Abstract
At low micromolar concentrations, polyunsaturated fatty acids (PUFAs) alter the function of many membrane proteins. PUFAs exert their effects on unrelated proteins at similar concentrations, suggesting a common mode of action. Because lipid bilayers serve as the common “solvent” for membrane proteins, the common mechanism could be that PUFAs adsorb to the bilayer/solution interface to promote a negative-going change in lipid intrinsic curvature and, like other reversibly adsorbing amphiphiles, increase bilayer elasticity. PUFA adsorption thus would alter the bilayer deformation energy associated with protein conformational changes involving the protein/bilayer boundary, which would alter protein function. To explore the feasibility of such a mechanism, we used gramicidin (gA) analogues of different lengths together with bilayers of different thicknesses to assess whether docosahexaenoic acid (DHA) could exert its effects through a bilayer-mediated mechanism. Indeed, DHA increases gA channel appearance rates and lifetimes and decreases the free energy of channel formation. The appearance rate and lifetime changes increase with increasing channel-bilayer hydrophobic mismatch and are not related to differing DHA bilayer absorption coefficients. DHA thus alters bilayer elastic properties, not just lipid intrinsic curvature; the elasticity changes are important for DHA's bilayer-modifying actions. Oleic acid (OA), which has little effect on membrane protein function, exerts no such effects despite OA's adsorption coefficient being an order of magnitude greater than DHA's. These results suggest that DHA (and other PUFAs) may modulate membrane protein function by bilayer-mediated mechanisms that do not involve specific protein binding but rather changes in bilayer material properties.
Keywords: bilayer material properties, bilayer stiffness, gramicidin channels, hydrophobic mismatch, polyunsaturated fatty acid
Polyunsaturated fatty acids (PUFAs) modulate a wide variety of biological processes (1–3), and alter the function of a diverse group of unrelated membrane proteins (Table 1, for additional examples, see ref. 4), whereas saturated or mono-unsaturated fatty acids such as oleic acid (OA) are relatively inert. Among the acute effects of PUFAs is the reversal of the arrhythmias underlying sudden cardiac death in rats (5), dogs (6), and humans (7), most likely due to inhibition of cardiac sodium and L-type calcium channels. The mechanism(s) underlying the reversal remain unclear, but it occurs at the low micromolar PUFA concentrations where PUFAs are general modulators of membrane protein function. Because PUFAs avidly adsorb to biological membranes (8, 9), and the commonality among the proteins in Table 1 is that they are imbedded in lipid bilayers, PUFAs may act through some common, bilayer-mediated mechanism.
Table 1.
Membrane proteins that are modulated by DHA and other PUFAs
| Protein | PUFA | Action | Ref. |
|---|---|---|---|
| Cardiac Na+ channel | AA, EPA, DHA | Inhibit | 10 |
| L-type Ca2+ channel | ALA, LA, AA, EPA, DHA | Inhibit | 11 |
| Kv1.5 channel | AA, DHA | Inhibit | 12 |
| HERG channel | AA, DHA | Inhibit | 13 |
| TRAAK-1 channel | AA, EPA, DHA | Activate | 14 |
| TRPV1 | EPA, DHA | Activate | 15 |
| nAChR channel | DHA | ↑ Desensitization | 16 |
| GABAa channel | DHA | ↑ Desensitization | 17 |
| GluR6 glutamate receptor | AA, DHA | Inhibit | 18 |
| Connexin43 channel | GLA, AA, EPA, DHA | Inhibit | 19 |
| Na+,K+-ATPase | EPA, DHA | Inhibit | 20 |
AA, arachidonic acid; ALA, α-linolenic acid; EPA, eicosapentaenoic acid; GLA, γ-linolenic acid
Bilayer-dependent regulation of membrane function can occur when membrane proteins undergo conformational changes that involve the protein/bilayer boundary (for example, see ref. 21). Because lipid bilayers are elastic bodies (22) and bilayer-spanning proteins are coupled to the host bilayer through hydrophobic interactions (23), membrane protein conformational changes incur an energetic cost (24–26), the bilayer deformation energy (ΔGdef0), which causes protein function to be modulated by the lipid bilayer (27, 28).
For a given protein, ΔGdef0 varies as a function of the mismatch between the protein's hydrophobic length (l) and the bilayer's hydrophobic thickness (d0), the intrinsic curvature (c0) of the bilayer-forming lipids, and the bilayer compression and bending moduli. To a first approximation, ΔGdef0 can be expressed as a biquadratic form in the hydrophobic mismatch, d0 − l, and c0 (28):
where the coefficients HB, Hx, and Hc are determined by the protein geometry, the bilayer thickness and elastic moduli (29). The elastic moduli (30–32) and intrinsic curvature (33, 34) may be altered by reversibly adsorbing amphiphiles, which would provide a basis for the acute effects of PUFAs on membrane protein function, although the relative importance of changes in elastic moduli and curvature would need to be established. PUFAs and micelle-forming amphiphiles, for example, have opposite effects on lipid intrinsic curvature (34, 35), yet both shift the steady-state inactivation curve for voltage-dependent sodium channels in the same (hyperpolarizing) direction (35, 36), which would suggest that changes in elastic moduli dominate over changes in curvature.
