Abstract
In marsupials, dosage compensation involves silencing of the father's X-chromosome. Because no XIST orthologue has been found, how imprinted X-inactivation occurs is unknown. In eutherians, the X is subject to meiotic sex chromosome inactivation (MSCI) in the paternal germ line and persists thereafter as postmeiotic sex chromatin (PMSC). One hypothesis proposes that the paternal X is inherited by the eutherian zygote as a preinactive X and raises the possibility of a similar process in the marsupial germ line. Here we demonstrate that MSCI and PMSC occur in the opossum. Surprisingly, silencing occurs before X–Y association. After MSCI, the X and Y fuse through a dense plate without obvious synapsis. Significantly, sex chromosome silencing continues after meiosis, with the opossum PMSC sharing features of eutherian PMSC. These results reveal a common gametogenic program in two diverse clades of mammals and support the idea that male germ-line silencing may have provided an ancestral form of mammalian dosage compensation.
Keywords: meiosis, X-inactivation
In mammals, sex is determined by the differential inheritance of the X and Y chromosomes, with the female inheriting two X chromosomes (XX) and the male inheriting an X and a Y (XY). With some variation, the XY scheme of sex determination can be seen in all three extant clades of mammals, including the prototherians (monotremes) that evolved some 300 million years ago, the metatherians (marsupials) that evolved 150–200 million years ago, and the eutherians (placental mammals) that evolved 100–150 million years ago. In addition to the sexually dimorphic development of males and females, this system of sex determination has important consequences for other aspects of mammalian development: one relating to the inequality of sex chromosome gene dosage and the other to the behavior of sex chromosomes in the germ line. Both stem from the fact that genetic content on the Y has gradually eroded over 300 million years of evolution (1–3).
Because the Y carries only a fraction of the genetic material found on the X, females have nearly twice the sex chromosome gene dosage as males, often necessitating coevolution of dosage compensation. In mammals, dosage compensation is achieved by the transcriptional inactivation of one X in the female. Three forms of X-chromosome inactivation (XCI) have been reported. In marsupials XCI is imprinted to occur exclusively on the paternal X, although the degree of silencing varies among somatic tissues (1, 4, 5). In contrast, eutherian XCI can be either imprinted or random (6, 7). In somatic tissues, XCI is random and can occur on either the maternal or paternal X. However, in the placental tissues of some eutherian mammals (e.g., mouse and cow), the paternal X resembles that in marsupials and is preferentially inactivated (7, 8). A third form of XCI is known to occur in the male germ line of eutherian mammals. During the first meiotic prophase, the X and Y become transcriptionally silenced in a process known as meiotic sex chromosome inactivation (MSCI) (9, 10).
The process of MSCI is the second significant consequence of adopting the XY method of sex determination in mammals. MSCI has so far been documented only in eutherian mammals in which, during prophase I of meiosis, homologous chromosomes pair and exchange genetic material. In the male germ line, however, the X and Y can pair only through their remaining homologous sequence, the pseudoautosomal region (11). For the mouse it was recently shown that asynapsed regions of the X and Y become transcriptionally inactivated simply by virtue of their being unpaired during pachytene of prophase I (12, 13), in a process that is termed meiotic silencing by unpaired chromatin (14). Several recent studies have also shown that the effects of male MSCI unexpectedly extend beyond meiosis I and continue through the end of spermatogenesis (15–17).
MSCI has long led to questions regarding its raison d'etre. The enrichment of spermatogenesis genes on the X and Y (18–20) despite meiotic and postmeiotic silencing raises one of the major paradoxes in the field. One idea is that MSCI and meiotic silencing by unpaired chromatin exist only as an evolutionary relic of meiotic silencing of unpaired DNA, a host defense mechanism first described in Neurospora crassa (21) with analogies in metazoans such as Caenorhabditis elegans (22). Other ideas suggest that silencing is obligatory for the suppression of recombination between nonhomologous regions of the X and Y (23), or for preventing asynapsed XY regions from triggering the meiotic checkpoint (24).
Meiotic and postmeiotic silencing may subserve yet another purpose: the problem of dosage compensation in the earliest mammals as the Y-chromosome lost genetic material. Imprinted paternal X silencing in the early eutherian embryo may at least in part be built on MSCI and its aftereffects in the male germ line (25–27), a hypothesis proposed earlier for the marsupial embryo (5, 28–30). In support of this, one study finds that the paternal X is already silent at conception and may be preinactivated (26). Because meiotic silencing of unpaired DNA/meiotic silencing by unpaired chromatin would silence any portion of the X that no longer has homology with the Y (12, 13), MSCI and its aftereffects would provide an immediate stop-gap measure of dosage compensation at a time of rapid change on the sex chromosomes (27). However, this hypothesis is opposed by the view that XCI takes place de novo at the four- to eight-cell stage, which would therefore argue against dosage compensation as a beneficiary of meiotic and postmeiotic silencing (see ref. 31 for full discussion). Thus, the basis of imprinted XCI in eutherians is currently controversial.
