Abstract
Large interindividual variance has been observed in sensitivity to drugs. To comprehensively decipher the genetic contribution to these variations in drug susceptibility, we present a genome-wide model using human lymphoblastoid cell lines from the International HapMap consortium, of which extensive genotypic information is available, to identify genetic variants that contribute to chemotherapeutic agent-induced cytotoxicity. Our model integrated genotype, gene expression, and sensitivity of HapMap cell lines to drugs. Cell lines derived from 30 trios of European descent (Center d'Etude du Polymorphisme Humain population) and 30 trios of African descent (Yoruban population) were used. Cell growth inhibition at increasing concentrations of etoposide for 72 h was determined by using alamarBlue assay. Gene expression on 176 HapMap cell lines (87 Center d'Etude du Polymorphisme Humain population and 89 Yoruban population) was determined by using the Affymetrix GeneChip Human Exon 1.0ST Array. We evaluated associations between genotype and cytotoxicity, genotype and gene expression and correlated gene expression of the identified candidates with cytotoxicity. The analysis identified 63 genetic variants that contribute to etoposide-induced toxicity through their effect on gene expression. These include genes that may play a role in cancer (AGPAT2, IL1B, and WNT5B) and genes not yet known to be associated with sensitivity to etoposide. This unbiased method can be used to elucidate genetic variants contributing to a wide range of cellular phenotypes induced by chemotherapeutic agents.
Keywords: HapMap, pharmacogenomics, toxicity, whole-genome association
Candidate gene and genome-wide approaches have been used to identify genes important in cellular sensitivity to drugs. Although candidate gene approaches have had reasonable success in identifying genes important in the mechanisms of action of drugs, the multigenic nature of the drug effect has limited the ability of these approaches to explain much of the interindividual variation in drug effect. Genome-wide approaches open up the possibility to identify multiple components or pathways that contribute to cell susceptibility to drugs. It is particularly challenging to study genes that contribute to cellular sensitivity to chemotherapeutic drugs, because their antitumor effect is dictated by somatic mutation in the tumor and toxic effects controlled by host genomic effects. Furthermore, chemotherapy cannot be given to noncancerous family members for classical genetic studies. Recently, the International HapMap Consortium genotyped cell lines derived from trios of European and Yoruban descent, providing an extremely rich data set for genotype–drug effect correlations (1).
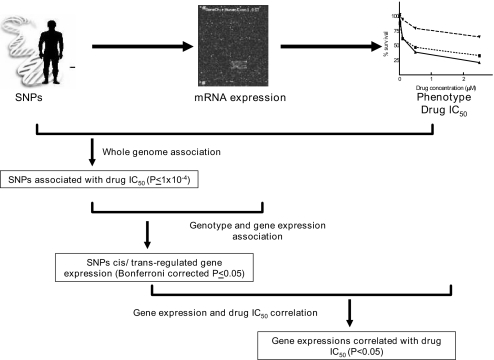
Using data generated on the HapMap cell lines, we designed a three-way model, correlating genotype, gene expression, and cytotoxicity data, with the aim of identifying potentially functional SNPs and/or haplotypes associated with chemotherapeutic agent-induced cytotoxicity (Fig. 1). Cell lines derived from individuals of African and European descent allowed us to define a set of genetic variants that contribute to chemotherapeutic-induced cytotoxicity through their effects on gene expression in two different populations. The long-term goal is to identify gene polymorphisms that influence chemotherapeutic-induced toxicity in patients, to identify those “at risk” for adverse events associated with these agents.
Fig. 1.
Genome-wide model to identify genetic variants important in drug cytotoxicity via expression levels. Model includes whole genome association between SNP genotype and gene expression, association between genotype and gene expression and linear regression between gene expression and etoposide IC50. The image to display SNPs is courtesy of the U.S. Department of Energy's Joint Genome Institute (Walnut Creek, CA; www.jgi.doe.gov).
Etoposide, a topoisomerase II inhibitor (2), was chosen to illustrate the utility of our model because of its wide usage in the treatment of disseminated testicular carcinomas, lung cancer, germinal malignancies, non-Hodgkin's lymphoma, acute myelogenous leukemia, and Kaposi's sarcoma. Etoposide is associated with bone marrow suppression, fatigue, skin rash, and diarrhea and can cause a severe delayed toxicity, treatment-related acute myeloid leukemia or myelodysplastic syndrome (3, 4). Treatment-induced toxicity has hindered the use of this agent to its full potential. Therefore, the focus of this paper is to demonstrate our global genome approach in the context of identifying genetic variants important in response or toxicities associated with etoposide through their effect on gene expression.
Results
Cell Cytotoxicity.
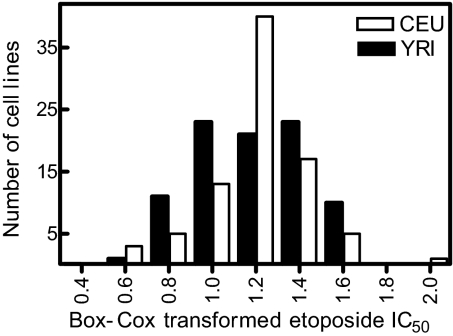
Using the alamarBlue cytotoxicity assay, 87 and 89 cell lines derived from Center d'Etude du Polymorphisme Humain population (CEU) and Yoruban population (YRI) trios were exposed to increasing concentrations of etoposide (0.02–2.5 μM) for 72 h. Although our intention was to evaluate 90 CEU and 90 YRI lines, two CEU and one YRI cell lines failed to reach 85% viability on the experiment day on more than three attempts and therefore were not further evaluated. Additionally, one CEU cell line (GM12236) was not available from Coriell at the time of phenotyping. Similar dose-dependent etoposide cell growth inhibition was observed in cell lines from both populations. Interindividual variation in the IC50 was 433- and 222-fold in CEU and YRI cell lines, respectively (Fig. 2). The median IC50 in CEU and YRI cell lines was 0.43 and 0.40 μM, respectively (5).