To address these questions we used gramicidin (gA) channels of different lengths to monitor how PUFAs modulate lipid bilayer properties. gA channels are formed by transbilayer association of two monomers (37). When the bilayer's hydrophobic thickness, thickness to match the protein's hydrophobic length, differs from the channel's hydrophobic length, l, the bilayer will adjust its d0 resulting in a local bilayer deformation with energetic cost ΔGdef0 (cf. Eq. 1). The bilayer responds by applying a disjoining (restoring) force to the channel dimer:
Changes in this disjoining force are reflected as changes in channel lifetime (τ), meaning that gA channels can be used as force transducers (38) to report changes in bilayer elasticity and lipid curvature. It is thus possible to assess whether docosahexaenoic acid (DHA) alters bilayer properties and thus might exert its effects on membrane protein function through an indirect (bilayer-mediated) mechanism.
Results
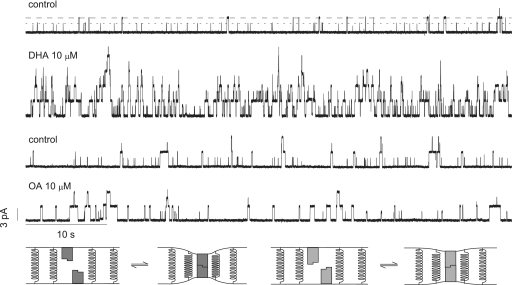
DHA is a potent modifier of gramicidin channel activity, whereas OA is not (Fig. 1). One can examine the role of channel-bilayer hydrophobic mismatch by comparing the relative changes in lifetimes and appearance rates for the shorter and longer channels (Fig. 1 Lower). 10 μM DHA§, but not 10 μM OA, increases the appearance rates of both gA(13) and AgA−(15) channels with the larger effect on the shorter channels (Figs. 2 and 3). [The enantiomeric gA(13) and AgA−(15) were used to prevent hybrid channel formation, which simplifies the analysis.]
Fig. 1.
Effect of OA and DHA on gA channel activity. Current traces before and after addition of 10 μM DHA (top two traces) or OA (bottom two traces) to both sides of a DC18:1PC/n-decane bilayer containing gA(13) and AgA−(15). (Results from two different experiments.) The interrupted lines denote the current levels for gA(13) (short dash) and AgA−(15) (long dash). 1 M NaCl, 10 mM Hepes, pH 7, ± 200 mV, 500 Hz. The cartoons at the bottom of the figure illustrate the differences in bilayer deformation with differing hydrophobic mismatch between channel (shaded blocks) and lipid bilayer (represented by springs).
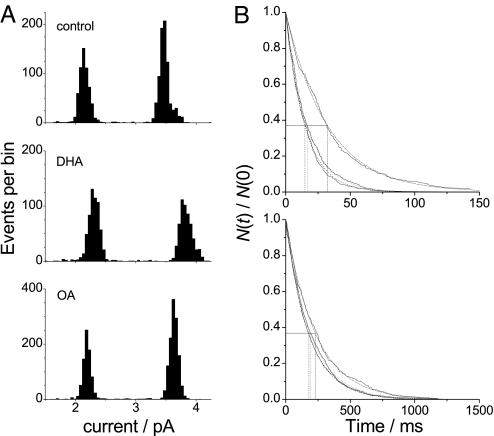
Fig. 2.
Effect of OA and DHA on gA single-channel current transitions and lifetimes. (A) Current transition amplitude histograms of gA(13) (left peak) and AgA−(15) (right peak) channels in DC18:1PC bilayers in the absence or presence of 10 μM DHA or OA. (B) Normalized single-channel survivor histograms for gA(13) (Upper) and AgA−(15) (Lower) fitted with single exponential distributions; note the 10-fold difference in the scale of the abscissae. The vertical dotted lines indicate the average channel lifetimes of (from left to right) control, 10 μM OA, and 10 μM DHA.
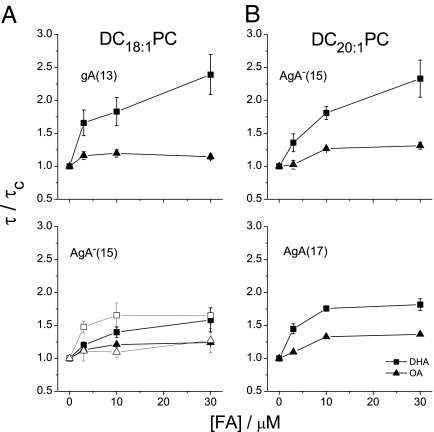
Fig. 3.
Concentration dependence of OA's and DHA's effects on channel lifetimes. Changes in the lifetime relative to control are plotted as normalized dose-response curves for DHA (filled square) and OA (filled triangle). (A) gA(13) and AgA−(15) channels in DC18:1PC bilayers, as well as AgA−(15) channels in DC18:1PC:Chol (1:1) bilayers [DHA (open squares) and OA (open triangles) in Lower]; (B) AgA−(15) and AgA(17) channels in DC20:1PC bilayers.
DHA also increases the single-channel current transition amplitudes (i), whereas OA has little or no effect (Fig. 2A). The lifetime distributions in the absence or presence of DHA (or OA) can be fit by single exponential distributions (Fig. 2B), meaning that DHA modulates the function of an existing channel type, rather than promoting the appearance of a new channel population. (In the absence of gA, neither DHA nor OA caused channel-like activity.)
When DHA was added to only one side of a bilayer, the single-channel currents and lifetimes were similar at both polarities (results not shown), indicating that transmembrane DHA flux (for example, see refs. 39 and 40) caused the DHA mole fraction to be similar in the two leaflets.