Much remains unknown regarding XCI in marsupials and to what extent mechanisms might be shared with those in eutherians. Eutherian XCI is regulated by the X-inactivation center (XIC/Xic), which contains the noncoding genes Xist (32–34), Tsix (35–37), and Xite (38). Repeated attempts to find the XIC orthologue in marsupials have failed and instead find that the syntenic region is rearranged (39–41). Interestingly, a recent report suggests that the ancestral XIST was vertebrate LNX3, a protein-coding gene with functions still extant but unrelated to dosage compensation in the marsupial (42). Without an XIC, how would XCI be achieved in the marsupial? One view holds that dosage compensation may have evolved independently in the marsupial and eutherian (42). Yet the classic view proposes that marsupial XCI results from preprogramming events in the paternal germ line (5, 28, 29). Therefore, a strictly germ-line-driven process, such as one proposed for imprinted XCI in the early mouse embryo (25, 26), might function in the marsupial (5, 27, 29, 30). Significantly, MSCI in the mouse does not require XIST (43, 44). However, there has been no formal evidence of MSCI so far in the marsupial, although a condensed X and Y has been reported in prophase I (45). Here we sought to determine whether a germ-line-driven mechanism might be feasible for marsupial XCI by investigating spermatogenic events in the South American opossum, Monodelphis domestica. We report that meiotic and postmeiotic events are surprisingly well conserved between metatherian and eutherian mammals.
Results and Discussion
Sex Chromosome Behavior During Opossum Meiosis.
During meiosis a primary spermatocyte proceeds through two division rounds (meiosis I and II) to generate four haploid spermatids that bear either an X- or Y-chromosome in addition to a haploid complement of autosomes. During meiosis I, homologous chromosomes pair and undergo homologous exchange during prophase (leptotene, zygotene, pachytene, and diplotene) and are segregated to distinct nuclei through metaphase, anaphase, and telophase. Primary spermatocytes at these various stages can be distinguished from each other by SCP3 staining of the axial elements and by their sex chromosome configuration. They differ from other cell types in a seminiferous tubular spread by their diploid chromosome constitution. It is during prophase I that MSCI takes place.
To determine the behavior of M. domestica chromosomes during meiosis I, we coimmunostained centromeric proteins and SCP3 and observed a 2n = 18 karyotype (Fig. 1), confirming a previous report of eight autosome pairs and a pair of sex chromosomes (XX female, XY male) (46). At leptotene, homologous chromosomes had yet to pair (pale SCP3 staining), but a bouquet-like arrangement of chromosomes was already evident with all homologous telomeres clustered at one pole of the nuclear membrane (data not shown). At zygotene, synapsis (as revealed by stronger SCP3 staining) first became evident at the telomeric poles of each autosome pair and moved inward along the axial elements (Fig. 1A). At this stage, the sex chromosomes became distinguishable for the first time (without chromosome painting by FISH), discernible as condensed acrocentric chromosomes (Fig. 1A′). Thus, through leptotene and zygotene, the behavior of the M. domestica X and Y paralleled that of the mouse X and Y.
Fig. 1.
Meiotic behavior of sex chromosomes in M. domestica. (A–E) Double immunostaining with anti-SCP3 (green) and anti-centromere (CEN; red) proteins. ∗, polarized direction of bouquet structure. (A–E and A′–D′) Multiple focal planes are projected. (A′–D′) Higher magnification of sex chromosomes. (E′) Higher magnification of DP shown in E′ (single z-sections). (F and G) DNA FISH (X-paint, green; Y-paint, red). Meiotic stages are noted above each panel. Arrowheads, DPs; arrows, XY bodies.
By early pachytene, homologous autosomes appeared fully synapsed, while the X and Y remained separate and became progressively more condensed, as evident by their decreased axial length and increased staining of axial elements (Fig. 1B). Interestingly, unlike the mouse X and Y, the opossum sex chromosomes folded into an arc, i.e., looped so that the two ends came in close contact (Fig. 1B′). Unlike observations in eutherians, X–Y association occurred late during pachytene and only after the autosomes had synapsed (Fig. 1 C and C′), confirming delayed sex chromosome association described previously in M. domestica (47) and other marsupials (48). This contrasts with eutherian pachytene, during which partial homology between the X and Y enables synaptonemal complex formation and true homologous pairing, akin to what is observed among autosomal pairs.
Some measure of X–Y association could be observed at mid-pachytene in the opossum with the formation of an XY body resembling that in eutherian spermatocytes (Fig. 1 C and C′). However, the absence of any obvious homology between the sex chromosomes appeared to preclude the formation of synaptonemal complex (48). Instead, the X and Y associate through a dense plate (DP) (49) between the X and Y arcs (Fig. 1 C and C′). SCP3 staining of the XY body became very intense, whereas staining of autosomes became weaker (Fig. 1C). By late pachytene, the axial elements of the sex chromosomes (SCP3 staining) became thin and entangled, whereas the DP became increasingly prominent (Fig. 1 D and D′). The DP appeared to be attached to the nuclear envelope, consistent with previous description (48). At diplotene, SCP3 staining of the DP remained intense but became extremely weak on autosomes (Fig. 1 E and E′). By DAPI staining alone, the XY body could easily be identified in the diplotene nucleus as a bright, condensed structure [Fig. 1E and supporting information (SI) Fig. 6]. By contrast, the DP was not DAPI-intense, suggesting that the DP is a proteinaceous structure with little if any chromosomal DNA (Fig. 1E′) (49). As confirmed by X- and Y-painting, the sex chromosomes were clearly distinguishable as DAPI-intense structures from early pachytene to diplotene (Fig. 1 F and G).