Fig. 2.
Frequency distribution of Box–Cox transformed etoposide IC50 in the CEU (n = 87) and YRI (n = 89).
Quantitative Transmission Disequilibrium Test (QTDT) Genotype–Cytotoxicity Association.
SNPs (387,417), which represent 22,667 genes, were evaluated for their association with etoposide IC50. This covered ≈85% annotated genes of the whole human genome. Using an arbitrary P value threshold (P ≤ 0.0001), 49, 122, and 51 SNPs were found to significantly associate with etoposide IC50 in the combined, CEU and YRI and individual CEU and YRI, respectively. The binomial tests between the probability of our significant findings and the probability of random discovery showed P = 0.046, <10−8, 0.024 in the combined, CEU, and YRI, respectively, which indicates our association test results are not likely to be random effects. These SNPs were located in or within 10 kb up- and downstream of 26, 35, and 22 genes (Table 1). A list of these SNPs can be found in supporting information (SI) Table 3. Gene ontology (GO) analysis performed on these significant genes in comparison to 22,667 total genes tested indicated that these genes are enriched in cell organization and biogenesis, endocytosis, cell adhesion, intracellular protein transport, intracellular signaling, and cell differentiation process (Table 2).
Table 1.
Three-way model significant results in combined, CEU, or YRI
| Model | Combined populations | CEU | YRI |
|---|---|---|---|
| SNP associated with etoposide IC50 (P ≤ 0.0001) | 49 SNPs (in or near 26 genes) | 122 SNPs (in or near 35 genes) | 51 SNPs (in or near 22 genes) |
| SNP associated with etoposide IC50 and with gene expression (Bonferroni corrected P < 0.05) | 7 trans-acting events (6 genes) | 2 cis- and 132 trans-acting events (21 genes) | 45 trans-acting events (40 genes) |
| Gene expression correlated with etoposide IC50(P < 0.05) | 3 genes (associated with 4 SNPs) | 18 genes (associated with 54 SNPs) | 24 genes (associated with 6 SNPs) |
Table 2.
Biological process of GO for host and target genes obtained from association studies of genotype and etoposide IC50 or of genotype and gene expression for combined, CEU, and YRI
| Gene symbol* | GO biological function | Corrected P value |
|---|---|---|
| Determined from association analysis between genotype and IC50 | ||
| FHOD3, FMN2 | Cell organization and biogenesis | 2 × 10−4 |
| GATA3, PCDH15 | Sensory perception of sound | 5.5 × 10−3 |
| DNM3, AP4S1 | Endocytosis | 5.9 × 10−3 |
| FBN1, PCDH15, GRM8 | Visual perception | 6.6 × 10−3 |
| FHOD3, FMN2 | Actin cytoskeleton organization and biogenesis | 6.6 × 10−3 |
| PCDH15, CDH2 | Homophilic cell adhesion | 7.7 × 10−3 |
| DANJC6, PTPRD | Protein amino acid dephosphorylation | 7.8 × 10−3 |
| SPON1, PCDH15, CNTN5, HNT, CDH2 | Cell adhesion | 8.3 × 10−3 |
| STX18, AP4S1 | Intracellular protein transport | 9.3 × 10−3 |
| AP4S1, SORBS2, NUP205, SLC35C2 | Transport | 9.7 × 10−3 |
| KCNN3, SLC24A4 | Potassium ion transport | 9.9 × 10−3 |
| GATA3, KLF13 | Transcription from RNA polymerase II promoter | 0.01 |
| KCNN3, GRM8 | Synaptic transmission | 0.01 |
| ADCY2, FMN2, KSR2 | Intracellular signaling cascade | 0.01 |
| SPON1, FBN1, FMN2 | Development | 0.02 |
| KCNN3, SLIT1 | Nervous system development | 0.03 |
| SLIT1, BMP7 | Cell differentiation | 0.03 |
| SLC24A4, GRID1 | Ion transport | 0.03 |
| Determined from association analysis between genotype and expression | ||
| CAPNS1, CAPN1, TCIRG1, POU3F2 | Positive regulation of cell proliferation | 2.3 × 10−4 |
| TCIRG1, ATP13A1, ATP2A3 | Proton transport | 2.4 × 10−4 |
| TUBA1, C9orf48 | Microtubule-based movement | 6.3 × 10−3 |
| SLC27A4, ATP13A1, ATP2A3, AGPAT2 | Metabolism | 7.6 × 10−3 |
| ATP13A1, ATP2A3 | Cation transport | 0.01 |
| GBP2, GBP4, GBP7, NOTCH1 | Immune response | 0.01 |
| IGSF8, ACTN4 | Cell motility | 0.01 |
| CASP10, CAPN1, PEPD, ANPEP | Proteolysis | 0.01 |
| RECQL4, WNT5B, HOXB9, PTMA | Development | 0.01 |
| RECQL4, UHRF1 | DNA repair | 0.02 |
| NEU3, PYGB | Carbohydrate metabolism | 0.03 |
| NOTCH1, ANPEP | Cell differentiation | 0.03 |
| PTMA, IL1B | Regulation of progression through cell cycle | 0.03 |
| IGSF8, IL1B | Cell proliferation | 0.03 |
| IL1B, TNFRSF6B | Apoptosis | 0.04 |
*Represents both host genes (the genes hosting the SNPs of cis-/trans-acting regulations) and target genes (the genes whose mRNA levels regulated by the cis-/trans-acting regulators)
QTDT Genotype and Gene Expression Association.