As will be important below, DHA is more active in altering the function of the shorter gA(13) channels. This difference is not due to the difference in channel chirality because DHA has equal effect on the function of left- and right-handed channels of the same length [10 μM DHA increases the lifetimes of the enantiomeric AgA(15) and AgA−(15) channels by 1.51- and 1.55-fold, respectively]. To a first approximation, gA channels' conformational preference does not vary as a function of bilayer thickness (41); the length dependence thus suggests that DHA alters gA channel function by a bilayer-mediated mechanism dependent on channel-bilayer hydrophobic mismatch (35, 42). To explore this further, we examined how the FA-induced changes in gA channel function varied as a function of bilayer thickness, channel length, and the presence of cholesterol, which increases the bilayer elastic moduli (43).
Fig. 3 shows changes in gA channel lifetime (τ) for different bilayer compositions at nominal [FA]s between 3 μM and 30 μM. At all [FA], DHA is a more effective modulator than OA. In cholesterol-free bilayers, the changes in channel function increase with increasing channel-bilayer hydrophobic mismatch, whether the channel length or the bilayer thickness is varied. In the presence of cholesterol, at a cholesterol:DC18:1PC molar ratio of 1:1, DHA is an even more effective modulator of channel function than in DC18:1PC bilayers, whereas OA remains inert. DHA also caused larger changes in the single-channel current than was the case in the absence of cholesterol. A 10 μM concentration of DHA caused a 15% increase in the absence and a 65% increase in the presence of cholesterol. Even OA increased the current: 10 μM OA caused a 5% increase in the absence and a 15% increase in the presence of cholesterol.
The different effects of DHA (and of OA) do not reflect different FA adsorption to the bilayers. The lipid bilayer/electrolyte partition coefficients (Kp) of both FAs were measured using ADIFAB (44). In cholesterol-free bilayers, Kp for OA was 10-fold greater than for DHA, with no variation as a function of acyl chain length (see Table 4). The greater “potency” of DHA, relative to OA, therefore is not due to greater adsorption to the bilayer; the bilayer thickness-dependent effects of DHA on the lifetime of AgA− (15) channels are due to changes in hydrophobic mismatch, not to changes in adsorption. Cholesterol had no effect on Kp for DHA, which is consistent with the results of Anel et al. (8) on other FAs, including OA.
Table 4.
Partition coefficients of OA and DHA into different bilayers
Dimeric gA channels (D) form by transmembrane association of two nonconducting monomers (M):
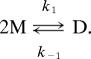 |
DHA increases both the channel appearance rate (f = k1·[M]2) and lifetime (τ = 1/k−1), shifting the distribution between M and D (45):
where KD is the dimerization constant, ΔGtotM→D is the total free energy of dimerization, R the gas constant and T temperature in kelvin. ΔGtotM→D can be expressed as the sum of two contributions: the intrinsic free energy change of dimerization (or protein “conformational change,” ΔGdefM→D) and the free energy change of the bilayer deformation (ΔΔGdefM→D). Because the effect of DHA increases whether the bilayer thickness is increased or the channel length is decreased, we conclude that the changes in [D] result from changes in ΔΔGdefM→D rather than from changes in ΔGprotM→D. It thus becomes possible to estimate the DHA-dependent changes in ΔΔGdefM→D from the changes in f and τ because [D] = KD·[M]2 = f·τ (Fig. 4).
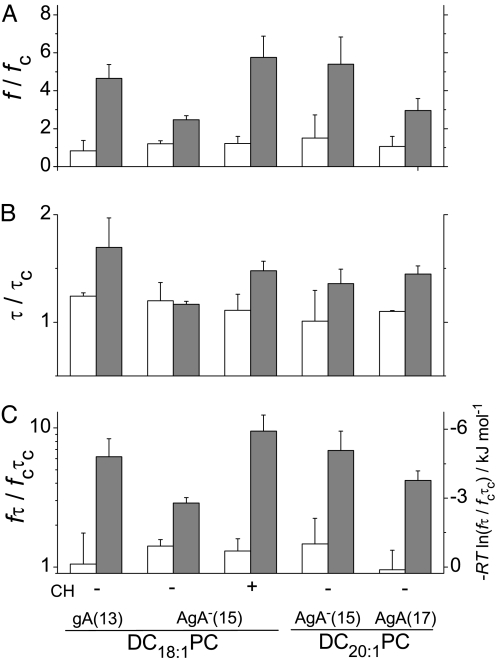
Fig. 4.
Changes in kinetics and energetics of channel formation. (A) The relative increase in channel appearance rate (f) induced by 3 μM OA (white) or DHA (gray) in DC18:1PC, DC18:1PC:Chol (1:1) and DC20:1PC bilayers. (B) Corresponding changes in gA channel lifetimes (τ). (C) Changes in dimerization constant (left axis) and free energy of dimerization (right axis) for gA channel formation induced by 3 μM OA or DHA. τc, lifetime in the absence of fatty acid; τ, lifetime in the presence of fatty acid; fc, channel appearance rate in the absence of fatty acid; f, appearance rate in the presence of fatty acid.
