Abstract
The T cell antigen receptor (TCR) mediates recognition of peptide antigens bound in the groove of major histocompatibility complex (MHC) molecules. This dual recognition is mediated by the complementarity-determining residue (CDR) loops of the α and β chains of a single TCR which contact exposed residues of the peptide antigen and amino acids along the MHC α helices. The recent description of T cells that recognize hydrophobic microbial lipid antigens has challenged immunologists to explain, in molecular terms, the nature of this interaction. Structural studies on the murine CD1d1 molecule revealed an electrostatically neutral putative antigen-binding groove beneath the CD1 α helices. Here, we demonstrate that α/β TCRs, when transferred into TCR-deficient recipient cells, confer specificity for both the foreign lipid antigen and CD1 isoform. Sequence analysis of a panel of CD1-restricted, lipid-specific TCRs reveals the incorporation of template-independent N nucleotides that encode diverse sequences and frequent charged basic residues at the V(D)J junctions. These sequences permit a model for recognition in which the TCR CDR3 loops containing charged residues project between the CD1 α helices, contacting the lipid antigen hydrophilic head moieties as well as adjacent CD1 residues in a manner that explains antigen specificity and CD1 restriction.
Keywords: CD1, antigen presentation, T cell receptor, Mycobacterium tuberculosis, mycolic acid
Acentral event in the recognition of microbial pathogens is the interaction of specific TCRs with their ligands (1–3). In most cases, these ligands are perceived to be complexes between MHC molecules and peptide antigens. Recently, we identified human α/β T cells that respond to lipid and glycolipid antigens from mycobacteria and that are restricted by CD1a, b, or c. These three CD1 isoforms are distantly related to MHC molecules and are expressed on professional APCs, including B cells, macrophages, and dendritic cells, as well as transiently on thymocytes during development (4–8). The expression of CD1a, b, and c on APCs and the bacterial origin of the lipid antigens they present suggest a role for lipid-specific, CD1-restricted T cells in host defense.
The appreciation that human T cells can detect lipid antigens and be restricted by molecules other than those encoded in the MHC raises fundamental questions about how nonpeptide antigens and CD1 molecules are recognized. The atomic structure of mouse CD1d1 resembles class I MHC molecules in overall topography, but in place of the peptide-binding groove of MHC, CD1d1 possesses an antigen-binding super domain composed of a hydrophobic and nonpolar cavity potentially capable of binding the acyl chains of lipid antigens (9). This finding suggests that CD1 molecules may have evolved to function as lipid-binding antigen-presenting molecules. In support of this hypothesis, recent data demonstrate a direct interaction between purified glycolipid antigens and CD1b (10) and indicate that glycosylphosphatidylinositol is a self lipid bound by murine CD1d1 (11).
The recognition of human CD1d and mouse CD1d1 has been linked to a population of T cells expressing an invariant, germline-encoded Vα24-Jα18 TCR α chain paired primarily with Vβ11 TCR β chains (12, 13). T cells bearing the invariant Vα24-Jα18 TCR α chain are stimulated by CD1d+ APCs, suggesting that the TCR on these cells may interact directly with CD1d, or with CD1d containing bound self-lipids. In contrast to CD1d-restricted T cells, which respond to APCs in the absence of a foreign antigen, the CD1a-, b-, and c-restricted T cells examined in this report only respond in the presence of foreign mycobacterial lipid antigens. Nothing is known about the TCRs that recognize CD1a-, b-, and c-restricted foreign lipid antigens. Thus far, it has not been possible to visualize the molecular basis for recognition of long chain mycolic acids and glycolipids like lipoarbinomannan. No data are available as to whether only invariant TCRs recognize these foreign lipid antigens and no model has been proposed for how the TCR might interact with an amphipathic lipid antigen–CD1 complex.
To address these questions, we have cloned the TCRs from a panel of CD1a-, b-, or c-restricted, lipid antigen– specific T cells. By gene transfer, we demonstrate that the TCR confers specificity for both foreign lipid antigen and CD1 isoform. Sequence analyses revealed notable diversity in TCR gene segment usage, indicating that invariant TCRs that recognize CD1d are not predictive of the TCRs against microbial lipid antigens. Importantly, frequent usage of basic amino acids in the complementarity-determining residue (CDR)31 regions suggests a model for the interaction of αβ TCR with lipid–CD1 complexes in which these TCR loops project directly between the CD1 α helices and mediate electrostatic interactions with the polar functions of amphipathic lipid and glycolipid antigens.
Materials and Methods
Cell Lines and mAbs.
The T cell lines used in this study, DN1, CD8-1, CD8-2, DN.POTT, and LDN5, have been described previously (4, 6, 14). T cells were cultured in 24-well Linbro® tissue culture plates (ICN Biomedicals, Inc.) in RPMI 1640 supplemented with 10% FCS, Hepes, l-glutamine, essential and nonessential amino acids, sodium pyruvate, beta-mercaptoethanol, and penicillin/streptomycin (complete medium) and containing 1.5 nM recombinant human IL-2 (rhIL-2) (Ajinomoto Co.). T cell lines were restimulated every 2 wk with CD1+ monocytes and a chloroform/methanol extract of Mycobacterium tuberculosis H37Ra (prepared from a 200 mg dry bacteria/ml suspension; Difco Laboratories) at a 1:5,000 dilution. CD1+ monocytes were prepared as described previously (15). In brief, adherent cells from random donor Leukopacks were cultured for 3 d in complete medium containing 300 U/ml GM-CSF (Immunex Corp.) and 200 U/ml IL-4 (gift of Schering Corp.). J.RT3-T3.5 cells, derivatives of Jurkat cells that have defective endogenous TCR β chain expression, were obtained from the American Type Culture Collection and were cultured in complete medium supplemented with G418 (1 mg/ml; GIBCO-BRL) and Hygromycin B (0.5 mg/ml; GIBCO-BRL) for selection of transfectants. IL-2–dependent HT-2 cells were obtained from the American Type Culture Collection and were cultured in complete medium supplemented with 2 nM rhIL-2. The following purified mAbs were used: P3 (nonbinding control, mouse IgG1) (16), 10H3.9.3 (anti-CD1a, mouse IgG2a) (17), BCD1b3.1 (anti-CD1b, mouse IgG1) (18), F10/21A3.1 (anti-CD1c, mouse IgG1; Porcelli, S.A., unpublished results), and SPVT3b (anti-CD3ε, mouse IgG2a) (19).
Preparation of Lipid Antigens.
The lipids recognized by the CD8-1 and CD8-2 T cell lines were partially purified as follows. M. tuberculosis H37Ra (Difco Laboratories) was sonicated in PBS and extracted with chloroform/methanol in a 2:1 ratio. The organic phase was recovered, dried by rotary evaporation, and resuspended in chloroform. The organic extract was then fractionated using silica gel chromatography. The organic extract equivalent of 10 mg dry bacteria was loaded in chloroform onto a 1 g silica gel (Selecto Scientific) column. The column was then eluted with a step gradient of methanol in chloroform from 0 to 100% methanol in 10% increments. The lipid(s) recognized by CD8-1 were found to fractionate predominantly in the 60:40 chloroform/methanol fraction and the lipid(s) recognized by CD8-2 were found predominantly in the 90:10 chloroform/ methanol fraction (hereafter referred to as silica fraction 60:40 and silica fraction 90:10, respectively; reference 19a).
To isolate mycolic acids from M. tuberculosis H37Ra, dry bacteria were suspended in chloroform/methanol in a 2:1 ratio to extract the majority of the lipids from the cell wall. The delipidated cell walls were pelleted and dried. The pellet was saponified with a methanol/potassium hydroxide solution to release mycolic acids, which were then extracted with hexane. Mycolic acids were then precipitated with ether/ethanol and dissolved in chloroform. The presence of alpha, methoxy, and keto mycolic acids was confirmed by thin-layer chromatography (data not shown).