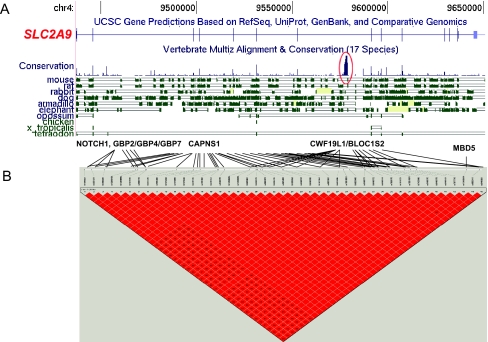
To obtain equally enriched gene expression data, we generated expression data by using the Affymetrix GeneChip Human Exon 1.0 ST Array (exon array). This exon array contains ≈1.4 million probe sets designed to represent all known and predicted exon regions within the human genome (Build 34). Gene expression analysis was performed on 176 lymphoblastoid cell lines (LCLs) (87 CEU and 89 YRI) by using the exon array. The QTDT association analysis was conducted between gene expression and the SNPs that were significantly associated with etoposide IC50 (Table 2). After normalization and robust multiarray average summary by the core sets of exon probes, we obtained gene/transcript cluster signal intensity ranging from 4 to 13. Only 14,722 genes that had the mean sample gene expression intensity of greater than five, indicating expression in both CEU and YRI, were included in the analysis. We found 7 trans-acting relationships in the combined populations, 2 cis- and 132 trans-acting relationships in CEU, and 45 trans-acting relationships in YRI (Bonferroni corrected P < 0.05; Table 1). These cis–trans-acting relationships involved 7 SNPs located in or within 10 kb up- or downstream of 3 genes and associated with 6 genes in the combined population; 56 SNPs located in or within 10 kb up- or downstream of 9 genes and associated with 21 genes in CEU; and 6 SNPs located in or within 10 kb up- or downstream of 5 genes and associated with 40 genes in YRI (SI Table 3). We identified 45 SNPs in SLC2A9 gene (chromosome 4) that were significantly associated with gene expression of CAPNS1 (chromosome 19), GBP7/GBP2/GBP4 (chromosome 1), NOTCH1 (chromosome 9), MBD5 (chromosome 2), and CWF19L1/ BLOC1S2 (chromosome 10) in CEU (Fig. 4). We use the host genes to refer to the genes hosting the SNPs of cis- or trans-acting regulations and target genes to refer to the genes whose mRNA levels are regulated by the cis- or trans-acting regulators. The GO test on 65 target genes of >14,722 reference genes showed that these target genes are enriched in biological processes, including cell proliferation regulation, cell metabolism, development, cell motility, DNA repair, proteolysis, and apoptosis (Table 2).
Fig. 4.
Relationship between SNPs located in SLC2A9 gene, gene expression and etoposide IC50 in CEU population. (A) SLC2A9 gene conservation in 17 other species (obtained from University of California Santa Cruz Genome Browser on Human March 2006 Assembly; http://genome.ucsc.edu). The circle indicates the highly conserved region. (B) HapMap LD plot of 45 significant SNPs associated with etoposide IC50 and gene expression. The black lines show significant associations between SNPs in SLC2A9 and potential target genes.
Linear Regression of Gene Expression and Etoposide IC50.
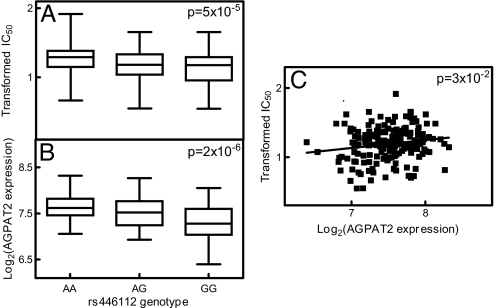
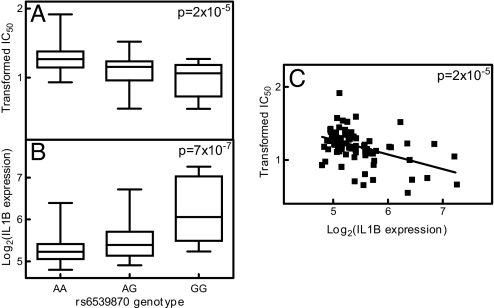
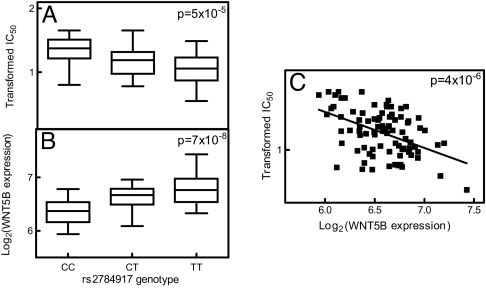
We examined the correlation between gene expression and etoposide IC50 by using a general linear model that was constructed to reflect the trio relationship in our data. Six gene expressions were evaluated in the combined population. Three had significant correlation to etoposide IC50 (P < 0.05). They were TSPAN7, CAPNS1, and AGPAT2 (Fig. 3 and SI Table 3). We also found 18 and 24 genes whose expression significantly correlated to etoposide IC50 in CEU and YRI, respectively (P < 0.05; SI Table 3). We identified 4, 54, and 6 SNPs that were significantly associated with etoposide IC50 through regulation of 3, 18, and 24 gene expressions in the combined CEU and YRI, respectively. After taking into consideration linkage disequilibrium (LD), our final findings consist of 3, 7, and 5 representative SNPs that significantly associated with 3, 18, and 24 gene expressions. One example was the significant association between the genotype of rs446112 (located in intron 1 of ZNF663 gene on chromosome 20) and etoposide IC50 (P = 5 × 10−5). This SNP was associated with the expression of AGPAT2 gene (located on chromosome 9, P = 2 × 10−6), whose expression significantly correlated to etoposide IC50 (P = 0.03; Fig. 3). In the individual CEU population, we identified 45 significant SNPs (located in SLC2A9 gene on chromosome 4 and in high LD) whose genotypes were associated with etoposide IC50 and expression of 8 genes (Fig. 4). The expression of these 8 genes was significantly correlated with etoposide IC50 (SI Table 3). Also in the CEU population, we identified rs6539870 (located in the 5′-tail end of SLC6A15 gene on chromosome 12) as associated with etoposide IC50 (P = 2 × 10−5) and IL1B gene expression (P = 7 × 10−7). We also found a strong correlation between the IL1B expression and etoposide IC50 (P = 2 × 10−5 and Fig. 5). In the YRI, we identified a strong association between the genotype of SNP rs2784917, located in SLIT1 gene on chromosome 10, and the expression of WNT5B gene, located on chromosome 12 (P = 7 × 10−8) along with a significant SNP genotype and etoposide IC50 association (P = 5 × 10−5). The TT genotype of rs2784917 was associated with higher WNT5B gene expression and lower etoposide IC50. This was further indicated by the inverse correlation found between WNT5B gene expression and etoposide IC50 (P = 4 × 10−6 and Fig. 6).