Fig. 4A shows changes in f induced by 3 μM DHA or OA for gA channels of varying lengths in bilayers of varying thicknesses (and in the absence and presence of cholesterol). (At 3 μM, bilayer breakage is relatively infrequent such that changes in channel appearance rate can be quantified in the same bilayer.) In parallel with the increase in f (Fig. 1), DHA (but not OA) also increases τ (Fig. 4B). Fig. 4C shows the relative increases in the time-averaged channel concentrations [D] (left ordinate), along with the corresponding changes in ΔΔGdefM→D (right ordinate). DHA increases KD by a factor of 3–8, depending on the channel-bilayer hydrophobic mismatch, corresponding to decreases in ΔΔGdefM→D between 3 and 5 kJ/mol. In the stiffer DC18:1PC:Chol (1:1) bilayers, ΔΔGdefM→D decreases by 6 kJ/mol, twice the change in DC18:1PC bilayers.
The changes in ΔΔGdefM→D could occur (cf. Eq. 1) due to changes in bilayer thickness (d0), bilayer elastic coefficients HB, HX, and HC, lipid intrinsic curvature (c0), or any combination thereof. We can exclude changes in d0 because the bilayer specific capacitance does not vary by the addition of DHA or OA (being 4.1 ± 0.3 nF/mm2 in the absence, and 4.3 ± 0.1 or 4.2 ± 0.2 nF/mm2 in the presence of 10 μM DHA or OA, respectively). The effect of DHA on τ increases with increasing hydrophobic mismatch; we conclude that DHA decreases the disjoining force the bilayer imposes on the channels (cf. Eq. 2) by reducing the magnitude of HB (increasing bilayer elasticity).
DHA alters not only gA channel appearance rates and lifetimes, it also increases i (Fig. 2). These current changes could arise because adsorption of DHA at the bilayer/solution interface imparts a negative surface charge to increase [Na+] at the channel entrance (and thus the single-channel current). DHA adsorption also may increase the “dielectric constant” due to the presence of the polyunsaturated acyl chains into the bilayer core, as the dielectric constant of hydrocarbons increase by ≈0.2 per double bond (46) (the single-channel current transition amplitudes are ≈10% higher in DC18:2PC as compared with DC18:1PC bilayers; ref. 47). Whatever the mechanism, the current changes provide insight into how DHA alters gA channel function.
Like other amphiphiles, DHA would be expected to alter global bilayer properties: to decrease the bilayer compression and bending moduli (30–32). In addition to these global effects, the gA-induced bilayer deformation may cause a local enrichment (or depletion) of DHA adjacent to the channel (ref. 48, cf. Fig. 5A), which would further reduce Fdis and ΔΔGdefM→D. We can distinguish between these possibilities, enrichment vs. depletion, using the single-channel current changes as a measure of the local DHA surface density and examine how DHA increases i and τ in bilayers of different thickness. [As noted previously, the gA channel structure (41) does not vary with bilayer thickness.]
Fig. 5.
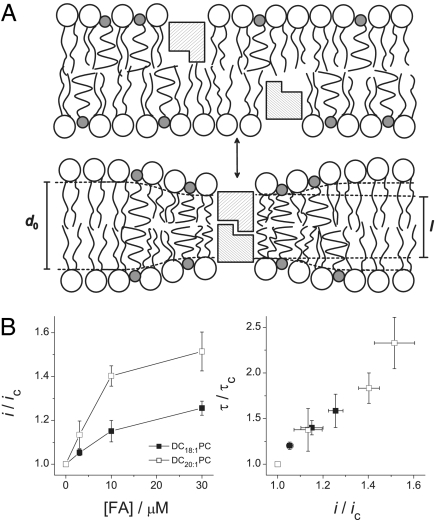
DHA alteration of bilayer mechanical properties. (A) Schematic model in which DHA is enriched around the channel in the area of bilayer deformation. l is the channel hydrophobic length and d0 the bilayer hydrophobic thickness. (B) Changes in AgA−(15) channel properties by DHA in DC18:1PC (filled squares) and DC20:1PC (open squares). (Left) Normalized single-channel current (i) changes. (Right) Changes in τ as a function of changes in i.
In the absence of DHA, changes in channel-bilayer hydrophobic mismatch do not affect the single-channel current transitions: in the case of AgA−(15) channels, 3.40 ± 0.04 pA in DC18:1PC vs. 3.36 ± 0.04 pA in DC20:1PC bilayers (mean ± SEM, n = 6). The DHA-induced current changes, however, are larger in DC20:1PC as compared with DC18:1PC bilayers (Fig. 5B), indicating that the local [DHA] in the vicinity of the channels is higher in the thicker DC20:1PC bilayers; the relative lifetime changes vary as a linear function of the relative current changes, indicating that the changes in channel function depend, at least in part, on changes in local bilayer properties. Similar results were obtained with gA(13).
That the effect of DHA on both i and τ is larger in the thicker DC20:1PC bilayers provides additional support for the conclusion that the effect of DHA is not due to direct binding to the channels; if that were the case, the current (and relative lifetime) changes should not depend on bilayer thickness. We conclude that the DHA-induced changes in the free energy of gA channel formation arise because DHA modifies lipid bilayer material properties, in particular bilayer elasticity, and that the effect is enhanced by the local accumulation of DHA in the vicinity of the channel.
Discussion
In this article, we extend the number of channels that are modulated by DHA and find, analogous to the situation for many other membrane proteins, that OA has little effect on gA channel function. The DHA-induced changes in gA channel function result from changes in bilayer elasticity, which stabilize the conducting dimer. Changes in material properties similarly may modulate the function of other membrane proteins; whether the modulation is activating or inhibiting depends on the ΔΔGdef0 for the particular conformational change. In the present context, we note that maneuvers that stabilize gA channels promote inactivation of voltage-dependent channels (35).