T Cell Cloning.
T cell clones were isolated from cell lines by limiting dilution. T cells were plated at 0.3, 1, or 5 cells/well in 96-well round-bottomed microtiter plates. Normal human PBMC (105/well) and a 1:1 mixture of two EBV-transformed human B cell lines (5 × 104/well) were irradiated (5,000 rad) and added as feeder cells. PHA-P (Difco Laboratories) was added to a final concentration of 1:4,000 in a total volume of 150 μl/well in complete medium containing 1.5 nM rhIL-2. Plates were fed after 7 d of culture at 37°C and then every 3–4 d thereafter. Clones were expanded by restimulation with PHA and feeder cells every 2–3 wk.
T Cell Proliferation Assays.
T cells (5–50 × 103/well) were cultured in triplicate in 96-well flat-bottomed microtiter plates with or without irradiated (5,000 rad) CD1+ monocytes (5 × 104/well) in the presence or absence of antigen. To assess CD1 restriction, purified mAbs specific for CD1a, b, or c were added at a final concentration of 20 μg/ml. Cells were cultured for 3 d at 37°C, pulsed with [3H]thymidine (1 μCi/well, 6.7 Ci/mmol, New England Nuclear) and incubated for 5–6 h. The plates were harvested on a Tomtec 96-well plate harvester (Wallac, Inc.) and thymidine incorporation was measured with a Betaplate liquid scintillation counter (Wallac Inc.).
Inverse PCR.
Inverse PCR was used to determine the TCR α and β gene usage as described previously (20, 21). In brief, total RNA was isolated from 2 × 105–2 × 106 T cells mixed with 106 EBV-transformed B cell line DG-EBV as carrier using guanidinium thiocyanate (22). Oligo(dT)-primed double-stranded cDNA was synthesized from total RNA, and incubated with T4 DNA polymerase to form blunt-ended cDNA. The blunt-ended cDNA was then circularized with T4 DNA ligase and used as a template for PCR using a pair of Cα- or Cβ-specific primers oriented in opposite directions. The primers were Cα-forward: 5′-gggtcgacgacctcatgtctagcacagt-3′; Cα-inverse: 5′-gcatgcggccgccctgctatgctgtgtgtct-3′; Cβ-forward: 5′-gggtcgacacacagcgacctcgggtggg-3′; Cβ-inverse: 5′-gcatgcggccgccatggtcaagagaaagga-3′. The amplified TCR genes (700–800 bp in length) that include V(D)J sequences were cloned into pBluescript II (Stratagene) and sequenced using the Sequenase v. 2.0 sequencing kit (United States Biochemical) according to the manufacturer's instructions.
TCR Cloning.
To isolate full-length cDNAs encoding the TCRs, we designed Vα- and Vβ-specific primers for the appropriate V genes identified by sequence analysis of inverse PCR products. The following primers were used: BV5S1: 5′-gggctcgagccctgagcacagacacagtg-3′; BV2S1: 5′-gggctcgagaaggtggtgtgaggccat-3′; BV9S1: 5′-gggctcgagctgcagaccagaatcctgccc-3′; CBETA2: 5′-gggggatcctgggtgaggatgaagaatg-3′; AV8S2: 5′-gggctcgagtatgactgatcctatttggg-3′; AV1S3: 5′-ggggtaccagctcaaggtcctgca-3′; AV16S1: 5′-gggctcgagcttagctggagccatggc-3′; CALPHA: 5′-gggggatccaggctgtcttacaatcttgca-3′. RNA isolated from T cell clones or lines was used to synthesize single-stranded cDNA with Superscript II reverse transcriptase (GIBCO-BRL) and an oligo(dT) primer. This cDNA was used as a template for PCR with the appropriate primers using the following reaction conditions: 95°C, 30 s; 52°C, 1 min; 72°C, 1 min; 30 cycles. PCR products were digested with appropriate restriction endonucleases, gel purified, and ligated into pREP7 (TCR α chains) or pREP9 (TCR β chains) (Invitrogen).
TCR Transfection.
α/β TCR transfectants of J.RT3-T3.5 cells were produced as described (23). J.RT3-T3.5 cells were pelleted and resuspended at 4 × 107/ml in complete medium, and 0.3 ml of cells was aliquoted to each electroporation cuvette. 20 μg of each plasmid (pREP7-TCR-α and REP9-TCR-β) was added to the cells and incubated at room temperature for 10 min. The cells were electroporated (960 μF, 250 V) with a Gene Pulser (Bio-Rad Laboratories, Inc.) and incubated at room temperature for an additional 10 min. The transfectants were cultured for 48 h at 37°C in complete medium and then transferred to medium containing G418 (1 mg/ml) and Hygromycin B (0.5 mg/ml). After 2–4 wk of culture, transfectants were analyzed by flow cytometry and used in T cell transfectant stimulation assays.
Flow Cytometry.
Transfectants were analyzed for cell surface expression of TCR–CD3 complexes as follows. Cells were incubated for 45 min on ice in the presence of 20 μg/ml SPVT3b (anti-CD3ε) or control antibodies in 50 μl PBS/5% FCS at 106 cells/ml. The cells were washed with PBS/FCS and resuspended in 20 μg/ml FITC-goat F(ab′)2 anti–mouse IgG/IgM (Biosource International) in 50 μl PBS/FCS at 106 cells/ml and incubated for 45 min on ice. Cells were washed, resuspended in PBS/FCS and analyzed on a FACSORT® flow cytometer (Becton Dickinson and Co.). Dead cells were excluded based on forward and side scatter.
T Cell Transfectant Stimulation Assay.
J.RT3-T3.5/TCR-α /β transfectants (105/well) were cultured in 96-well flat-bottomed microtiter plates either in the presence of 10 ng/ml PMA alone, or in the presence of PMA plus CD1+ APCs with or without different lipid antigens (mycolic acids isolated from M. tuberculosis H37Ra or silica column fractions of an organic extract of M. tuberculosis H37Ra). Human PB CD1+ monocytes (5 × 104/well) were used as APCs with or without the following mAbs (at a concentration of 20 μg/ml): P3 (isotype control), 10H3.9.3 (anti-CD1a), BCD1b3.1 (anti-CD1b), or F10/21A3.1 (anti-CD1c). Cultures were set up in a total volume of 200 μl/well in triplicate. 25-μl aliquots of the culture supernatants were collected after 20–24 h and diluted 1:4 with culture medium. To measure the amount of IL-2 released into the supernatants, HT-2 cells (5,000/well) were added and cultured for 25–30 h. [3H]Thymidine (1 μCi/well, 6.7 Ci/mmol) was added during the final 5–6 h of culture. [3H]Thymidine incorporation was measured with a liquid scintillation counter.
TCR–CD1b Modeling.
Alignment of amino acid sequences was performed using the programs PILEUP and BESTFIT, part of the Wisconsin Package Version 9.1 (Genetics Computer Group). The primary structure of human CD1b was aligned with the murine CD1d1 molecule (46.9% identity). Residues differing between proteins were mutated using the program O (24), and the conformation for different side chains was assigned based on the rotamer library and the main chain conformation (25). The model was subjected to 250 steps of energy minimization as implemented in the program X-PLOR (26). Three cycles carried out after assignment of different initial velocities resulted in virtually identical models. Bad contacts between side chains were checked manually.
The sequences of the DN1 α and β chains were individually aligned with the corresponding chains of TCRs whose structures were available in the Protein Data Bank (1TCR [27], 1A07 [28], 1KB5 [29], 1NFD [30], 2CKB [31]). The variable α and β domains of DN1 were constructed based on the corresponding chains of the 2C TCR in complex with the MHC molecule H-2Kb (31). The pairing between the α and β chains was modeled as in 2C because of the high sequence homology (43.7% identity for the α chain, 56.8% for the β chain). The CDR3 regions were modeled based on the loop library implemented in O. Side chains were mutated and the model was minimized as described above. Additionally, the TCR framework residues were restrained using an harmonic strength constant of 5 kcal/molÅ2, whereas the CDR loops were allowed to move freely to explore more conformational space.