Fig. 3.
Relationship between rs446112, AGPAT2 gene expression and etoposide IC50 in combined CEU and YRI. (A) rs446112 genotype and Box–Cox transformed etoposide IC50 association. (B) rs446112 genotype and log2 transformed AGPAT2 expression association. (C) Log2 transformed AGPAT2 expression and Box-Cox transformed etoposide IC50 correlation.
Fig. 5.
Relationship among rs6539870, IL1B gene expression, and etoposide IC50 in CEU. (A) rs6539870 genotype and Box–Cox-transformed etoposide IC50 association. (B) rs6539870 genotype and log2 transformed IL1B expression association. (C) Log2-transformed IL1B expression and Box–Cox-transformed etoposide IC50 correlation.
Fig. 6.
Relationship among rs2784917, WNT5B gene expression, and etoposide IC50 in YRI. (A) rs2784917 genotype and Box–Cox-transformed etoposide IC50 association. (B) rs2784917 genotype and log2-transformed WNT5B expression association. (C) Log2-transformed WNT5B expression and Box–Cox-transformed etoposide IC50 correlation.
Multivariate Models to Predict Etoposide IC50 with Genotypes.
To examine the overall contributions of our selected genetic variants to sensitivity of etoposide, additional general linear models were constructed. All SNP genotypes that were significantly associated with etoposide IC50 through their effects on gene expression were included as the independent variables to predict etoposide IC50 as the dependent variable in each tested population. The backwards elimination approach was applied for models reduction. In the combined population, three of the four tested SNPs were included in the final model (P < 0.0003 for all SNPs). Specifically, rs460869, rs6588131, and rs16965867 were all significant predictors of etoposide IC50. Computing a weighted sum of R2 from each group of unrelated individuals gives an overall estimate of R2 = 0.30, indicating 30% of the variation in etoposide IC50 can be explained by these three SNPs in the combined populations. In the CEU population, rs10018204, rs11222869, rs16965867, rs1846644, and rs6539870 were included in the final model (P < 0.015 for all five SNPs). The indicator of gender is a significant predictor (P < 0.0077), which is in agreement with our previous finding of a significant difference in etoposide IC50 between female and male within the YRI (5). The overall estimate of R2 = 0.55, indicating 55% of the etoposide IC50 variation can be explained by these five SNPs in the CEU population. In the YRI, four of the six tested SNPs were included in the final model (P < 0.015 for all SNPs). Specifically, rs10061997, rs12190776, rs2784917, and rs9730073 were all significant predictors of etoposide IC50. The overall estimate of R2 = 0.40, indicates 40% of the etoposide IC50 variation can be explained by these 4 SNPs in the YRI.
Discussion
We have developed a genome-wide approach to identify genetic variants that are important in chemotherapy-induced cytotoxicity as well as other quantitative traits that can be measured in HapMap cell lines. Our model includes complementary approaches merging whole-genome association between genotype and phenotype (sensitivity to drug) and association between genotype and gene expression, as well as linear regression analysis between gene expression and phenotype to identify genetic variants that are important to drug-induced cytotoxicity through modulation of gene expression.
Previously, our laboratory used cell lines derived from large pedigrees to demonstrate that a significant genetic component contributed to susceptibility to the cytotoxic effects of cisplatin, a chemotherapeutic agent (6). Variation in susceptibility to two additional chemotherapeutic agents, fluorouracil (5-FU) and docetaxel, were also shown to have a significant genetic component (7). Cheung's group has shown that expression of a considerable number of genes is directly controlled by cis- or trans-acting elements of the genotype (8–10). Many studies have also demonstrated the correlation between gene expression and treatment effect (11–19). The present study links genotype to gene expression to cell line treatment response and ultimately identified genetic variants that influence drug-induced cytotoxicity.