The DHA-induced changes in gA channel function occur at [FA] similar to those used in studies on integral membrane proteins (nominal concentrations of 1–10 μM, cf. the examples in Table 1). These concentrations are high compared with the unbound [FA] in plasma (49). The actual [FA] in the electrolyte solutions, however, are two orders of magnitude less [see supporting information (SI) Text]. At 3 μM DHA (or OA), for example, the FA mole fraction (mFA) in the bilayer is ≈0.01 in the unperturbed bilayer, although mDHA will be higher in the perturbed bilayer adjacent to the channel (cf. Fig. 5).
FAs promote the propensity to form inverse hexagonal phases (50) and cause negative-going changes in c0 (34). Changes in c0, per se, may cause changes in membrane protein function (34); ΔGdef0, however, varies as a function of not only c0 but also d0 − l and the elastic moduli, as expressed in the H coefficients (Eq. 1). If the changes in gA channel function predominantly were due to changes in c0, then molecules that promote positive-going changes in c0, such as Triton X-100 (35), should have effects opposite to those of DHA. That is not the case: both DHA and Triton X-100 (51) increase f and τ, and changes in c0 cannot be the dominant mechanism underlying the changes in channel function.
The potency of DHA as a modifier of gA channel function increases with increasing channel-bilayer hydrophobic mismatch, whether one alters channel length or bilayer thickness (Figs. 3–5). The DHA-induced changes in channel function therefore do not result from direct channel-DHA interactions. The reduction in bilayer stiffness, as reported by the increases in f and τ, arises because DHA decreases the bilayer elastic moduli. Together with results on voltage-dependent sodium channels (35, 52), the present results provide further support for the notion that changes in bilayer elasticity constitute a general mechanism for modulating membrane protein function. Moreover, both PUFAs and lysophospholipids, which promote positive-going changes in c0, are activators of the 2P domain potassium channels (53), providing additional support for the importance of changes in bilayer elasticity.
Finally, the results in Fig. 5 show that DHA is enriched in the vicinity of gA channels, whereas OA is not. Such local enrichment results because the free energy cost of the enrichment is balanced by a decreased bilayer deformation energy, and could occur because DHA with its six double bonds is more flexible than saturated or mono-unsaturated FAs (54, 55), which should enable DHA to pack more efficiently than OA into the perturbed bilayer region adjacent to a gA channel (membrane protein). This suggestion is consistent with the results of Feller et al. (56) who, based on molecular dynamics simulations on rhodopsin imbedded in 1-stearoyl-2-docosahexaenoyl-PC bilayers, found the docosahexaenoyl chain on average to be closer to rhodopsin than the stearoyl chain. Taken together, these findings highlight the difficulties associated with distinguishing “binding,” due to short-range protein–ligand interactions, from “enrichment” due longer-range interactions, such as protein-induced bilayer perturbations. Indeed, even though PUFAs modulate gA channel function by a bilayer-mediated mechanism, the modulation is “specific” in the sense that some closely related molecules, such as OA, are inert. This “specificity,” however, does not reflect chemically specific interactions, but rather the difference in physical properties of polyunsaturated vs. monounsaturated or saturated acyl chains. The distinctions among different FA effects therefore reflect the complex interplay between bilayer perturbations and local amphiphile accumulation in determining the energetic cost associated with protein conformational changes and the ensuing bilayer deformations.
Materials and Methods
Materials.
gA analogues of different length and helix sense (Tables 2–4) were synthesized and purified as described (57). Stock solutions (≈10−6 M) were stored in at −40°C and were diluted to 10−8 M to 10−9 M in absolute ethanol. The fatty acids (Table 4) were from Sigma (St. Louis, MO). For the electrophysiology experiments, stock solutions (0.5–5 mM plus an equimolar amount of NaOH) were made in absolute ethanol, stabilized with 25 μM butylated hydroxytoluene (BHT) and stored under argon. They were stable against oxidation for 2–3 weeks. For the determination of partition coefficients, stock solutions (0.5–5 mM) were made daily in 0.01 M NaOH without BHT and kept on ice. Acrylodated intestinal fatty acid binding protein (ADIFAB) was from Invitrogen (Carlsbad, CA) and was dissolved in 50 mM Tris, 1 mM EGTA, 0.05% azide, pH 8.0.
Table 2.
gA sequences
| Analogue | Sequence* | Hydrophobic Channel Length (nm)† |
|---|---|---|
| gA(13) | f-ALAVVVWLWLWLW-e | 1.9 |
| AgA(15) | f-AGALAVVVWLWLWLW-e | 2.2 |
| AgA(17) | f-AAAGALAVVVWLWLWLW-e | 2.5 |
*The underlined residues are D-amino acids; f = formyl; e = ethanolamine.
†The hydrophobic length of the 15-aa gA channel is from ref. 58; the lengths of the other channels are adjusted by 0.3 nm per L-D pair of residues.
Table 3.
Hydrophobic thickness of bilayers formed by different phospholipids
| Phospholipid | Hydrophobic thickness, nm* |
|---|---|
| Dioleoylphosphatidylcholine (DC18:1PC) | 4.8 |
| Dieicosenoylphosphatidylcholine (DC20:1PC) | 5.4 |
*Results from ref. 59.
Bilayer Formation and Single-Channel Measurements.