Thus, although the opossum also develops an XY body at mid-pachytene, it differs from the eutherian counterpart by an absence of any true synapsis and by its association through a DP. Furthermore, we noted that, although it resides at a peripheral nuclear location, the opossum XY body does not protrude out of the nucleus at mid-late pachytene as is characteristic of the mouse XY body. Overall, the behavior of the M. domestica sex chromosomes during male meiosis is similar to what has been described for other marsupials (48).
MSCI in the Opossum.
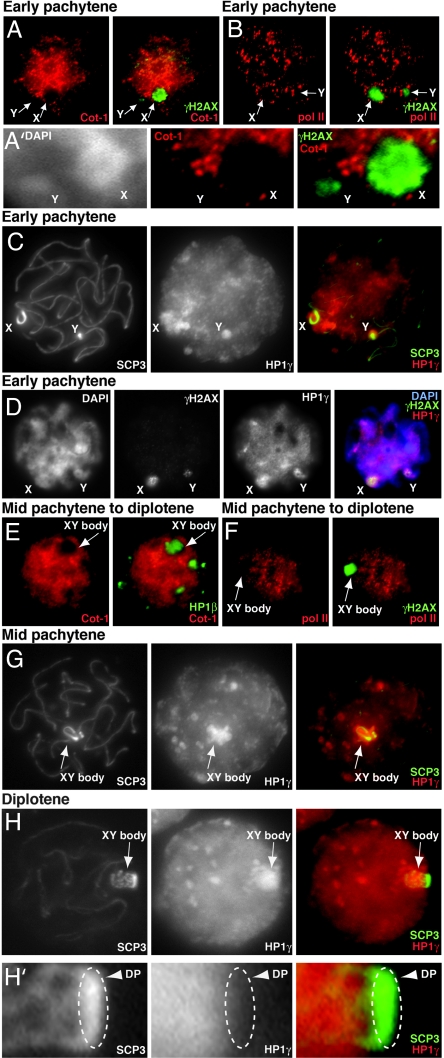
Although MSCI is well established in eutherians, this phenomenon has not previously been documented in marsupials. Because some aspects of the opossum XY body are reminiscent of that in eutherians, we next asked whether the opossum X and Y are also subject to MSCI. We performed Cot-1 RNA FISH, a technique whereby new RNA synthesis can be detected through hybridization to highly repetitive elements (Cot-1 fraction) found in the 3′ untranslated regions and introns before splicing (15). Indeed, Cot-1 RNA FISH showed that the X and Y excluded Cot-1 hybridization by early pachytene (Fig. 2 A and A′). Immunostaining for the RNA polymerase II (Pol-II) showed a dearth of Pol-II (Fig. 2B). At pachytene, the sex chromosomes became decorated with the heterochromatin-associated proteins, HP1β and HP1γ (Fig. 2C and data not shown), and also by γH2AX, a protein associated with the repair of double-strand breaks that is known to be an early mark of MSCI (50) (Fig. 2D). During this time, the X and Y appeared to be the prominent DAPI-intense structure in the early pachytene spermatocyte (Figs. 1F and 2D), even when the cells were prepared under relatively harsh conditions (hypotonically swollen nuclei) (SI Fig. 6). These characteristics persisted through diplotene, as Cot-1 and Pol-II signals continued to be excluded from the XY body (Fig. 2 E and F) and as HP1β, HP1γ, and γH2AX continued to be enriched on the XY body (Fig. 2 G and H; data not shown for HP1β). [Note that whereas the DP was consistently labeled by SCP3, other proteins such as HP1β, HP1γ, and γH2AX could not be detected on this structure at any time (Fig. 2 H and H′, SI Fig. 7, and data not shown).] Thus, we concluded that the opossum sex chromosomes are indeed subject to MSCI during pachytene in a manner similar to that observed for eutherian sex chromosomes.
Fig. 2.
MSCI in M. domestica. (A and E) Cot-1 RNA FISH (red) and immunostaining with anti-γH2AX (green). (B and F) Double immunostaining with anti-Pol-II (red) and γH2AX (green). (C, G, and H) Double immunostaining with anti-SCP3 (green) and HP1γ (red). (D) Double immunostaining with anti-γH2AX (green) and HP1γ (red). (A′ and H′) Higher magnification of sex chromosome and DP shown in A and H, respectively. (A, A′, B, E, F, and H′) Single z-sections. (C, D, G, and H) Multiple focal planes are projected. Meiotic stages are noted above panels. Arrows, sex chromosomes; arrowheads, DPs.