The CD1–TCR model was constructed by superimposing the models of CD1b and DN1 variable domains on the 2C/H-2Kb-dEV8 complex structure (31), using the lsq_explicit option in O. The model was manually adjusted to remove unreasonable contacts, and subjected to a final minimization as described for the DN1 TCR. The quality of the model was assessed using PROCHECK (32). The figures were generated with MIDAS (33).
Results
Isolation of T Cell Clones and Inverse PCR Analysis.
Five lipid-specific human T cell lines have been characterized as restricted by CD1a (CD8-2), CD1b (DN1, DN.POTT, LDN5), and CD1c (CD8-1) (Table I) (4, 6, 14). To study the nature of the TCR in recognition of CD1-presented lipid antigens, several of these T cell lines were cloned by limiting dilution, and the specificities of the T cell clones were confirmed in proliferation assays using purified lipid/ glycolipid antigens and CD1+ monocyte APCs. The CD1 restriction of these clones was confirmed by blocking proliferation with mAbs specific for CD1a, b, or c. A clone isolated from the CD8-2 T cell line (designated CD8-2.1) proliferated when cultured with CD1+ monocytes and the lipid fraction of M. tuberculosis strain H37Ra designated silica fraction 90:10 (see Materials and Methods). Proliferation was blocked by 96% by the inclusion of the anti-CD1a mAb 10H3.9, but not by control mAb P3, anti-CD1b mAb BCD1b3.1, or anti-CD1c mAb F10/21A3.8 (Fig. 1 A). Similarly, clones isolated by limiting dilution from the DN1 (designated DN1.F9, Fig. 1 B) and CD8-1 (designated CD8-1.A2.3, Fig. 1 C) T cell lines proliferated in the presence of M. tuberculosis strain H37Ra mycolic acids or H37Ra-derived silica fraction 60:40, respectively. Proliferation of DN1.F9 cells was blocked by 57% with anti-CD1b, but only by 1–24% by control or anti-CD1a or c mAbs, whereas proliferation of CD8-1.A2.3 cells was blocked by 88% with anti-CD1c mAb, but not by control or anti-CD1a or b mAbs. Thus, the specificity of these T cell clones matches that of the published parent T cell lines shown in Table I, and, consequently, they were used as sources of RNA for TCR molecular cloning. Repeated T cell cloning attempts with DN.POTT and LDN5 cell lines failed to yield clones that expanded adequately for complete analysis. Consequently, RNA was extracted from these two long-term homogeneous, but uncloned, T cell lines for TCR analysis. The specificity of the transfectants derived was assessed to assure that they displayed the specificity of the parental cell lines, indicating that the correct antigen-specific TCR α and β chain pairs had been isolated.
Table I.
Specificity of T Cell Lines Analyzed
| T cell line | Restriction element | Antigen specificity | ||
|---|---|---|---|---|
| CD8-1 | CD1c | Phospholipid | ||
| CD8-2 | CD1a | Lipid | ||
| DN1 | CD1b | Mycolic acid | ||
| DN.POTT | CD1b | Mycolic acid | ||
| LDN5 | CD1b | Glucose monomycolate |
CD8-1 and CD8-2 have specificity for two different lipid antigens present in M. tuberculosis H37Ra extracts. Both antigens are extracted from M. tuberculosis in chloroform/methanol (2:1), and can be resolved by step gradient silica chromatography into fractions eluting in 60:40 (chloroform/methanol) (CD8-1) and 90:10 (chloroform/methanol) (CD8-2). Preliminary characterization of the CD8-1 antigen shows that it is likely to be a phospholipid (Moody, D.B., W. Muchlecker, and S.A. Porcelli, unpublished observations).
Figure 1.
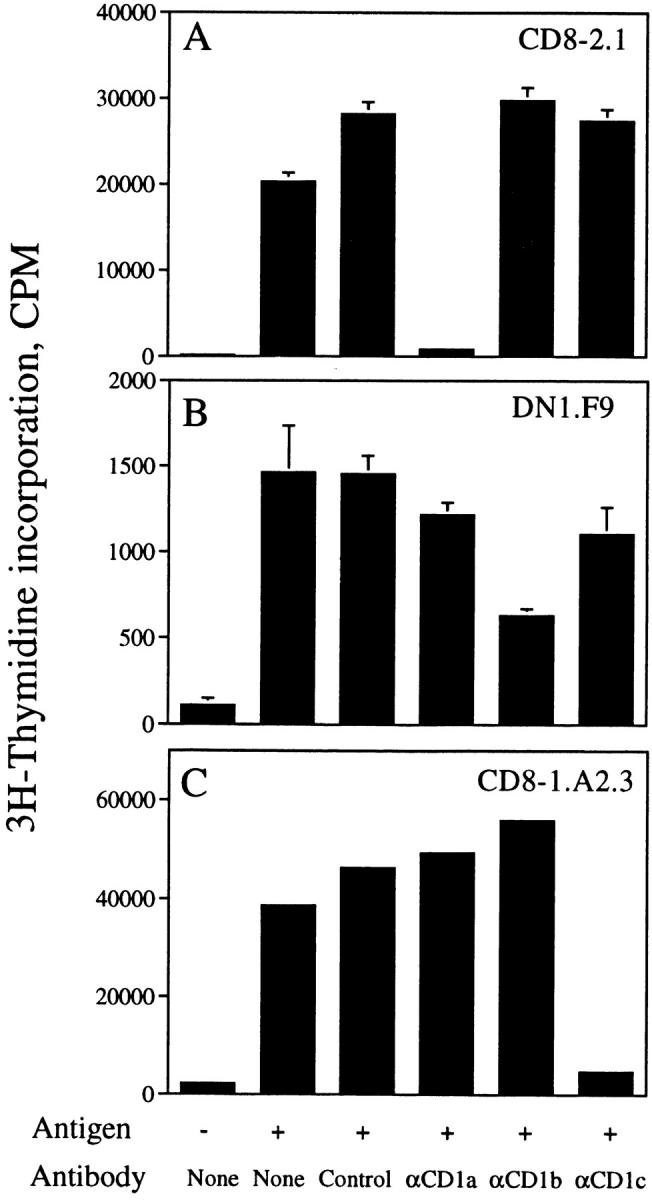
Specificity of T cell clones isolated from the CD8-2, DN1, and CD8-1 lines. CD8-2.1 (A), DN1.F9 (B), or CD8-1.A2.3 (C) T cells (5 × 104/well) were cultured with CD1+ monocytes (5 × 104/well) with or without antigen (silica fraction 90:10 at 2 μg/ml [A], mycolic acid at 40 μg/ml [B], or silica fraction 60:40 at 5 μg/ml [C]) for 72 h. The following mAbs were added at a final concentration of 20 μg/ml: P3 (IgG control), 10H3.9.3 (αCD1a), BCD1b3.1 (αCD1b), or F10/21A3.8 (αCD1c). [3H]Thymidine was added during the final 6 h of culture, after which the plates were harvested and [3H]thymidine incorporation measured in a liquid scintillation counter. Results are expressed as mean cpm ± SD of triplicate cultures (A and B). In C, results are from an experiment in which short-term clones were screened in singlicate. The pattern of antigen/CD1 recognition observed for these clones matches numerous experiments performed with the T cell lines.
To identify the TCR gene usage of these T cell lines and clones, we used inverse PCR analysis, which allows cloning of any TCR without prior knowledge of V or J gene segment usage. Double-stranded cDNA was synthesized from total cellular RNA and circularized by blunt-ended ligation. The circular cDNAs were then used as templates in PCR reactions with pairs of Cα- or Cβ-specific primers oriented in opposite directions. The resulting amplicons thus included the TCR V, N/D/N, and J regions of the TCR genes, which allowed the identification of TCR gene segment usage. The inverse PCR products obtained from the CD1-restricted T cell cDNAs were of the expected size (∼700 bp), and these products were cloned into pBluescript II and sequenced.