For etoposide, our model allowed us to uncover previously unknown genetic variants important in drug cytotoxicity. We applied our model to the combined populations (CEU + YRI), which provided more power to detect significant genetic variants and because both populations showed similar sensitivity to etoposide at all concentrations studied. However, the possibility exists that, even though both populations showed similar sensitivity to etoposide, this phenotype could be controlled by different genetic variants in each population; therefore, we also applied our model independently to CEU and YRI. One challenge in the application of this genome-wide approach is that the multiple testing may result in false discovery. To decrease the false discovery rate, a conventional approach is to use a stringent statistical cutoff (20). However, these arbitrarily chosen cutoff values may not hold true biological meaning. Our stepwise approach linked SNP genotype, gene expression, and sensitivity to drugs together. We limited our search to identify genetic variants that associated sensitivity to drugs through regulation of gene expression. Without the gene expression element of the model, the QTDT analysis between genotype and etoposide IC50 yielded 49, 122, and 51 significant SNPs (P ≤ 0.0001) associated with etoposide IC50 from the combined CEU- and YRI-derived cell lines, respectively. Taking into account these associations composed of those SNPs associated and not associated with expression, we further reduced our list to only those associated with expression to obtain 4, 54, and 6 SNPs through the expression of 3, 18, and 24 genes in combined, CEU and YRI, respectively. The stepwise approach allows us to narrow down genes whose expression is correlated to etoposide cytotoxicity and provides us with greater confidence in the SNPs identified for further validation.
All SNP genotypes and gene expressions found through this model can be considered relevant to sensitivity to etoposide. Functional studies are currently underway to confirm the role of these genetic variants contribution. Of the 63 genetic variants we identified, some of them have not been described previously, whereas others are supported by literature to play a role in the cell sensitivity to etoposide. For example, we identified a strong association among genetic variants of SNP rs446112, gene expression of AGPAT2, and susceptibility to etoposide-induced cytotoxicity in the combined populations. Niesporek et al. (11) have shown the increased expression of AGPAT2 was significantly linked to reduced overall survival time as well as to shorter progression-free survival time in ovarian cancer patients. Our study showed that the AA genotype of SNP rs446112 was associated with higher AGPAT2 gene expression, which correlated with lower sensitivity to etoposide. Further studies to confirm AGPAT2 as a mechanism of resistance to etoposide are warranted.
An association between genetic variants of SNP rs6539870, gene expression of IL1B, and etoposide IC50 was identified in CEU. IL1B is an important cytokine, which mediates the inflammatory response and is involved in a variety of cellular activities, including cell proliferation, differentiation, and apoptosis (21). It has been shown that IL-1α, a close member of the same interleukin protein family, can dramatically increase the sensitivity of etoposide in osteosarcoma cells (22). The synergistic antitumor effects of IL-1α and etoposide have also been observed in melanoma and ovarian cell lines (23, 24). Our study showed that the GG genotype of SNP rs6539870 associated with higher IL1B gene expression and greater sensitivity to etoposide. Another association was identified among genetic variants of rs2784917, gene expression of WNT5B, and etoposide IC50 in the YRI. WNT signaling pathway plays a key role in carcinogenesis and embryogenesis and has been found to be up-regulated in gastric, esophageal, pancreatic, breast cancer cell line, and uterine leiomyoma cells (25, 26). Our study showed that the TT genotype of SNP rs2784917 was associated with higher WNT5B gene expression and greater sensitivity to etoposide. These genes may be targets for synergizing tumor cells to etoposide. Additionally, many genes identified through our model were not previously studied and could also be targets for etoposide treatment induced toxicity.
We also found that the expression levels of many genes shared the same regulatory region. For example, we found 45 SNPs with high LD located in the SLC2A9 gene. These SNPs are significantly associated with the expression of eight genes. Among the 45 SNPs, we identified haplotypes associated with NOTCH1 and GBP2/4/7 gene expression as well as haplotypes associated with CWF19L1/BLOC1S2 gene expression. An intronic region located in SLC2A9 gene that has been shown to be highly conserved across 17 species (University of California Santa Cruz Human Genome Browser, March 2006 assembly; http://genome.ucsc.edu) contains two of our significant SNPs (rs6449178 and rs6449179). Further study is needed to evaluate the role of this region, because it may host a new gene or contain an alternative splicing site or noncoding, micro-, or siRNA.
The ultimate goal of the model is to identify genetic variants important in drug response or toxicity, so patients at risk for nonresponse or toxicities can be given alternative therapy. However, one limitation of the model is that it represents only one type of cell line and may not represent protein expression in a tissue of known toxicity or tumor. Furthermore, candidate genes known to contribute to the pharmacokinetics of etoposide, e.g., CYP3A, UGT1A1, and ABCB1, are not expressed or are expressed at very low levels in LCLs (data not shown). The advantage to the model is that genes important in the pharmacodynamics of the drug can be identified without confounding variables from pharmacokinetic variables. Although the model allowed us to identify several genetic variants significantly associated with etoposide-induced cytotoxicity through gene expression, functional studies are required to confirm the contribution of these genetic variants to etoposide-induced cytotoxicity. There are also other genetic variants that are not associated through expression that may contribute to the variation in cytotoxicity that will require further study.
In summary, this genome-wide approach successfully integrated genotype, gene expression, and sensitivity to drug information to identify genetic variants that are important in drug treatment. It can be used to uncover important genetic variants contributing to a wide range of phenotypes that can be measured in LCLs.
Methods
Cell Lines.
EBV-transformed B LCLs derived from 30 Center d'Etude du Polymorphisme Humain trios (mother, father, and child) from Utah residents with ancestry from Northern and Western Europe (HAPMAPPT01, CEU) and 30 trios collected from the Yoruba in Ibadan, Nigeria (HAPMAPPT03, YRI) were purchased from the Coriell Institute for Medical Research (Camden, NJ). Cell lines were maintained in RPMI medium 1640 (Mediatech, Herndon, VA) supplemented with 15% FBS (HyClone, Logan, UT) and 1% l-glutamine (Invitrogen, Carlsbad, CA). Cell lines were passaged three times per week and seeded at a concentration of 350,000 cells/ml at 37°C in a 95% humidified 5% CO2 atmosphere.
