Soc London A. 2005;363:433–450. doi: 10.1098/rsta.2004.1501. [DOI] [PubMed] [Google Scholar]
- 20.Dedmon MM, Lindorff-Larsen K, Christodoulou J, Vendruscolo M, Dobson CM. J Am Chem Soc. 2005;127:476–477. doi: 10.1021/ja044834j. [DOI] [PubMed] [Google Scholar]
- 21.Lindorff-Larsen K, Best RB, DePristo MA, Dobson CM, Vendruscolo M. Nature. 2005;433:128–132. doi: 10.1038/nature03199. [DOI] [PubMed] [Google Scholar]
- 22.Clore GM, Schwieters CD. J Mol Biol. 2006;355:879–886. doi: 10.1016/j.jmb.2005.11.042. [DOI] [PubMed] [Google Scholar]
- 23.Rohl CA, Baker D. J Am Chem Soc. 2002;124:2723–2729. doi: 10.1021/ja016880e. [DOI] [PubMed] [Google Scholar]
- 24.Simons KT, Kooperberg C, Huang E, Baker D. J Mol Biol. 1997;268:209–225. doi: 10.1006/jmbi.1997.0959. [DOI] [PubMed] [Google Scholar]
- 25.Bradley P, Misura KMS, Baker D. Science. 2005;309:1868–1871. doi: 10.1126/science.1113801. [DOI] [PubMed] [Google Scholar]
- 26.Schueler-Furman O, Wang C, Bradley P, Misura K, Baker D. Science. 2005;310:638–642. doi: 10.1126/science.1112160. [DOI] [PubMed] [Google Scholar]
- 27.Delaglio F, Kontaxis G, Bax A. J Am Chem Soc. 2000;122:2142–2143. [Google Scholar]
- 28.Bowers PM, Strauss CEM, Baker D. J Biomol NMR. 2000;18:311–318. doi: 10.1023/a:1026744431105. [DOI] [PubMed] [Google Scholar]
- 29.Meiler J, Baker D. Proc Natl Acad Sci USA. 2003;100:15404–15409. doi: 10.1073/pnas.2434121100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hooft RWW, Vriend G, Sander C, Abola EE. Nature. 1996;381:272. doi: 10.1038/381272a0. [DOI] [PubMed] [Google Scholar]
- 31.Cornilescu G, Marquardt JL, Ottiger M, Bax A. J Am Chem Soc. 1998;120:6836–6837. [Google Scholar]
- 32.Bax A, Kontaxis G, Tjandra N. Methods Enzymol. 2001;339:127–174. doi: 10.1016/s0076-6879(01)39313-8. [DOI] [PubMed] [Google Scholar]
- 33.Zweckstetter M, Bax A. J Am Chem Soc. 2000;122:3791–3792. [Google Scholar]
- 34.Chandonia J-M, Brenner SE. Science. 2006;311:347–351. doi: 10.1126/science.1121018. [DOI] [PubMed] [Google Scholar]
- 35.Chandonia J-M, Hon G, Walker NS, Conte LL, Koehl P, Levitt M, Brenner SE. Nucleic Acids Res. 2004;32:D189–D192. doi: 10.1093/nar/gkh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frishman D, Argos P. Proteins. 1995;23:566–579. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
- 37.Schueler-Furman O, Wang C, Bradley P, Misura K, Baker D. Science. 2005;310:638–642. doi: 10.1126/science.1112160. [DOI] [PubMed] [Google Scholar]
- 38.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. J Comput Chem. 1983;4:187–217. [Google Scholar]
- 39.Lazaridis T, Karplus M. Proteins. 1999;35:133–152. doi: 10.1002/(sici)1097-0134(19990501)35:2<133::aid-prot1>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 40.Zhou H, Zhou Y. Protein Sci. 2002;11:2714–2726. doi: 10.1110/ps.0217002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simons KT, Ruczinski I, Kooperberg C, Fox BA, Bystroff C, Baker D. Proteins. 1999;34:82–95. doi: 10.1002/(sici)1097-0134(19990101)34:1<82::aid-prot7>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 42.Keasar C, Levitt M. J Mol Biol. 2003;329:159–174. doi: 10.1016/S0022-2836(03)00323-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dunbrack RL, Cohen FE. Protein Sci. 1997;6:1661–1681. doi: 10.1002/pro.5560060807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gong H, Fleming PJ, Rose GD. Proc Natl Acad Sci USA. 2005;102:16227–16232. doi: 10.1073/pnas.0508415102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kortemme T, Morozov AV, Baker D. J Mol Biol. 2003;326:1239–1259. doi: 10.1016/s0022-2836(03)00021-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.