Single-channel measurements were done as described (57) using 20 mg/ml solutions of dioleoylphosphatidylcholine (DC18:1PC) or dieicosenoylphosphatidylcholine (DC20:1PC) (Avanti Polar Lipids, Alabaster, AL) in n-decane (99.9% pure; Chemsampco, Trenton, NJ) across a ≈1.6 mm diameter hole in a 0.1-mm-thick Teflon partition separating two compartments filled with 1 M NaCl, 10 mM Hepes, pH 7.0 (Sigma). The current signal was recorded with an AxoPatch 1C patch clamp (Axon Instruments, Foster City, CA), filtered at 2,000 Hz, digitized and digitally filtered to between 100 and 500 Hz. Single-channel current transitions were detected as described (60).
The gA analogues were added to both sides of the bilayer. We used pairs of analogues having opposite helical sense to prevent heterodimerization. Except where noted, the FAs were added to both sides of the bilayer; the solutions were stirred for five min after addition and equilibrated for 10 min. (The final [BHT] was <0.1 μM; 0.5 μM BHT has no effect on channel function, results not shown.)
The current transitions for each channel type appear as a single peak in current transition amplitude histograms constructed as in ref. 60. For each channel type, lifetime histograms then were constructed for channels with current transitions falling within the characteristic peak in the amplitude histogram (see Fig. 2). The average channel lifetimes, τ, were determined by fitting a single exponential distribution N(t) = N(0)·e−t/τ [where N(t) is the number of channels open longer than time t] to the lifetime distributions. The reported results are averages from at least three independent experiments, each with 300–1,000 channel events. Changes in channel appearance rate, f, were determined as the ratio of channel appearance rates from two 5- to 10-min recordings before and 10–20 min after OA or DHA addition (using only experiments where the bilayer did not break after FA addition).
The membrane capacitance was measured using a sawtooth potential waveform (61).
Fatty Acid Partition Into Lipid Vesicles.
Lipid bilayer/electrolyte FA partition coefficients (Kp) were determined in 1 M NaCl, 10 mM Hepes, pH 7.0 at 25 ± 1°C by ADIFAB (44) using a PerkinElmer (Salem, MA) 650–40 fluorescence spectrophotometer, exciting at 386 nm and measuring the fluorescence emission at 432 and 505 nm. After correcting for FA binding to the cuvette walls, the FA-ADIFAB dissociation constant (KD) was determined.
The lipid-FFA partition coefficients were calculated as
where [FA]t is total FA concentration, [FFA] is the free FA concentration, and Vm and Va are the volumes of the membrane and aqueous solutions (Vm/Va ≈ 10−3 per mM phospholipid; ref. 44). Lipid vesicles were prepared by miniextrusion (62) and diluted to ≈100 μM phospholipid and 0.2 μM ADIFAB was added; after each increase in [FA]t (3–30 μM), the system was equilibrated for 3–5 min and the fluorescence emission was measured. Each sample was assayed for the actual phospholipid concentration (63), and Vm/Va was adjusted accordingly. For each [FA], Kp was determined in triplicate. Kp did not vary as a function of [FA]; the reported Kp values are averages over [FA] = 3–30 μM.
To estimate the FA adsorption of to the bilayer chamber, DHA or OA (3–30 μM) were added to a lipid-free Teflon chamber, stirred for 5 min and 0.5 ml was removed for [FFA] determination as above. The reported results are averaged over all measurements.
Supplementary Material
Acknowledgments
We thank Md. Ashrafuzzaman, Helgi Ingolfsson, and Jens A. Lundbæk for helpful discussions. This work was supported by National Institutes of Health Grants GM021342 (to O.S.A.) and RR15569 (to R.E.K.) and the Jacques Cohenca Predoctoral Fellowship (to M.J.B.).
Abbreviations
- PUFA
polyunsaturated fatty acid
- OA
oleic acid
- gA
gramicidin
- DHA
docosahexaenoic acid.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0701015104/DC1.
FAs absorb to surfaces, and the nominal FA concentrations (based on the total amount added) are higher than the actual concentrations (see Discussion).