Reconstitution of TCR Expression.
Although α/β TCRs clearly interact with peptide–MHC complexes, it is not known what role the TCR plays in the recognition of lipid antigens and CD1 molecules. To test whether α/β TCRs mediate lipid antigen–specific, CD1-restricted recognition, we cloned full-length cDNAs encoding the TCR α and β chains from the DN1.F9, CD8-1.A2.3, and CD8-2.1 T cell clones. The TCR gene segment usage identified by inverse PCR analysis allowed design of V gene–specific 5′ untranslated region (UTR) primers for the appropriate Vα and Vβ genes, identified for each clone, together with Cα or Cβ 3′ UTR primers. The TCR-α and TCR-β cDNA PCR products were cloned into the pREP7 (α chains) and pREP9 (β chains) episomal mammalian expression vectors. TCR-β–deficient Jurkat T cells (J.RT3-T3.5) were cotransfected with the TCR-α and TCR-β cDNAs from DN1, CD8-1, or CD8-2, and selected for 3–4 wk in medium supplemented with G418 and Hygromycin B, yielding transfectant cell lines designated DN1/J.RT3, CD8-1/J.RT3, and CD8-2/J.RT3, respectively. Flow cytometric analysis of the resulting T cell transfectant lines showed that DN1/J.RT3 was 48% CD3+ (with an MFI of 180), CD8-1/J.RT3 was 80% CD3+ (with an MFI of 264), and CD8-2/J.RT3 was 60% CD3+ (with an MFI of 219), whereas mock-transfected J.RT3-T3.5 cells were essentially CD3− (6% positive with an MFI of 60) (Fig. 2, A–D), indicating that transfected TCR genes were successfully expressed in association with endogenous CD3.
Figure 2.
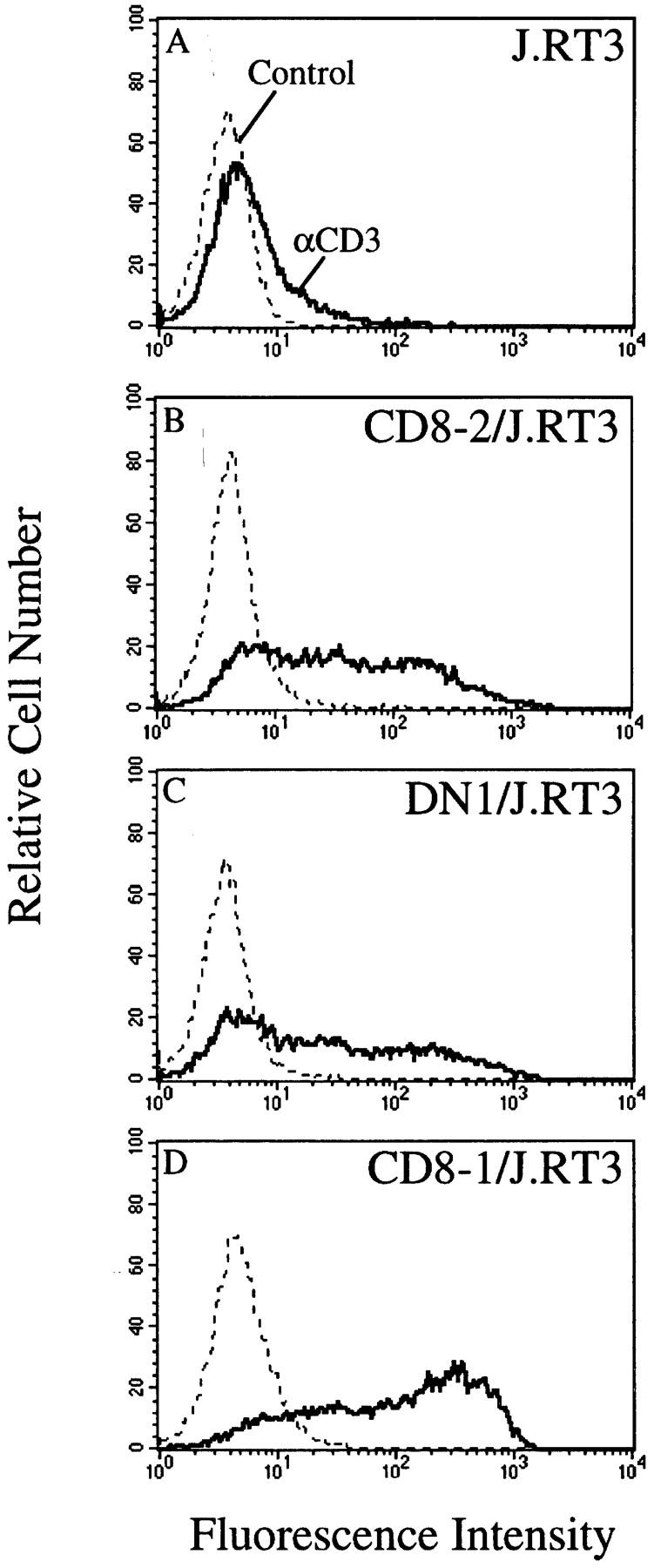
Expression of TCR– CD3 complex by J.RT3-T3.5 transfectants. J.RT3-T3.5 cells were transfected with CD8-2 (B), DN1 (C), or CD8-1 (D) α/β TCR cDNAs in pREP7 (α chains) and pREP9 (β chains) expression vectors or with vectors alone (A) and selected in medium containing G418 (1 mg/ml) and Hygromycin B (0.5 mg/ml) for 3–4 wk. Transfectant lines were labeled with the anti-CD3 mAb SPVT3b (solid line) or with control mouse IgG (broken line) followed by FITC goat anti–mouse Ig (Fab′)2 and analyzed on a flow cytometer.
TCR Transfectants Display Lipid Antigen Specificity.
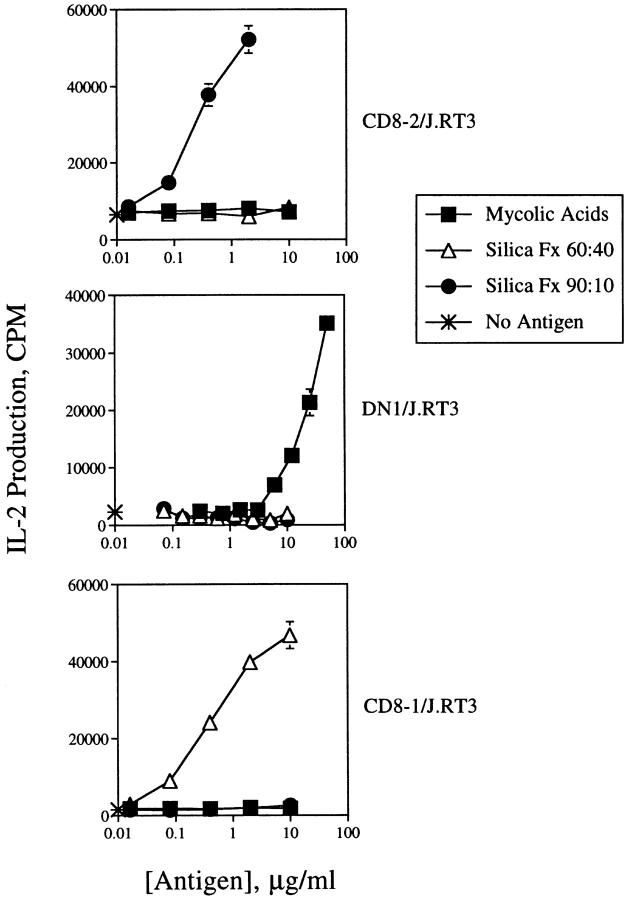
To test the TCR transfectants for recognition of lipid/glycolipid antigens and CD1, DN1/J.RT3, CD8-1/J.RT3, and CD8-2/J.RT3, cells were cultured with CD1+ monocytes and lipid antigens and IL-2 production was measured as an index of antigen-specific activation. CD8-2/J.RT3 cells produced IL-2 in a dose-dependent manner when cultured with the appropriate lipid fraction of M. tuberculosis H37Ra (silica fraction 90:10; see Materials and Methods), but not with other lipid preparations that did not contain the specific antigen recognized by CD8-2 (silica fraction 60:40 or purified mycolic acids) (Fig. 3 top). In contrast, DN1/J.RT3 and CD8-1/J.RT3 cells were specifically activated to secrete IL-2 only in the presence of purified M. tuberculosis mycolic acids or silica fraction 60:40, respectively (Fig. 3, middle and bottom). Thus, the lipid antigen specificity of these TCR transfectants matched precisely the antigen specificities of the original T cell lines. The sensitivity of the DN1/J.RT3 cells for antigen was ∼50-fold lower than the sensitivities of CD8-1/J.RT3 and CD8-2/J.RT3 cells for their respective antigens. It is not clear if this difference reflects variability in TCR expression levels, antigen uptake or loading efficiency, affinity of the TCR for different ligands, or some other parameter.