Drug.
Etoposide (NSC-141540) was provided by the Drug Synthesis and Chemistry Branch, Division of Cancer Treatment, National Cancer Institute, Bethesda, MD. PBS (pH 7.4) was from Invitrogen, and DMSO was from Sigma–Aldrich (St. Louis, MO).
Cytotoxicity Assay.
The cytotoxic effect was determined by using the nontoxic colorimetric-based assay, alamarBlue (Biosource, Camarillo, CA). Exponentially growing lymphoblastoid cells with >85% viability, as determined by the trypan blue dye exclusion method by using Vi-Cell XR viability analyzer (Beckman Coulter, Fullerton, CA), were plated in triplicate at a density of 1 × 105 cells/ml in 96-well round-bottom plates (Corning, Corning, NY) for 24 h. Etoposide was initially dissolved in DMSO and further diluted with media. Cells were treated with either vehicle (0.025% DMSO) or 0.02, 0.1, 0.5, and 2.5 μM etoposide for 72 h. AlamarBlue was added 24 h before absorbance reading at wavelengths 570 and 600 nm by using the Synergy-HT multidetection plate reader (BioTek, Winooski, VT). Percent survival was quantified by using the manufacturer's protocol. Final percent survival was averaged from at least six replicates from two independent experiments. IC50 was determined for each cell line by curve fitting of percent cell survival against concentrations of drug.
Genotype and Cytotoxicity Association Analysis.
SNP genotypes were downloaded from the International HapMap database (www.HapMap.org) (release 21). To perform a high-quality whole-genome association study, several data filters were used. To reduce possible genotyping errors, we excluded 100,536 and 138,533 SNPs with Mendelian allele transmission errors on 22 autosomes in the 30 CEU and 30 YRI HapMap trios, respectively. To exclude the extreme outliers and increase the power of the association studies within our limited number of samples, we included only the SNPs that met the criteria of having three genotypes (homozygous wild type, heterozygous, and homozygous variant) and containing a minimum of two samples for each genotype in the unrelated individuals of each population. To obtain functionally relevant SNPs, we further filtered the SNPs by location. Only SNPs located within 10-kb up- or downstream and within a gene were included. Thus, our final data set consisted of 387,417 very informative SNPs covering 22,667 well annotated genes.
Eighty-seven CEU and 89 YRI HapMap samples were phenotyped for etoposide cytotoxicity. A Box–Cox transformation was applied to these 176 IC50 values by using MINITAB14 (Minitab, State College, PA) followed by a Kolmogorov–Smirnov test of normality. The transformed data with P > 0.05 were considered normally distributed. The QTDT was performed to identify any genotype–cytotoxicity association by using QTDT software (27) (www.sph.umich.edu/csg/abecasis/QTDT). Because of the possible heterogeneity between and within each population, we performed QTDT studies in these two ethnic groups separately by using sex as a covariate and together by using sex and race as covariates. P ≤ 0.0001 was considered statistically significant. The binomial tests were performed between the probability of our significant findings and the probability of random discovery.
Gene Expression Assessment.
Before sample collection, LCLs were diluted to 500,000 cells/ml in RPMI growth media four times over 2 wk. LCLs (8.5 million cells) were in log-growth phase at the time of collection with ≥85% viability. Trypan blue exclusion test was used to assess viability and concentration. RNA from 87 CEU and 89 YRI cell lines were extracted at the fourth dilution by using RNeasy Plus Mini kits (Qiagen, Valencia, CA). RNA quality was assessed by using the RNA 6000 Nano Assay (Agilent Technologies, Palo Alto, CA). RNA samples were further purified and prepared according to the manufacturer's protocol by using Affymetrix's GeneChip Whole Transcript Sense Target Labeling Assay with reagents specifically designed for the Human Exon arrays. Gene expression profiles were assessed by using Affymetrix GeneChip Human Exon 1.0 ST array. Probe-signal intensities were sketch-normalized by using a subset of the 1.4 million probe sets. Transcript cluster expression was summarized by using a robust multiarray average method (28) with a core set of well annotated exons (≈200,000). The ExACT program developed by Affymetrix with its Integrated Genome Browser was used to determine gene signal.
Genotype and Gene Expression Association Analysis.
A second QTDT test that integrated candidate SNPs with mRNA level gene expression was performed to identify possible genotype-expression association. Significant SNPs generated from the genotype–cytotoxicity association in CEU, YRI, or combined populations were tested for their association with gene expression in the same population. Genes with average intensity greater than five from Affymetrix GeneChip Human Exon 1.0 ST Array analysis were considered as expressed genes and included in this association analysis. The QTDT test was carried out by using gene expression in CEU and YRI separately and in combined CEU and YRI with sex and race (in the combined samples) as covariates. We examined not only the cis-acting gene, defined as gene expression associated with SNP(s) within 2.5 Mb on the same chromosome, but also the trans-acting gene, defined as gene expression associated with SNP(s) on different chromosome(s). A Bonferroni correction (P = 0.05) was used to adjust raw P values after QTDT analysis.
GO Analysis.
To further characterize sets of functionally related genes that may affect drug response, we used Onto Express (29–32) to classify genes according to their GO annotations (33). The genes hosting SNPs that were significantly associated with etoposide IC50, and those genes that were regulated by SNPs in a cis or trans manner were tested for their potential enrichment in a particular GO term by using biological process function. Hypergeometric distribution (Bioconductor Vignette, http://bioconductor.org/docs/vignettes.html) was used with each GO term having an associated P value relating to the number of genes that are annotated at that term. GO terms enriched in our sets of genes relative to the references (as total number of genes tested) was indicated at a Benjamin–Hochberg FDR of 5%.