However, we also observed several interesting differences between eutherian and marsupial MSCI. First, although it is thought that eutherian MSCI occurs after the X and Y have partially synapsed through the pseudoautosomal region (10, 51), MSCI in the opossum occurs by early pachytene long before X–Y association (Fig. 2 A–D). This implied that MSCI does not require partial synapsis of the X and Y, consistent with the idea that MSCI is induced by unpaired DNA (meiotic silencing of unpaired DNA/meiotic silencing by unpaired chromatin) rather than by paired elements (12). A second significant difference is that HP1β and HP1γ association in the opossum takes place earlier than in the mouse. Whereas these proteins are found on the mouse XY body only late in pachytene (15), they could be observed on the opossum sex chromosomes by early pachytene as the chromosomes become looped (Fig. 2C) and decorated by γH2AX (Fig. 2D).
PMSC in the Opossum.
MSCI was previously believed to be specific to meiosis I. Because it is now known that most genes on the eutherian sex chromosomes do not reactivate at the end of meiosis I (15–17), we asked whether silencing also persists into spermiogenesis in opossum. We first examined secondary spermatocytes undergoing meiosis II. Secondary spermatocytes could usually be observed as two attached or closely juxtaposed daughter cells, one carrying only the X and the other only the Y. They can be distinguished from round spermatids (which also have segregated Xs and Ys) by their larger size and presence of sister chromatids (by centromeric staining; data not shown).
In secondary spermatocytes, we were surprised to find that γH2AX remained (Fig. 3 A and B; 94% of nuclei with γH2AX on sex chromosome, n = 70), in contrast to its disappearance from the eutherian XY after diplotene I (15). Because γH2AX is thought to be involved specifically with events during prophase I in eutherian spermatocytes, its continued presence on the segregated X and Y seemed rather puzzling. Analysis by Cot-1 RNA FISH, Pol-II immunostaining, and DAPI staining showed that the sex chromosomes remained undertranscribed in the secondary spermatocytes (Fig. 3 A and B). These results demonstrated that the sex chromosomes remained relatively suppressed in the secondary spermatocyte.
Fig. 3.
Continuity of silencing into PMSC in M. domestica. (A) Cot-1 RNA FISH (red) and immunostaining with anti-γH2AX (green). (B) Double immunostaining with anti-Pol-II (red) and γH2AX (green), followed by DNA FISH (X-paint, green; Y-paint, red). (C and D) Cot-1 RNA FISH (red) and immunostaining with anti-HP1β (green). (E) DNA FISH (X-paint, green). (F) DNA FISH (Y-paint, green). All images are single z-sections. Cell types: secondary spermatocytes in A and B and round spermatids in C–F.
To determine whether silencing persisted after the completion of meiosis, we investigated round spermatids. Round spermatids could be distinguished from secondary spermatocytes by their smaller size, tendency to cluster into groups of X-bearing and Y-bearing daughter cells, and a clear nine-chromosome constitution (e.g., nine centromeric signals) (Fig. 4 K and L). Indeed, both sex chromosomes remained within Cot-1 holes (Fig. 3 C and D), retained their DAPI-intense staining, and continued to be enriched for HP1β and HP1γ (Fig. 3 C–F and SI Fig. 8). These data showed that, just as in eutherians, the postmeiotic X of the opossum is transcriptionally suppressed. Their epigenetic profiles were also similar. Trimethylation of H3-K9 (H3–3meK9) was initially observed on the sex chromosomes (as marked by HP1γ) in primary spermatocytes. This occurred by the time of XY body formation in pachytene (meiosis I) (Fig. 4A and SI Fig. 9) and continued in round spermatids (Fig. 4 B and C and SI Fig. 9). In mice, H3–3meK9, HP1β, and HP1γ persist longer than any other chromatin marks and are present until genome-wide chromosome condensation at the end of spermiogenesis (SI Fig. 10) (15, 17). H3-K27 trimethylation (H3–3meK27), a marker of the inactive X in the mouse soma (52), was present at relatively low levels on the XY body as compared with the rest of the genome in opossum primary spermatocyte (Fig. 4G) and also on the X in round spermatids (Fig. 4H). These data demonstrated that the postmeiotic X continues to be transcriptionally suppressed in marsupials, reminiscent of the PMSC described in eutherians (15–17). Thus, the silencing initiated by marsupial MSCI in meiosis I persists through meiosis II and the postmeiotic period.
Fig. 4.
Characterization of M. domestica PMSC. (A–E, G, and H) Double immunostaining of various chromatin marks (green) as indicated along with HP1γ (red) in mid-pachytene to diplotene (A, D, and G), round spermatids (B, C, E, F, H, and I), mouse round spermatid (J), and Monodelphis round spermatids (K and L). Arrows, XY bodies. (F and I) Double immunostaining of H3–3meK9 (green) and γH2AX (red) in the round spermatids. All images in A–I are single z-sections. (J–L) Double immunostaining with anti-HP1β (green) and CEN (red) of round spermatid in mouse (J) and in M. domestica (K and L). Multiple focal planes of CEN staining are superimposed on single z-sections of DAPI and HP1β. ∗, chromocenter; arrowheads, mouse PMSCs. All images except J are of M. domestica.