Figure 3.
Lipid antigen specificity of J.RT3/α/β TCR transfectants. CD8-2/J.RT3 (top), DN1/J.RT3 (middle), or CD8-1/J.RT3 (bottom) transfectants (105/well) were cultured in triplicate for 24 h in the presence of PMA (10 ng/ml) and CD1+ monocytes (5 × 104/well) with no antigen (asterisk), or with a titration of M. tuberculosis H37Ra mycolic acids (▪), silica fraction 60:40 (▵), or silica fraction 90:10 (•). To measure IL-2 production by the transfectants, an aliquot of the supernatants was removed and diluted 1:4 with medium. HT-2 cells (5 × 103/well) were added to the supernatants and cultured for 16–24 h. [3H]Thymidine was added during the final 5–6 h culture, after which the plates were harvested and [3H]thymidine incorporation measured with a liquid scintillation counter. Results are expressed as the mean cpm ± SD of triplicate cultures.
Similarly, full-length TCR-α and TCR-β cDNAs were cloned from the LDN5 cell line using the sequence information obtained by inverse PCR analysis. Transfection of the LDN5 αβ TCR into J.RT3-T3.5 cells conferred the ability to respond to purified Mycobacterium phlei glucose monomycolate but not to other lipid or glycolipid antigens (data not shown). Although the TCR from DN.POTT was not reconstituted in J.RT3-T3.5 cells, 6/6 inverse PCR products representing in-frame TCR-α and 6/6 PCR products representing in-frame TCR-β transcripts were identical in sequence to those shown in Table II, suggesting that the DN.POTT T cell line is essentially clonal and providing primary sequence data on its TCR (data not shown).
Table II.
TCR Gene Usage and Predicted CDR3 Region Protein Sequences
| TCR-α T cell line | Gene usage | Vα | CDR3 | Jα | ||||
|---|---|---|---|---|---|---|---|---|
| CD8-1 | AV1S3, AJ17 | ...CAV | RRAAGNKLTF | GGG... | ||||
| CD8-2 | AV16S1, AJ34 | ...CAV | RHNTDKLIF | GTG... | ||||
| DN1 | AV8S2, AJ57 | ...CAE | PLSLPGGSEKLVF | GKG... | ||||
| DN.POTT | AV16S1, AJ31 | ...CAV | RDEGWARLMF | GDG... | ||||
| LDN5 | AV3S1, AJ9 | ...CAS | MYTGGFKTIF | GAG... | ||||
| TCR-β T cell line | Gene usage | Vβ | CDR3 | Jβ | ||||
| CD8-1 | BV2S1, BJ2S7 | ...CSA | RTYPGTGFYEQYF | GPG... | ||||
| CD8-2 | BV9S1, BJ2S1 | ...CAS | SSMYNEQFF | GPG... | ||||
| DN1 | BV5S1, BJ2S7 | ...CAS | SLVRRYEQYF | GPG... | ||||
| DN.POTT | BV6S1, BJ2S1 | ...CAS | SLTRSSHNEQFF | GPG... | ||||
| LDN5 | BV7S1, BJ2S1 | ...CAS | SQPIGGGEQFF | GPG... |
The TCR V and J gene usage for each T cell line is indicated. The predicted amino acid sequences from the CDR3 junctional regions of each TCR α and TCR β chain are shown with basic amino acid residues encoded by N nucleotides underscored. These sequence data are available from EMBL/GenBank/DDBJ under accession numbers AF107998–AF108007.
Since J.RT3-T3.5 cells express an endogenous TCR α chain with which the transfected TCR β chains could potentially form a second functional heterodimer, we made control J.RT3 cells transfected with only the TCR β chains of each of the TCRs studied. Although TCR β transfectants typically expressed levels of TCR at the cell surface similar to TCR-α/β transfectants and could be stimulated with anti-CD3, no stimulation of IL-2 production in response to CD1+ APCs with or without lipid antigens was observed (data not shown). Thus, both TCR α and β chains from the lipid/CD1-specific T cell lines were required to reconstitute a TCR capable of lipid-specific recognition.
TCR Transfectants Display Specific CD1 Isoform Restriction.
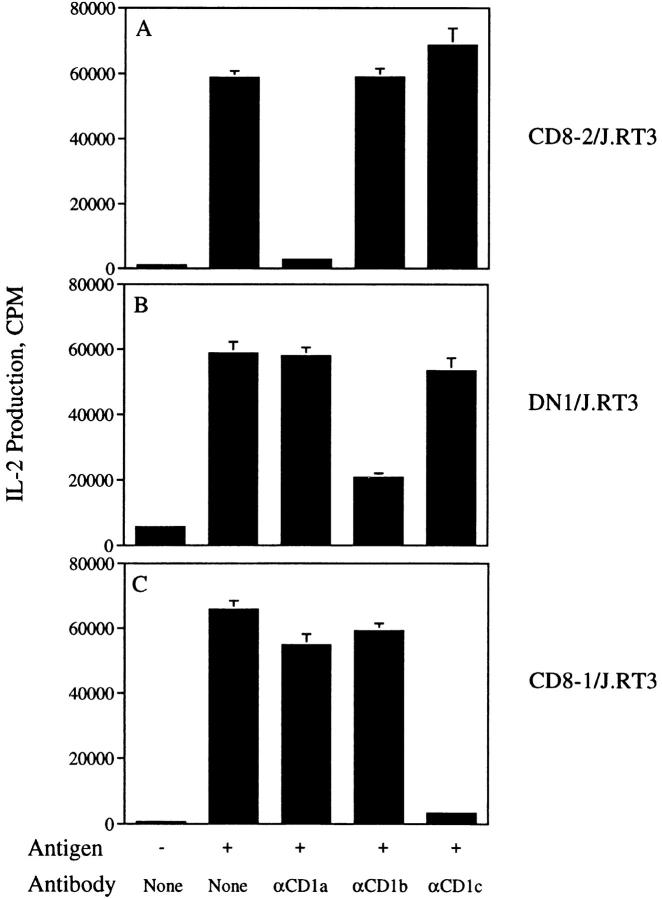
The CD1 dependence of the activation of the transfectants was determined using blocking mAbs specific for CD1a, b, or c. IL-2 production by CD8-2/J.RT3 transfectant cells was blocked by 95% with mAb 10H3.9.3 (anti-CD1a), but was slightly increased by mAbs BCD1b3.1 (anti-CD1b) and F10/ 21A3.1 (anti-CD1c). Similarly, the activation of DN1/J.RT3 and CD8-1/J.RT3 transfectants was specifically blocked by anti-CD1b and anti-CD1c, respectively, but not by mAbs to the other two CD1 isoforms tested (Fig. 4, A–C). Therefore, reconstitution of the TCRs from three different CD1- restricted T cells in TCR-deficient J.RT3-T3.5 cells confers recognition of both the specific antigens and the particular CD1 isoforms the original T cells were defined to recognize. These results support a model in which the TCR interacts with both lipid/glycolipid antigen and CD1 analogous to conventional TCR–MHC–peptide interactions.