Gene Expression and Etoposide IC50 Linear Regression Analysis.
To examine the relationship between gene expression and sensitivity to etoposide, a general linear model was constructed with etoposide IC50 (transformed by using the Box–Cox transformation) as the dependent variable and robust multiarray average-summarized log2-transformed gene expression level together with an indicator for gender as the independent variables. The dependent variable was transformed to satisfy the assumption of normality. Trios were treated as units of analysis, and members of different families were considered independent. The covariance structure within a trio was modeled by using a Toeplitz structure with two diagonal bands, such that the trios were ordered father, offspring, and then mother. With this covariance structure, mother and father IC50 values were independent, but the offspring's value was allowed to covary with both father's and mother's values. If a SNP was significantly associated with etoposide IC50, and the same SNP was significantly associated with gene expression, then the above approach was used to test whether gene expression significantly predicted IC50. Sixteen genes/transcripts were tested for their expression correlation with etoposide IC50 in the CEU population, and 33 genes were tested in the YRI. Only six genes were tested by using combined CEU and YRI. With the combined approach a predictor of population was included in the model. P < 0.05 was considered statistically significant. The model was programmed by using the PROC MIXED procedure in SAS/STAT software (Version 9.1, SAS Institute, Cary, NC). The REPEATED statement was used to model the Toeplitz covariance structure. The LD of significant SNPs within each population was evaluated by using Haploview version 3.32 (www.broad.mit.edu/mpg/haploview).
Multivariate Model to Predict Etoposide IC50 with Genotypes.
To examine the overall genetic variant contributions to sensitivity of etoposide, additional general linear models were constructed with transformed etoposide IC50 as the dependent variable. The independent variables included all of the significant SNP genotypes (assuming an additive genetic effect) that were selected from the three-way model in the combined populations and the two populations independently. These SNP genotypes are significantly associated with etoposide IC50 through their effect on gene expression. For the model of combined populations, indicators of race and sex were also included as predictors. Trios were analyzed as independent units. The covariance was modeled as described above. Models were reduced by using a backwards elimination approach. SNPs included in each of the final models were statistically significant at the α = 0.05 level. Using the final model, predicted transformed IC50 values were computed. Within the unrelated individuals (parents from the trios and separately offspring from the trios) an R2 was estimated between observed IC50 and the predicted IC50 from the final model. Last, a weighted average of the two R2 estimates was computed to quantify the amount of variation in etoposide IC50 explained by the selected SNP genotypes.
Supplementary Material
Acknowledgments
We thank Dr. Jeong-Ah Kang for excellent technical support in maintaining the cell lines and Cheryl Roe for critical review of the manuscript. This Pharmacogenetics of Anticancer Agents Research Group (http://pharmacogenetics.org) study was supported by the National Institutes of Health (NIH)/National Institute of General Medical Sciences (NIGMS) (Grant GM61393). This research is supported by the NIH/NIGMS Pharmacogenetics Research Network and Database, U01GM61374, Russ Altman, principal investigator (www.pharmgkb.org).
Abbreviations
- LCLs
lymphoblastoid cell lines
- QTDT
quantitative transmission disequilibrium test
- GO
gene ontology
- LD
linkage disequilibrium
- CEU
Center d'Etude du Polymorphisme Humain population
- YRI
Yoruban population.
Footnotes
Conflict of interest statement: T.A.C., T.X.C., A.C.S., and J.E.B. are employees of Affymetrix, Inc., 3420 Central Expressway, Santa Clara, CA 95051.
Data deposition: The expression data reported in this paper have been deposited in the National Center for Biotechnology Information Gene Omnibus database (GEO accession no. GSE 7792), and the phenotype data have been deposited in the PharmGKB database, www.pharmgkb.org (accession no. PS206922).
This article contains supporting information online at www.pnas.org/cgi/content/full/0703736104/DC1.