Some interesting differences between the mouse and opossum PMSC could also be observed. First, murine PMSC can be recognized as a DAPI-bright structure attached to the spermatid's single chromocenter, the DAPI-intense focal cluster of centric heterochromatin (Fig. 4J) (15–17). By contrast, we did not observe a chromocenter in the opossum, because immunostaining for HP1β and centromere proteins showed that the centromeres were distributed all over the nuclei and were not DAPI-intense (Fig. 4 K and L). The opossum PMSC was therefore uniquely DAPI-intense (Fig. 4 K and L). Other differences occurred in the profiles of various chromatin-associated proteins. For example, dimethylation of H3-K9 (H3–2meK9) was present in primary spermatocytes by pachytene/diplotene of both species (Fig. 4D) (15–17, 53) but was curiously absent from (or undetectable on) the X and Y of the opossum spermatid (Fig. 4E). Furthermore, γH2AX persisted longer in the opossum, decorating the X and Y in 94% of secondary spermatocytes (n = 70) (Fig. 3 A and B). A further surprise was that γH2AX was even enriched on the X and Y in 4% of early round spermatids (n = 451) (Fig. 4F and SI Fig. 9) before disappearing completely in later stages (Fig. 4I and SI Fig. 9). Thus, although PMSC is conserved in the marsupial, its epigenetic profile, subject to the vagaries of immunostaining, appears to differ slightly from that of eutherians.
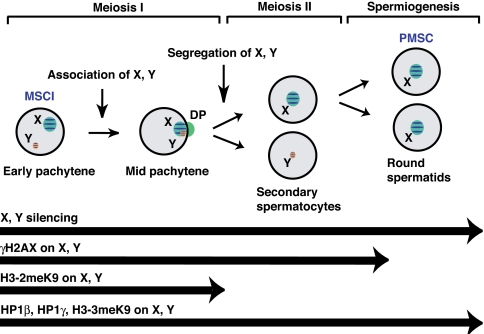
In conclusion, our study and related work (J. Hornecker, P. Samollow, E. Robinson, J.L.V., and J.R.M., unpublished data) show that MSCI and PMSC occur in the marsupial and that the silencing initiated during the first meiotic prophase continues through meiosis II and into the postmeiotic period (Fig. 5). Thus, spermatogenic events regulating transcriptional activity of the sex chromosomes are very well conserved in the marsupial and eutherian. This is in striking contrast to the absence of conservation in XIC elements that regulate XCI in the eutherian soma (39–42). These data are consistent with a mechanism of imprinted XCI that would occur independent of XIST in the marsupial and make possible a mechanism that relies instead on inheritance of a silent X derived from the male germ line (5, 25, 28–30).
Fig. 5.
Continuity of silencing from MSCI to PMSC in marsupial. The silencing takes place by MSCI at early pachytene and is maintained in spermatids as PMSC with modifications similar to those seen in eutherian. Barred chromosomes represent transcriptionally suppressed chromatin.
The state of the paternal X upon arrival in the opossum zygote requires further study. In the absence of any significant cytoplasm and the replacement of histones for protamines, how might epigenetic information be transmitted from the sperm to the zygote? Previous studies of X-linked genes silenced by MSCI in eutherians have shown that hypermethylation of DNA is not involved (54, 55). Studies of the active and inactive X-chromosomes in female kangaroos have indicated that DNA methylation is also not involved in somatic XCI (56). Our studies have highlighted three persistent marks of meiotic silencing (H3–3meK9, HP1β, and HP1γ) that are shared between eutherians and metatherians (Fig. 4 and SI Fig. 10) (15, 17). Interestingly, H3–3meK9, HP1β, and HP1γ are also the last chromatin-associated marks to be detected before protamine-mediated compaction during mouse spermiogenesis (SI Fig. 10). These marks are therefore candidates for transgenerational inheritance of epigenetic programming associated with the paternal X. Until recently, spermatozoa were believed to deliver little more than DNA into the oocyte. However, several studies now lend credence to paternal inheritance of both RNA and nonprotamine proteins that may be critical to early embryonic development (57–60). Given the apparent absence of the XIC, could epigenetic programming initiated by MSCI and maintained by PMSC survive protamine packaging to establish imprinted XCI in the marsupial embryo?
Materials and Methods
Slide Preparation.
M. domestica seminiferous tubules were prepared with slight modifications from previous methods (15). Testes were dissected in PBS on ice. Several pieces of seminiferous tubule were placed in 4% paraformaldehyde in 1× PBS plus 0.5% Triton X-100 for 10 min at room temperature, rinsed in 1× PBS, shredded between two forceps, cytospun onto a glass slide at 2,000 rpm (Cytospin 4; Thermo Fisher Scientific, Waltham, MA) for 10 min, and air-dried. Hypotonic treatment was performed as described (61). Mouse testis slides were prepared as described (15).
Fractionation of M. domestica Cot-1 DNA.
Fractionation of highly repetitive Cot-1 DNA from M. domestica was performed according to ref. 62. M. domestica genomic DNA (100 μg/ml) was denatured in 250 mM phosphate buffer (pH 6.8), then annealed for 24 h at 55°C. The double-stranded/repetitive fraction was purified by hydroxyapatite chromatography.