Figure 4.
CD1 specificity of J.RT3/α/β TCR transfectants. CD8-2/ J.RT3 (A), DN1/J.RT3 (B), or CD8-1/J.RT3 (C) transfectants (105/ well) were cultured in triplicate for 24 h in the presence of PMA (10 ng/ml) and CD1+ monocytes (5 × 104/well) with no antigen, or with 50 μg/ml M. tuberculosis H37Ra mycolic acids (DN1/J.RT3), 1 μg/ml silica fraction 60:40 (CD8-1/J.RT3), or 1 μg/ml silica fraction 90:10 (CD8-2/ J.RT3). Cultures with antigen were set up with or without the following mAbs at a final concentration of 20 μg/ml: 10H3.9.3 (anti-CD1a), BCD1b3.1 (anti-CD1b), or F10/21A3.8 (anti-CD1c). IL-2 production was measured as described for Fig. 3. Results are expressed as the mean cpm ± SD of triplicate cultures.
Analysis of TCR Sequences.
Having confirmed that α/β TCRs mediate lipid-specific, CD1-restricted recognition, and having defined that the specific TCR α/β pairs cloned from a panel of T cell lines were the correct ones for recognition, we next analyzed the TCR sequences to gain insight into the nature of TCR diversity for lipid-specific and CD1-restricted T cells. The DNA sequences were aligned with known TCR V and J segments using BLAST homology searches (34). Two of the TCRs, those from CD8-2 and DN.POTT, expressed the AV16S1 gene, and the other TCRs contained three different Vα genes, AV1S3, AV8S2, and AV3S1 (Table II). Five different Jα segments were detected in the TCRs from these cells (AJ17, AJ34, AJ31, AJ57, and AJ9). The TCR Vβ genes were similarly diverse, with BV2S1, BV9S1, BV6S1, BV5S1, and BV7S1 each represented once (Table II). However, significantly less diversity was observed in Jβ usage. Of the 13 functional human Jβ gene segments, only Jβ segments BJ2S1 or BJ2S7 were utilized by the 5 T cell lines. The deduced amino acid sequences of the TCR V-N/D/N-J junctions displayed in Table II indicate that the CDR3 regions of these TCRs have typical lengths (9–13 residues in CDR3α and 8–13 residues in CDR3β) and heterogeneous peptide sequences. Together, the combinatorial diversity of TCR Vα, Jα, and Vβ segment usage and the junctional diversity of CDR3-encoded residues make it clear that, unlike the invariant TCR-α genes of NKT cell recognition of CD1d, those of foreign lipid–specific, CD1a-, b-, or c-restricted TCR are diverse receptors similar to those of peptide-specific, MHC-restricted TCRs. Despite this general diversity, close scrutiny revealed an overrepresentation of basic residues, particularly arginine, concentrated near the NH2 terminal ends of the CDR3 regions. These basic residues appear in the α chain (CD8-2), in the β chain (DN1), or in both α and β chains (CD8-1 and DN.POTT) of the α/β TCR pairs (Table II), suggesting an important role in the specific recognition mediated by all of these TCRs (see below).
Modeling of the TCR–CD1 Interaction.
Given the overall structural similarity between MHC and the mouse CD1d1 crystal structures, together with evidence for lipid binding to human CD1b, we reasoned that lipid–CD1 complexes may form a TCR ligand analogous to peptide– MHC. Furthermore, our analysis of CD1-restricted TCR sequences indicates that they are comparable in primary structure to peptide–MHC-specific TCRs, using the same gene elements and having CDR3 lengths of comparable size to those found in MHC-restricted TCRs. Therefore, we modeled the DN1 TCR variable domains and human CD1b based on the known structures of other α/β TCRs and the mouse CD1d1 protein, respectively. We then modeled the DN1 TCR as a complex with human CD1b using the TCR/MHC/peptide ternary crystal structures to orient the molecules together (Fig. 5). This model suggests that the DN1 TCR could interact with CD1b in a manner that is similar to that of TCRα/β with peptide–MHC complexes. Strikingly, this would result in the positioning of the TCR CDR3 loops over the center of the putative CD1 antigen–binding site that is flanked by the CD1 α helices. Such an orientation would allow the basic residues in the DN1 TCR β chain CDR3 region to project into the central region of the groove that would provide direct interactions with exposed portions of a bound lipid or with the exposed carbohydrate of bound glycolipids. In particular, if the lipid antigen hydrophobic aliphatic chains are embedded in the electrostatically neutral CD1 pockets, the hydrophilic end of the lipid (or glycolipid) might be located between the CD1 α helices. This would orient the TCR CDR3 residues to interact with segments of the CD1 α helices and the charged CDR3 residues would project between the CD1 α helices in position to contact the polar head of the lipid (or glycolipid) (Fig. 5).
Figure 5.
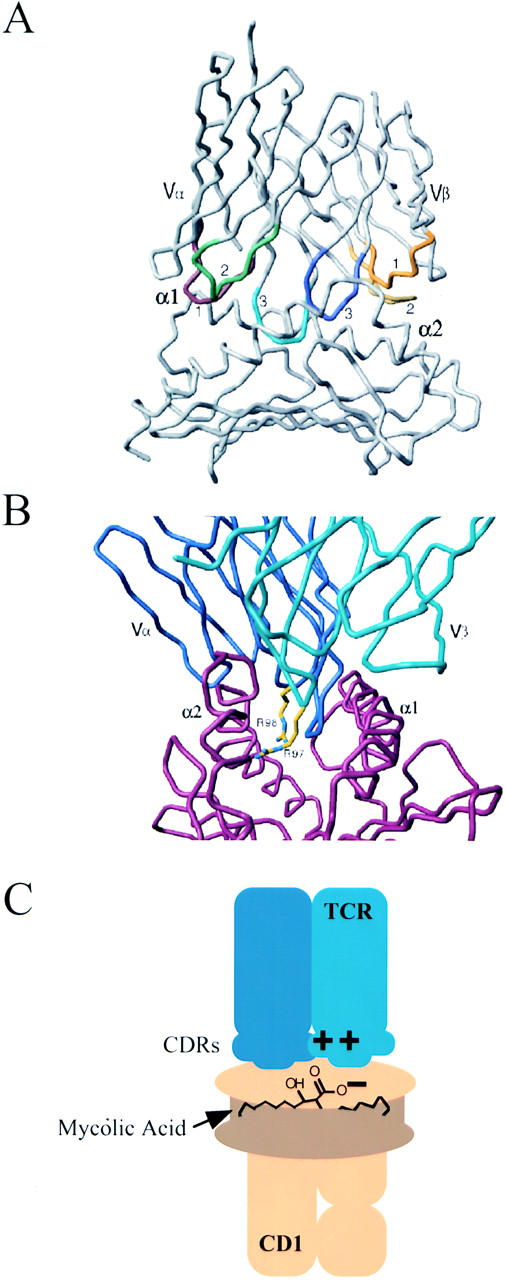
Molecular model of the DN1 TCR Vα/Vβ domains with the CD1b α1/α2 domains. The deduced amino acid sequences of the DN1 TCR and human CD1b were used to create a model of the protein structures based on the crystal structures for TCRs, mouse CD1d1, and TCR–MHC–peptide complexes. (A) Cα backbone of the TCR variable regions (top) complexed with the putative antigen binding region (α1 and α2 domains) of CD1b (bottom). The TCR CDR3 regions are colored: CDR1α, red; CDR2α, green; CDR3α, light blue; CDR1β, orange; CDR2β, yellow; CDR3β, dark blue. (B) View of the TCR (blue) and CD1b (red) complex, represented by the Cα backbone, looking down the putative antigen-binding site of CD1b, flanked to the left and right by the CD1 α2 and α1 helices, respectively. The side chains of R97 and R98 residues in the DN1 TCR CDR3β, indicated in yellow, project down into the hydrophobic cleft formed by the CD1 α helices. (C) Illustration of the proposed interaction between charged residues in the DN1 TCR CDR3 loops and polar or charged regions of mycolic acid bound to CD1b.