References
- 1.Bucher HC, Hengstler P, Schindler C, Meier G. Am J Med. 2002;112:298–304. doi: 10.1016/s0002-9343(01)01114-7. [DOI] [PubMed] [Google Scholar]
- 2.Kang JX. J Membr Biol. 2005;206:165–172. doi: 10.1007/s00232-005-0790-3. [DOI] [PubMed] [Google Scholar]
- 3.Hooper L, Thompson RL, Harrison RA, Summerbell CD, Ness AR, Moore HJ, Worthington HV, Durrington PN, Higgins JP, et al. BMJ. 2006;332:752–760. doi: 10.1136/bmj.38755.366331.2F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leaf A, Xiao YF. J Membr Biol. 2001;184:263–271. doi: 10.1007/s00232-001-0095-0. [DOI] [PubMed] [Google Scholar]
- 5.McLennan PL, Abeywardena MY, Charnock JS. Can J Physiol Pharmacol. 1985;63:1411–1417. doi: 10.1139/y85-232. [DOI] [PubMed] [Google Scholar]
- 6.Billman GE, Kang JX, Leaf A. Circulation. 1999;99:2452–2457. doi: 10.1161/01.cir.99.18.2452. [DOI] [PubMed] [Google Scholar]
- 7.Schrepf R, Limmert T, Claus Weber P, Theisen K, Sellmayer A. Lancet. 2004;363:1441–1442. doi: 10.1016/S0140-6736(04)16105-9. [DOI] [PubMed] [Google Scholar]
- 8.Anel A, Richieri GV, Kleinfeld AM. Biochemistry. 1993;32:530–536. doi: 10.1021/bi00053a018. [DOI] [PubMed] [Google Scholar]
- 9.Pound EM, Kang JX, Leaf A. J Lipid Res. 2001;42:346–351. [PubMed] [Google Scholar]
- 10.Xiao YF, Kang JX, Morgan JP, Leaf A. Proc Natl Acad Sci USA. 1995;92:11000–4. doi: 10.1073/pnas.92.24.11000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiao YF, Gomez AM, Morgan JP, Lederer WJ, Leaf A. Proc Natl Acad Sci USA. 1997;94:4182–4187. doi: 10.1073/pnas.94.8.4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Honore E, Barhanin J, Attali B, Lesage F, Lazdunski M. Proc Natl Acad Sci USA. 1994;91:1937–1941. doi: 10.1073/pnas.91.5.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guizy M, Arias C, David M, Gonzalez T, Valenzuela C. Am J Physiol. 2005;289:C1251–C1260. doi: 10.1152/ajpcell.00036.2005. [DOI] [PubMed] [Google Scholar]
- 14.Fink M, Lesage F, Duprat F, Heurteaux C, Reyes R, Fosset M, Lazdunski M. EMBO J. 1998;17:3297–3308. doi: 10.1093/emboj/17.12.3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matta JA, Miyares RL, Ahern GP. J Physiol. 2007;578:397–411. doi: 10.1113/jphysiol.2006.121988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bouzat CB, Barrantes FJ. Receptors Channels. 1993;1:251–258. [PubMed] [Google Scholar]
- 17.Nabekura J, Noguchi K, Witt MR, Nielsen M, Akaike N. J Biol Chem. 1998;273:11056–61. doi: 10.1074/jbc.273.18.11056. [DOI] [PubMed] [Google Scholar]
- 18.Wilding TJ, Chai YH, Huettner JE. J Physiol. 1998;513:331–339. doi: 10.1111/j.1469-7793.1998.331bb.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hii CS, Ferrante A, Schmidt S, Rathjen DA, Robinson BS, Poulos A, Murray AW. Carcinogenesis. 1995;16:1505–1511. doi: 10.1093/carcin/16.7.1505. [DOI] [PubMed] [Google Scholar]
- 20.Mayol V, Duran MJ, Gerbi A, Dignat-George F, Levy S, Sampol J, Maixent JM. Atherosclerosis. 1999;142:327–333. doi: 10.1016/s0021-9150(98)00253-6. [DOI] [PubMed] [Google Scholar]
- 21.Lee AG. Biochim Biophys Acta. 2003;1612:1–40. doi: 10.1016/s0005-2736(03)00056-7. [DOI] [PubMed] [Google Scholar]
- 22.Evans E, Needham D. J Phys Chem. 1987;91:4219–4228. [Google Scholar]
- 23.Owicki JC, Springgate MW, McConnell HM. Proc Natl Acad Sci USA. 1978;75:1616–1619. doi: 10.1073/pnas.75.4.1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mouritsen OG, Bloom M. Biophys J. 1984;46:141–153. doi: 10.1016/S0006-3495(84)84007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gruner SM. Proc Natl Acad Sci USA. 1985;82:3665–3669. doi: 10.1073/pnas.82.11.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang HW. Biophys J. 1986;50:1061–1070. doi: 10.1016/S0006-3495(86)83550-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sackmann E. In: Biological Membranes. Chapman D, editor. Vol 5. London: Academic; 1984. pp. 105–143. [Google Scholar]
- 28.Lundbæk JA, Birn P, Hansen AJ, Søgaard R, Nielsen C, Girshman J, Bruno MJ, Tape SE, Egebjerg J, et al. J Gen Physiol. 2004;123:599–621. doi: 10.1085/jgp.200308996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nielsen C, Andersen OS. Biophys J. 2000;79:2583–2604. doi: 10.1016/S0006-3495(00)76498-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Evans E, Rawicz W, Hofmann AF. In: Bile Acids in Gastroenterology Basic and Clinical Advances. Hofmann AF, Paumgartner G, Stiehl A, editors. Dordrecht, The Netherlands: Kluwer Academic; 1995. pp. 59–68. [Google Scholar]