FISH and Immunofluorescence.
Cot-1 RNA FISH was performed as described (15). For immunofluorescence, slides were incubated in PBT (0.15% BSA/0.1% Tween 20) plus 5% goat serum for 60 min before overnight incubation at 37°C with the following antibodies: SCP3 (Novus Biologicals, Littleton, CO), 1:100; centromere (Antibodies Incorporated, Davis, CA), 1:100; RNA Pol-II CTD 8WG16 (Upstate, Charlottesville, VA), 1:200; HP1β (Abcam, Cambridge, MA), 1:100; HP1γ (Chemicon, Temecula, CA), 1:1,000; H3–2meK9 (Upstate), 1:100; H3–3meK9 (Upstate; used unless otherwise designated), 1:200; H3–3meK9 (Abcam; used only in SI Fig. 10), 1:200; H3–3meK27 (Upstate), 1:200; γH2AX (kindly provided by R. Scully, Beth Israel Deaconess Medical Center, Boston, MA), 1:1,000; and γH2AX (Upstate), 1:5,000 (used in 1:1,000 at 4°C after Cot-1 RNA FISH). Thereafter, slides were washed three times for 5 min in PBS plus 0.1% Tween 20, incubated with secondary antibodies (Alexa dyes; Invitrogen, Carlsbad, CA) at 1:500 for 60 min in PBT, washed in PBS plus 0.1% Tween 20, and mounted in Vectashield with DAPI. For combined RNA FISH/immunostaining, we carried out RNA FISH first, followed by immunofluorescence. DNA FISH was performed by using chromosome painting for the M. domestica (kindly provided by W. Rens, Cambridge Resource Centre for Comparative Genomics, Cambridge, U.K.). All images were acquired with the Axioplan microscope (Zeiss, Thornwood, NY). Z-section images were acquired by using Openlab (Improvision, Lexington, MA).
Supplementary Material
Acknowledgments
We thank W. Rens and the Wellcome Trust for X and Y chromosome paints for M. domestica, J. Dennis for advice on Cot-1 DNA fractionation, and J. Hornecker and H. Yoshioka for help in preparation of M. domestica testes. This work was supported by a grant from the Japan Society for the Promotion of Science (to S.H.N.), a grant from the Robert J. Kleberg, Jr., and Helen C. Kleberg Foundation (to J.L.V.), and National Institutes of Health Grants HD-46637 (to J.R.M.) and GM-58839 (to J.T.L.). J.T.L. is also an Investigator of the Howard Hughes Medical Institute.
Abbreviations
- XCI
X-chromosome inactivation
- MSCI
meiotic sex chromosome inactivation
- PMSC
postmeiotic sex chromatin
- DP
dense plate
- XIC
X-inactivation center
- Pol-II
polymerase II.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0700323104/DC1.
References
- 1.Graves JA. Annu Rev Genet. 1996;30:233–260. doi: 10.1146/annurev.genet.30.1.233. [DOI] [PubMed] [Google Scholar]
- 2.Charlesworth B. Science. 1991;251:1030–1033. doi: 10.1126/science.1998119. [DOI] [PubMed] [Google Scholar]
- 3.Vicoso B, Charlesworth B. Nat Rev Genet. 2006;7:645–653. doi: 10.1038/nrg1914. [DOI] [PubMed] [Google Scholar]
- 4.Sharman GB. Nature. 1971;230:231–232. doi: 10.1038/230231a0. [DOI] [PubMed] [Google Scholar]
- 5.McCarrey JR. In: Gene Families: Studies of DNA, RNA, Enzymes, and Proteins. Xue G, Xue Z, Xu R, Holmes R, Hammond GL, Lim HA, editors. Teaneck, NJ: World Scientific; 2001. pp. 59–72. [Google Scholar]
- 6.Lyon MF. Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- 7.Takagi N, Sasaki M. Nature. 1975;256:640–642. doi: 10.1038/256640a0. [DOI] [PubMed] [Google Scholar]
- 8.Xue F, Tian XC, Du F, Kubota C, Taneja M, Dinnyes A, Dai Y, Levine H, Pereira LV, Yang X. Nat Genet. 2002;31:216–220. doi: 10.1038/ng900. [DOI] [PubMed] [Google Scholar]
- 9.Lifschytz E, Lindsley DL. Proc Natl Acad Sci USA. 1972;69:182–186. doi: 10.1073/pnas.69.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turner JM. Development (Cambridge, UK) 2007;134:1823–1831. doi: 10.1242/dev.000018. [DOI] [PubMed] [Google Scholar]
- 11.Burgoyne PS. Hum Genet. 1982;61:85–90. doi: 10.1007/BF00274192. [DOI] [PubMed] [Google Scholar]