References
- 1.The International HapMap Consortium. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sinha BK, Haim N, Dusre L, Kerrigan D, Pommier Y. Cancer Res. 1988;48:5096–5100. [PubMed] [Google Scholar]
- 3.Mistry AR, Felix CA, Whitmarsh RJ, Mason A, Reiter A, Cassinat B, Parry A, Walz C, Wiemels JL, Segal MR, et al. N Engl J Med. 2005;352:1529–1538. doi: 10.1056/NEJMoa042715. [DOI] [PubMed] [Google Scholar]
- 4.Ratain MJ, Kaminer LS, Bitran JD, Larson RA, Le Beau MM, Skosey C, Purl S, Hoffman PC, Wade J, Vardiman JW. Blood. 1987;70:1412–1417. [PubMed] [Google Scholar]
- 5.Huang RS, Kistner EO, Bleibel WK, Shukla SJ, Dolan ME. Mol Cancer Ther. 2007;6:31–36. doi: 10.1158/1535-7163.MCT-06-0591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dolan ME, Newbold KG, Nagasubramanian R, Wu X, Ratain MJ, Cook EH, Jr, Badner JA. Cancer Res. 2004;64:4353–6. doi: 10.1158/0008-5472.CAN-04-0340. [DOI] [PubMed] [Google Scholar]
- 7.Watters JW, Kraja A, Meucci MA, Province MA, McLeod HL. Proc Natl Acad Sci USA. 2004;101:11809–11814. doi: 10.1073/pnas.0404580101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheung V, Conlin L, Weber T, Arcaro M, Jen K, Morley M, Spielman R. Nat Genet. 2003;33:422–425. doi: 10.1038/ng1094. [DOI] [PubMed] [Google Scholar]
- 9.Cheung VG, Spielman RS, Ewens KG, Weber TM, Morley M, Burdick JT. Nature. 2005;437:1365–1369. doi: 10.1038/nature04244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morley M, Molony CM, Weber TM, Devlin JL, Ewens KG, Spielman RS, Cheung VG. Nature. 2004;430:743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niesporek S, Denkert C, Weichert W, Köbel M, Noske A, Sehouli J, Singer JW, Dietel M, Hauptmann S. Br J Cancer. 2005;92:1729–1736. doi: 10.1038/sj.bjc.6602528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirano T, Kizaki M, Kato K, Abe F, Masuda N, Umezawa K. Leuk Res. 2002;26:1097–1103. doi: 10.1016/s0145-2126(02)00052-8. [DOI] [PubMed] [Google Scholar]
- 13.Debret R, Le Naour R, Sallenave J, Deshorgue A, Hornebeck W, Guenounou M, Bernard P, Antonicelli F. J Invest Dermatol. 2006;126:1860–1868. doi: 10.1038/sj.jid.5700337. [DOI] [PubMed] [Google Scholar]
- 14.Breit S, Stanulla M, Flohr T, Schrappe M, Ludwig W-D, Tolle G, Happich M, Muckenthaler MU, Kulozik AE. Blood. 2006;108:1151–1157. doi: 10.1182/blood-2005-12-4956. [DOI] [PubMed] [Google Scholar]
- 15.Cohen L, Bourbonniere M, Sabbagh L, Bouchard A, Chew T, Jeannequin P, Lazure C, Sekaly R. Cell Death Differ. 2005;12:243–254. doi: 10.1038/sj.cdd.4401568. [DOI] [PubMed] [Google Scholar]
- 16.Nefedova Y, Cheng P, Alsina M, Dalton WS, Gabrilovich DI. Blood. 2004;103:3503–3510. doi: 10.1182/blood-2003-07-2340. [DOI] [PubMed] [Google Scholar]
- 17.Kakugawa Y, Wada T, Yamaguchi K, Yamanami H, Ouchi K, Sato I, Miyagi T. Proc Natl Acad Sci USA. 2002;99:10718–10723. doi: 10.1073/pnas.152597199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suzuki S, Takahashi S, Takahashi S, Takeshita K, Hikosaka A, Wakita T, Nishiyama N, Fujita T, Okamura T, Shirai T. Prostate. 2006;66:463–469. doi: 10.1002/pros.20385. [DOI] [PubMed] [Google Scholar]
- 19.Chung Y, Kim T, Kim D, Namkoong H, Kim H, Ha S, Kim S, Shin S, Kim J, Lee Y, et al. Leukemia. 2006;20:1542–1550. doi: 10.1038/sj.leu.2404310. [DOI] [PubMed] [Google Scholar]
- 20.Stranger BE, Forrest MS, Clark AG, Minichiello MJ, Deutsch S, Lyle R, Hunt S, Kahl B, Antonarakis SE, Tavaré S, et al. PLoS Genet. 2005;1:e78. doi: 10.1371/journal.pgen.0010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roy D, Sarkar S, Felty Q. Front Biosci. 2006;11:889–898. doi: 10.2741/1845. [DOI] [PubMed] [Google Scholar]
- 22.Jia S, Zwelling L, McWatters A, An T, Kleinerman E. J Exp Ther Oncol. 2002;2:27–36. doi: 10.1046/j.1359-4117.2002.01003.x. [DOI] [PubMed] [Google Scholar]
- 23.Usui N, Matsushima K, Pilaro A, Longo D, Wiltrout R. Biotherapy. 1996;9:199–208. doi: 10.1007/BF02620733. [DOI] [PubMed] [Google Scholar]
- 24.Monti E, Mimnaugh E, Sinha B. Biochim Biophys Acta. 1993;1180:231–235. doi: 10.1016/0925-4439(93)90043-z. [DOI] [PubMed] [Google Scholar]
- 25.Saitoh T, Katoh M. Int J Mol Med. 2002;10:345–349. [PubMed] [Google Scholar]
- 26.Mangioni S, Vigano P, Lattuada D, Abbiati A, Vignali M, Di Blasio AM. J Clin Endocrinol Metab. 2005;90:5349–5355. doi: 10.1210/jc.2005-0272. [DOI] [PubMed] [Google Scholar]
- 27.Abecasis G, Cardon L, Cookson W. Am J Hum Genet. 2000;66:279–292. doi: 10.1086/302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 29.Draghici S, Khatri P, Bhavsar P, Shah A, Krawetz SA, Tainsky MA. Nucleic Acids Res. 2003;31:3775–3781. doi: 10.1093/nar/gkg624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Draghici S, Khatri P, Martins R, Ostermeier G, Krawetz S. Genomics. 2003;81:98–104. doi: 10.1016/s0888-7543(02)00021-6. [DOI] [PubMed] [Google Scholar]
- 31.Khatri P, Draghici S, Ostermeier G, Krawetz S. Genomics. 2002;79:266–270. doi: 10.1006/geno.2002.6698. [DOI] [PubMed] [Google Scholar]
- 32.Khatri P, Sellamuthu S, Malhotra P, Amin K, Done A, Draghici S. Nucleic Acids Res. 2005;33:W762–W765. doi: 10.1093/nar/gki472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ashburner M, Ball C, Blake J, Botstein D, Butler H, Cherry J, Davis A, Dolinski K, Dwight S, Eppig J, et al. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.