Discussion
That T cells bearing α/β TCR mediate lipid and glycolipid-specific, CD1a-, b-, or c-restricted recognition raised questions about the role of the α/β TCR in this process. Previously, the α/β TCR was thought to interact solely with MHC class I or II molecules complexed with antigenic peptides or protein superantigens. Here we have demonstrated that the TCRs from a panel of lipid/CD1– specific T cells mediate the ability of those T cells to discriminate between foreign lipid antigens and CD1 isoforms. The recent crystal structure of mouse CD1d1 revealed the presence of a hydrophobic binding cavity potentially capable of accommodating the hydrophobic aliphatic portions of a lipid antigen (35). Together with these structural data and the recent evidence that glycolipid antigens can bind directly to human CD1b in vitro (10), our findings suggest that lipids and CD1 molecules form complexes analogous to peptide–MHC complexes that are recognized together by a single TCR.
Recognition of lipid–CD1 complexes by the α/β TCR greatly expands the universe of foreign antigens recognized by T cells and reveals that previous assumptions that α/β TCRs were exclusively peptide/MHC–specific were not entirely correct. Although the relative frequencies of CD1-restricted versus MHC-restricted T cells are unknown, the prominent expression of CD1a, b, and c on many professional APCs suggests that recognition of this lineage of antigen-presenting molecules may be substantial. Here, the TCRs were derived from both the major CD8+ and relatively minor CD4−CD8− T cell pools that contain foreign lipid/CD1–specific T cells. We have also recently demonstrated that γ/δ T cells can recognize CD1c via a TCR- dependent mechanism (Spada, F., E.P. Grant, D. Leslie, and M.B. Brenner, unpublished observations), indicating the ability of both TCR types to interact with CD1.
The sequence analysis of a cross-section of CD1- restricted T cells shows dramatic heterogeneity in Vα, Vβ, and Jα gene usage among the TCRs. This diversity is in marked contrast to human CD1d-restricted T cells, which use an invariant germline-encoded TCR α chain composed of a precise AV24S1-AJ18 rearrangement together with a restricted pattern of Vβ chains (36), and to murine CD1d1-restricted NK T cells which use a similar TCR (37). These CD1d-specific T cells recognize CD1d in the absence of exogenously supplied lipid antigens, and it is not clear if they respond to empty CD1d molecules or to CD1d complexed to endogenous lipids/glycolipids. In the analysis presented here, three CD1b-restricted TCRs (DN1, DN.POTT, and LDN5) are described that use three different Vα and Vβ genes and have sequence diversity in both TCR-α and -β CDR3 regions. Thus, the recognition of exogenous lipid antigens with CD1b is more like peptide–MHC recognition than the response to CD1d. By extension, we expect that T cells specific for CD1a- and CD1c-restricted lipids will also have significantly greater diversity than has been observed for CD1d-specific T cells. This hypothesis is supported by the presence of template-independent N nucleotides in the TCR-α and -β sequences of CD8-2 (CD1a-restricted) and CD8-1 (CD1c-restricted) TCRs.
Despite the diversity that we observed in TCR usage, we noted the frequent coding of basic residues in CDR3α and CDR3β, primarily as a result of N nucleotide additions. If the hydrophobic antigen–binding pocket of CD1 binds to the hydrophobic acyl portions of a lipid antigen, the polar phosphate, carboxylic acid, or carbohydrate regions of the antigens would be exposed at the opening in the CD1 groove between the CD1 α helices to facilitate direct contacts with the TCR. We speculate that in the case of the mycolic acid–specific TCRs (DN1 and DN.POTT) the positively charged residues in the TCR CDR3 regions are positioned to interact with negatively charged lipid antigen head groups that would be exposed between the CD1 α helices. It is noteworthy that the one TCR described here that is specific for an antigen which does not have a negatively charged head group, LDN5, lacks positively charged amino acids in the CDR3 regions. We generated a molecular model of the DN1 TCR variable domains together with a model of the CD1b α1 and α2 domains based on the crystal structures of TCR–MHC– peptide complexes (27, 28) and the murine CD1d1 molecule (9) in order to visualize the hypothetical mode of interaction of these proteins (Fig. 5 A). This model suggests that CD1-restricted TCRs could interact with CD1 in a way analogous to TCR–MHC. Importantly, this model readily positions the basic residues R97 and R98 in the DN1 CDR3β loop in a location favorable for direct interaction with any moieties protruding from the CD1b hydrophobic pockets and positioned between the CD1 α helices (Fig. 5 B). Given the likely possibility that lipid binding to CD1 would be primarily through hydrophobic interactions of acyl chains inside the hydrophobic cavity, we speculate that the polar regions of the lipid antigens would be most likely to be exposed at the opening to this putative antigen-binding site. In the case of mycolic acid, the most polar region of the molecule consists of a carboxylic acid group and a hydroxyl group. Therefore, one or both of the arginine residues in the DN1 CDR3β might directly interact with this region of a bound mycolic acid molecule, making contacts important for the specificity and strength of the interaction between TCR and CD1–lipid, as illustrated schematically in Fig. 5 C. Ultimately, crystallographic studies are required in order to determine whether this model is correct and to reveal any unexpected differences between the mechanisms by which TCR interacts with lipid–CD1 and peptide–MHC ligands.
All of the TCRs isolated in this study incorporate either the BJ2S1 or BJ2S7 gene segments. This finding is also in contrast to the TCR sequences reported for CD1d-reactive T cells, which show no obvious bias in Jβ segment usage. Interestingly, the TCRs expressed by a CD1 directly reactive jejunal IEL T cell line described by Balk et al. predominantly used BJ2S1, and a CD1c directly reactive T cell clone isolated from that line expressed BJ2S7 (38). Additionally, two other CD1a or CD1c directly reactive T cell lines derived in our laboratories also express BJ2S1 and BJ2S7 (Grant, E.P., S.A. Porcelli, and M.B. Brenner, unpublished observations). The BJ2S1 and BJ2S7 gene products are quite similar to one another and are distinguished from the other Jβ segments by the motif ‘EQ(Y/F)F', which is predicted to lie near the COOH-terminal end of the CDR3β. It is possible that this portion of the CDR3β mediates important interactions with CD1a, b, and c molecules themselves, accounting for their increased incidence in the TCRs analyzed here. Alternatively, these residues may be critical for the positioning of the CDR loops such that they adopt a conformation suitable for interaction with CD1a, b, or c.
In these analyses, we have examined the receptors from a panel of T cells with diverse specificities for lipid antigens and CD1 isoform restriction. Despite this diversity, we have identified two potentially important TCR structural features that may be involved in the interaction of these TCRs with their lipid–CD1 ligands. We did not observe any clear bias in V gene usage by these TCRs, which testifies to the diversity of the T cell repertoire capable of interacting with nonpolymorphic CD1 molecules and their lipid antigens. Nonetheless, it is possible that a more extensive analysis of T cells with specificity for a single lipid antigen and with a common CD1 isoform restriction will reveal additional antigen-specific or CD1a-, b-, or c-specific biases in V gene usage.