- 12.Turner JM, Mahadevaiah SK, Fernandez-Capetillo O, Nussenzweig A, Xu X, Deng CX, Burgoyne PS. Nat Genet. 2005;37:41–47. doi: 10.1038/ng1484. [DOI] [PubMed] [Google Scholar]
- 13.Baarends WM, Wassenaar E, van der Laan R, Hoogerbrugge J, Sleddens-Linkels E, Hoeijmakers JH, de Boer P, Grootegoed JA. Mol Cell Biol. 2005;25:1041–1053. doi: 10.1128/MCB.25.3.1041-1053.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schimenti J. Nat Genet. 2005;37:11–13. doi: 10.1038/ng0105-11. [DOI] [PubMed] [Google Scholar]
- 15.Namekawa SH, Park PJ, Zhang LF, Shima JE, McCarrey JR, Griswold MD, Lee JT. Curr Biol. 2006;16:660–667. doi: 10.1016/j.cub.2006.01.066. [DOI] [PubMed] [Google Scholar]
- 16.Turner JM, Mahadevaiah SK, Ellis PJ, Mitchell MJ, Burgoyne PS. Dev Cell. 2006;10:521–529. doi: 10.1016/j.devcel.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 17.Greaves IK, Rangasamy D, Devoy M, Marshall Graves JA, Tremethick DJ. Mol Cell Biol. 2006;26:5394–5405. doi: 10.1128/MCB.00519-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang PJ, McCarrey JR, Yang F, Page DC. Nat Genet. 2001;27:422–426. doi: 10.1038/86927. [DOI] [PubMed] [Google Scholar]
- 19.Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, Repping S, Pyntikova T, Ali J, Bieri T, et al. Nature. 2003;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- 20.Khil PP, Smirnova NA, Romanienko PJ, Camerini-Otero RD. Nat Genet. 2004;36:642–646. doi: 10.1038/ng1368. [DOI] [PubMed] [Google Scholar]
- 21.Shiu PK, Raju NB, Zickler D, Metzenberg RL. Cell. 2001;107:905–916. doi: 10.1016/s0092-8674(01)00609-2. [DOI] [PubMed] [Google Scholar]
- 22.Bean CJ, Schaner CE, Kelly WG. Nat Genet. 2004;36:100–105. doi: 10.1038/ng1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jablonka E, Lamb MJ. J Theor Biol. 1988;133:23–36. doi: 10.1016/s0022-5193(88)80022-5. [DOI] [PubMed] [Google Scholar]
- 24.Miklos GL. Cytogenet Cell Genet. 1974;13:558–577. doi: 10.1159/000130307. [DOI] [PubMed] [Google Scholar]
- 25.Huynh KD, Lee JT. Curr Opin Cell Biol. 2001;13:690–697. doi: 10.1016/s0955-0674(00)00272-6. [DOI] [PubMed] [Google Scholar]
- 26.Huynh KD, Lee JT. Nature. 2003;426:857–862. doi: 10.1038/nature02222. [DOI] [PubMed] [Google Scholar]
- 27.Huynh KD, Lee JT. Nat Rev Genet. 2005;6:410–418. doi: 10.1038/nrg1604. [DOI] [PubMed] [Google Scholar]
- 28.Cooper DW. Nature. 1971;230:292–294. doi: 10.1038/230292a0. [DOI] [PubMed] [Google Scholar]
- 29.Vandeberg JL. J Exp Zool. 1983;228:271–286. doi: 10.1002/jez.1402280211. [DOI] [PubMed] [Google Scholar]
- 30.Lyon MF. In: Results and Problems in Cell Differentiation. Ohlsson R, editor. Heidelberg: Springer; 1999. pp. 73–90. [DOI] [PubMed] [Google Scholar]
- 31.Heard E, Disteche CM. Genes Dev. 2006;20:1848–1867. doi: 10.1101/gad.1422906. [DOI] [PubMed] [Google Scholar]
- 32.Borsani G, Tonlorenzi R, Simmler MC, Dandolo L, Arnaud D, Capra V, Grompe M, Pizzuti A, Muzny D, Lawrence C, et al. Nature. 1991;351:325–329. doi: 10.1038/351325a0. [DOI] [PubMed] [Google Scholar]
- 33.Brockdorff N, Ashworth A, Kay GF, Cooper P, Smith S, McCabe VM, Norris DP, Penny GD, Patel D, Rastan S. Nature. 1991;351:329–331. doi: 10.1038/351329a0. [DOI] [PubMed] [Google Scholar]
- 34.Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, Willard HF. Nature. 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 35.Lee JT. Cell. 2000;103:17–27. doi: 10.1016/s0092-8674(00)00101-x. [DOI] [PubMed] [Google Scholar]
- 36.Sado T, Wang Z, Sasaki H, Li E. Development (Cambridge, UK) 2001;128:1275–1286. doi: 10.1242/dev.128.8.1275. [DOI] [PubMed] [Google Scholar]
- 37.Lee JT, Lu N. Cell. 1999;99:47–57. doi: 10.1016/s0092-8674(00)80061-6. [DOI] [PubMed] [Google Scholar]
- 38.Ogawa Y, Lee JT. Mol Cell. 2003;11:731–743. doi: 10.1016/s1097-2765(03)00063-7. [DOI] [PubMed] [Google Scholar]