- 31.Ly HV, Longo ML. Biophys J. 2004;87:1013–1033. doi: 10.1529/biophysj.103.034280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou Y, Raphael RM. Biophys J. 2005;89:1789–1801. doi: 10.1529/biophysj.104.054510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seddon JM. Biochim Biophys Acta. 1990;1031:1–69. doi: 10.1016/0304-4157(90)90002-t. [DOI] [PubMed] [Google Scholar]
- 34.Tate MW, Eikenberry EF, Turner DC, Shyamsunder E, Gruner SM. Chem Phys Lipids. 1991;57:147–164. doi: 10.1016/0009-3084(91)90073-k. [DOI] [PubMed] [Google Scholar]
- 35.Lundbæk JA, Birn P, Tape SE, Toombes GE, Sogaard R, Koeppe R. E., 2nd, Gruner SM, Hansen AJ, Andersen OS. Mol Pharmacol. 2005;68:680–689. doi: 10.1124/mol.105.013573. [DOI] [PubMed] [Google Scholar]
- 36.Xiao YF, Wright SN, Wang GK, Morgan JP, Leaf A. Proc Natl Acad Sci USA. 1998;95:2680–2685. doi: 10.1073/pnas.95.5.2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Connell AM, Koeppe RE, II, Andersen OS. Science. 1990;250:1256–1259. doi: 10.1126/science.1700867. [DOI] [PubMed] [Google Scholar]
- 38.Andersen OS, Nielsen C, Maer AM, Lundbæk JA, Goulian M, Koeppe RE., 2nd Methods Enzymol. 1999;294:208–224. doi: 10.1016/s0076-6879(99)94013-2. [DOI] [PubMed] [Google Scholar]
- 39.Kamp F, Zakim D, Zhang F, Noy N, Hamilton JA. Biochemistry. 1995;34:11928–37. doi: 10.1021/bi00037a034. [DOI] [PubMed] [Google Scholar]
- 40.Kleinfeld AM, Chu P, Romero C. Biochemistry. 1997;36:14146–58. doi: 10.1021/bi971440e. [DOI] [PubMed] [Google Scholar]
- 41.Wallace BA, Veatch WR, Blout ER. Biochemistry. 1981;20:5754–5760. doi: 10.1021/bi00523a018. [DOI] [PubMed] [Google Scholar]
- 42.Hwang TC, Koeppe RE, II, Andersen OS. Biochemistry. 2003;42:13646–58. doi: 10.1021/bi034887y. [DOI] [PubMed] [Google Scholar]
- 43.Needham D, Nunn RS. Biophys J. 1990;58:997–1009. doi: 10.1016/S0006-3495(90)82444-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Richieri GV, Ogata RT, Kleinfeld AM. Mol Cell Biochem. 1999;192:87–94. [PubMed] [Google Scholar]
- 45.Ashrafuzzaman MD, Lampson MA, Greathouse DV, Koeppe RE, II, Andersen OS. J Phys Condens Matter. 2006;18:S1235–S1255. [Google Scholar]
- 46.Lide DR, editor. Handbook of Chemistry and Physics. Vol 77. Boca Raton: CRC Press; 1996. [Google Scholar]
- 47.Girshman J, Greathouse DV, Koeppe RE, II, Andersen OS. Biophys J. 1997;73:1310–1319. doi: 10.1016/S0006-3495(97)78164-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Andersen OS, Sawyer DB, Koeppe RE., II . In: Biomembrane Structure and Function. Easwaran KRK, Gaber B, editors. Schenectady, NY: Adenine; 1992. pp. 227–244. [Google Scholar]
- 49.Richieri GV, Kleinfeld AM. J Lipid Res. 1995;36:229–240. [PubMed] [Google Scholar]
- 50.Epand RM, Epand RF, Ahmed N, Chen R. Chem Phys Lipids. 1991;57:75–80. doi: 10.1016/0009-3084(91)90051-c. [DOI] [PubMed] [Google Scholar]
- 51.Sawyer DB, Andersen OS. Biochim Biophys Acta. 1989;987:129–132. doi: 10.1016/0005-2736(89)90464-1. [DOI] [PubMed] [Google Scholar]
- 52.Leaf A, Xiao YF, Kang JX, Billman GE. Pharmacol Ther. 2003;98:355–377. doi: 10.1016/s0163-7258(03)00039-1. [DOI] [PubMed] [Google Scholar]
- 53.Patel AJ, Lazdunski M, Honore E. Curr Opin Cell Biol. 2001;13:422–428. doi: 10.1016/s0955-0674(00)00231-3. [DOI] [PubMed] [Google Scholar]
- 54.Rich MR. Biochim Biophys Acta. 1993;1178:87–96. doi: 10.1016/0167-4889(93)90113-4. [DOI] [PubMed] [Google Scholar]
- 55.Feller SE, Gawrisch K, MacKerell AD., Jr J Am Chem Soc. 2002;124:318–326. doi: 10.1021/ja0118340. [DOI] [PubMed] [Google Scholar]
- 56.Grossfield A, Feller SE, Pitman MC. Proc Natl Acad Sci USA. 2006;103:4888–4893. doi: 10.1073/pnas.0508352103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Greathouse DV, Koeppe RE, II, Providence LL, Shobana S, Andersen OS. Methods Enzymol. 1999;294:525–550. doi: 10.1016/s0076-6879(99)94031-4. [DOI] [PubMed] [Google Scholar]
- 58.Elliott JR, Needham D, Dilger JP, Haydon DA. Biochim Biophys Acta. 1983;735:95–103. doi: 10.1016/0005-2736(83)90264-x. [DOI] [PubMed] [Google Scholar]
- 59.Benz R, Fröhlich O, Läuger P, Montal M. Biochim Biophys Acta. 1975;394:323–334. doi: 10.1016/0005-2736(75)90287-4. [DOI] [PubMed] [Google Scholar]
- 60.Andersen OS. Biophys J. 1983;41:119–133. doi: 10.1016/S0006-3495(83)84414-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lundbæk JA, Birn P, Girshman J, Hansen AJ, Andersen OS. Biochemistry. 1996;35:3825–3830. doi: 10.1021/bi952250b. [DOI] [PubMed] [Google Scholar]
- 62.Hope MJ, Bally MB, Webb G, Cullis PR. Biochim Biophys Acta. 1985;812:55–65. doi: 10.1016/0005-2736(85)90521-8. [DOI] [PubMed] [Google Scholar]
- 63.Chen PS, Toribara TY, Warner H. Anal Chem. 1956;28:1756–1758. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.