- 39.Davidow LS, Breen M, Duke SE, Samollow PB, McCarrey JR, Lee JT. Chromosome Res. 2007;15:137–146. doi: 10.1007/s10577-007-1121-6. [DOI] [PubMed] [Google Scholar]
- 40.Hore TA, Koina E, Wakefield MJ, Marshall Graves JA. Chromosome Res. 2007;15:147–161. doi: 10.1007/s10577-007-1119-0. [DOI] [PubMed] [Google Scholar]
- 41.Shevchenko AI, Zakharova IS, Elisaphenko EA, Kolesnikov NN, Whitehead S, Bird C, Ross M, Weidman JR, Jirtle RL, Karamysheva TV, et al. Chromosome Res. 2007;15:127–136. doi: 10.1007/s10577-006-1115-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Duret L, Chureau C, Samain S, Weissenbach J, Avner P. Science. 2006;312:1653–1655. doi: 10.1126/science.1126316. [DOI] [PubMed] [Google Scholar]
- 43.Turner JM, Mahadevaiah SK, Elliott DJ, Garchon HJ, Pehrson JR, Jaenisch R, Burgoyne PS. J Cell Sci. 2002;115:4097–4105. doi: 10.1242/jcs.00111. [DOI] [PubMed] [Google Scholar]
- 44.McCarrey JR, Watson C, Atencio J, Ostermeier GC, Marahrens Y, Jaenisch R, Krawetz SA. Genesis. 2002;34:257–266. doi: 10.1002/gene.10163. [DOI] [PubMed] [Google Scholar]
- 45.Solari AJ. Int Rev Cytol. 1974;38:273–317. doi: 10.1016/s0074-7696(08)60928-6. [DOI] [PubMed] [Google Scholar]
- 46.Reig OA, Bianchi NO. Experientia. 1969;25:1210–1211. doi: 10.1007/BF01900283. [DOI] [PubMed] [Google Scholar]
- 47.Solari AJ, Bianchi NO. Chromosoma. 1975;52:11–25. doi: 10.1007/BF00285785. [DOI] [PubMed] [Google Scholar]
- 48.Page J, Berrios S, Rufas JS, Parra MT, Suja JA, Heyting C, Fernandez-Donoso R. J Cell Sci. 2003;116:551–560. doi: 10.1242/jcs.00252. [DOI] [PubMed] [Google Scholar]
- 49.Page J, Viera A, Parra MT, de la Fuente R, Suja JA, Prieto I, Barbero JL, Rufas JS, Berrios S, Fernandez-Donoso R. PLoS Genet. 2006;2:e136. doi: 10.1371/journal.pgen.0020136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mahadevaiah SK, Turner JM, Baudat F, Rogakou EP, de Boer P, Blanco-Rodriguez J, Jasin M, Keeney S, Bonner WM, Burgoyne PS. Nat Genet. 2001;27:271–276. doi: 10.1038/85830. [DOI] [PubMed] [Google Scholar]
- 51.Turner JM, Aprelikova O, Xu X, Wang R, Kim S, Chandramouli GV, Barrett JC, Burgoyne PS, Deng CX. Curr Biol. 2004;14:2135–2142. doi: 10.1016/j.cub.2004.11.032. [DOI] [PubMed] [Google Scholar]
- 52.Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, de la Cruz CC, Otte AP, Panning B, Zhang Y. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 53.Khalil AM, Boyar FZ, Driscoll DJ. Proc Natl Acad Sci USA. 2004;101:16583–16587. doi: 10.1073/pnas.0406325101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McCarrey JR, Berg WM, Paragioudakis SJ, Zhang PL, Dilworth DD, Arnold BL, Rossi JJ. Dev Biol. 1992;154:160–168. doi: 10.1016/0012-1606(92)90056-m. [DOI] [PubMed] [Google Scholar]
- 55.Grant M, Zuccotti M, Monk M. Nat Genet. 1992;2:161–166. doi: 10.1038/ng1092-161. [DOI] [PubMed] [Google Scholar]
- 56.Loebel DA, Johnston PG. Mamm Genome. 1997;8:146–147. doi: 10.1007/s003359900376. [DOI] [PubMed] [Google Scholar]
- 57.Krawetz SA. Nat Rev Genet. 2005;6:633–642. doi: 10.1038/nrg1654. [DOI] [PubMed] [Google Scholar]
- 58.Chu DS, Liu H, Nix P, Wu TF, Ralston EJ, Yates JR, III, Meyer BJ. Nature. 2006;443:101–105. doi: 10.1038/nature05050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gatewood JM, Cook GR, Balhorn R, Bradbury EM, Schmid CW. Science. 1987;236:962–964. doi: 10.1126/science.3576213. [DOI] [PubMed] [Google Scholar]
- 60.Bench GS, Friz AM, Corzett MH, Morse DH, Balhorn R. Cytometry. 1996;23:263–271. doi: 10.1002/(SICI)1097-0320(19960401)23:4<263::AID-CYTO1>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 61.Peters AH, Plug AW, van Vugt MJ, de Boer P. Chromosome Res. 1997;5:66–68. doi: 10.1023/a:1018445520117. [DOI] [PubMed] [Google Scholar]
- 62.Britten RJ, Kohne DE. Science. 1968;161:529–540. doi: 10.1126/science.161.3841.529. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.