Although the CD1 family of proteins is quite divergent from MHC at the sequence level, it is now clear that the two types of antigen-presenting molecules have sufficient structural similarities to allow the same set of TCR V and J segments to be combined to form TCR heterodimers that can interact with two extremely distinct classes of antigens. Thus, the previous paradigm that α/β TCRs are specific for peptide–MHC complexes must be modified. The TCR transfer studies presented here demonstrate directly that diverse TCR-α/β heterodimers instead mediate the recognition of foreign lipid and glycolipid antigens in the context of nonpolymorphic CD1a, b, and c antigen-presenting elements. Furthermore, although the CD8-1 and CD8-2 T cell lines are CD8α/β+, reconstitution of the TCRs in the J.RT3-T3.5 cell line, which is CD8α/β negative, confers the ability to productively interact with CD1, indicating that CD8 coreceptor expression is not essential to recognition of these presenting elements. These findings raise additional questions about CD1-restricted T cells, including how they are selected during thymic development and how α/β TCRs achieve specificity for CD1 versus MHC, given the use of the same families of germline TCR elements. Moreover, the relative frequency of CD1 versus MHC-reactive T cells during the course of infection and in autoimmune responses must now be evaluated to appreciate the relevance of these two antigen-presenting systems in various host responses.
Acknowledgments
We thank Dr. Masahiko Sugita for critical reading of the manuscript and Dr. D. Branch Moody for helpful discussions.
This work was supported by National Institutes of Health grants AI28973 (M.B. Brenner), CA58896 (I.A. Wilson), AI22553 (R.L. Modlin), AR40312 (R.L. Modlin), and the UNDP/World Bank/World Health Organization Special Program for Research and Training in Tropical Diseases (IMMLEP) (R.L. Modlin).
Abbreviations used in this paper
- CDR
complementarity-determining residues
- rhIL-2
recombinant human IL-2
- UTR
untranslated region
References
- 1.Allison JP, Ridge L, Lund J, Gross-Pelose J, Lanier L, McIntyre BW. The murine T cell antigen receptor and associated structures. Immunol Rev. 1984;81:145–160. doi: 10.1111/j.1600-065x.1984.tb01108.x. [DOI] [PubMed] [Google Scholar]
- 2.Haskins K, Kappler J, Marrack P. The major histocompatibility complex–restricted antigen receptor on T cells. Annu Rev Immunol. 1984;2:51–66. doi: 10.1146/annurev.iy.02.040184.000411. [DOI] [PubMed] [Google Scholar]
- 3.Meuer SC, Acuto O, Hercend T, Schlossman SF, Reinherz EL. The human T-cell receptor. Annu Rev Immunol. 1984;2:23–50. doi: 10.1146/annurev.iy.02.040184.000323. [DOI] [PubMed] [Google Scholar]
- 4.Beckman EM, Porcelli SA, Morita CT, Behar SM, Furlong ST, Brenner MB. Recognition of a lipid antigen by CD1-restricted αβ+T cells. Nature. 1994;372:691–694. doi: 10.1038/372691a0. [DOI] [PubMed] [Google Scholar]
- 5.Beckman EM, Melian A, Behar SM, Sieling PA, Chatterjee D, Furlong ST, Matsumoto R, Rosat JP, Modlin RL, Porcelli SA. CD1c restricts responses of mycobacteria-specific T cells—evidence for antigen presentation by a second member of the human CD1 family. J Immunol. 1996;157:2795–2803. [PubMed] [Google Scholar]
- 6.Moody DB, Reinhold BB, Guy MR, Beckman EM, Frederique DE, Furlong ST, Ye S, Reinhold VN, Sieling PA, Modlin RL, et al. Structural requirements for glycolipid antigen recognition by CD1b-restricted T cells. Science. 1997;278:283–286. doi: 10.1126/science.278.5336.283. [DOI] [PubMed] [Google Scholar]
- 7.Sieling PA, Chatterjee D, Porcelli SA, Prigozy TI, Mazzaccaro RJ, Soriano T, Bloom BR, Brenner MB, Kronenberg M, Brennan PJ, Modlin RL. CD1- restricted T cell recognition of microbial lipoglycan antigens. Science. 1995;269:227–230. doi: 10.1126/science.7542404. [DOI] [PubMed] [Google Scholar]
- 8.Porcelli SA. The CD1 family: a third lineage of antigen-presenting molecules. Adv Immunol. 1995;59:1–98. doi: 10.1016/s0065-2776(08)60629-x. [DOI] [PubMed] [Google Scholar]
- 9.Zeng Z-H, Castãno AR, Segelke BW, Stura EA, Peterson PA, Wilson IA. Crystal structure of mouse CD1: an MHC-like fold with a large hydrophobic binding groove. Science. 1997;277:339–345. doi: 10.1126/science.277.5324.339. [DOI] [PubMed] [Google Scholar]
- 10.Ernst WA, Maher J, Cho SG, Niazi KR, Chatterjee D, Moody DB, Besra GS, Watanabe Y, Jensen PE, Porcelli SA, et al. Molecular interaction of CD1b with lipoglycan antigens. Immunity. 1998;8:331–340. doi: 10.1016/s1074-7613(00)80538-5. [DOI] [PubMed] [Google Scholar]
- 11.Joyce S, Woods AS, Yewdell JW, Bennink JR, Desilva AD, Boesteanu A, Balk SP, Cotter RJ, Brutkiewicz RR. Natural ligand of mouse CD1d1-cellular glycosylphosphatidylinositol. Science. 1998;279:1541–1544. doi: 10.1126/science.279.5356.1541. [DOI] [PubMed] [Google Scholar]
- 12.Davodeau F, Peyrat MA, Necker A, Dominici R, Blanchard F, Leget C, Gaschet J, Costa P, Jacques Y, Godard A, et al. Close phenotypic and functional similarities between human and murine αβ T cells expressing invariant TCR α-chains. J Immunol. 1997;158:5603–5611. [PubMed] [Google Scholar]
- 13.Exley M, Garcia J, Balk SP, Porcelli S. Requirements for CD1d recognition by human invariant Vα24+ CD4−CD8−T cells. J Exp Med. 1997;186:109–120. doi: 10.1084/jem.186.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stenger S, Mazzaccaro RJ, Uyemura K, Cho S, Barnes PF, Rosat JP, Sette A, Brenner MB, Porcelli SA, Bloom BR, Modlin RL. Differential effects of cytolytic T cell subsets on intracellular infection. Science. 1997;276:1684–1687. doi: 10.1126/science.276.5319.1684. [DOI] [PubMed] [Google Scholar]
- 15.Porcelli S, Morita CT, Brenner MB. CD1b restricts the response of human CD4−8−T lymphocytes to a microbial antigen. Nature. 1992;360:593–597. doi: 10.1038/360593a0. [DOI] [PubMed] [Google Scholar]
- 16.Panchamoorthy G, McLean J, Modlin RL, Morita CT, Ishikawa S, Brenner MB, Band H. A predominance of the T cell receptor Vγ2/Vδ2 subset in human mycobacteria–responsive T cells suggests germline gene encoded recognition. J Immunol. 1991;147:3360–3369. [PubMed] [Google Scholar]
- 17.Olive D, Dubreuil P, Mawas C. Two distinct TL-like molecular subsets defined by monoclonal antibodies on the surface of human thymocytes with different expression on leukemia lines. Immunogenetics. 1984;20:253–264. doi: 10.1007/BF00364207. [DOI] [PubMed] [Google Scholar]
- 18.Behar SM, Porcelli SA, Beckman EM, Brenner MB. A pathway of costimulation that prevents anergy in CD28−T cells: B7-independent costimulation of CD1-restricted T cells. J Exp Med. 1995;182:2007–2018. doi: 10.1084/jem.182.6.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spits H, Keizer G, Borst J, Terhorst C, Hekman A, de Vries JE. Characterization of monoclonal antibodies against cell surface molecules associated with cytotoxic activity of natural and activated killer cells and cloned CTL lines. Hybridoma. 1983;2:423–437. doi: 10.1089/hyb.1983.2.423. [DOI] [PubMed] [Google Scholar]
- 19a.Rosat J-P, Grant EP, Beckman EM, Dascher CC, Sieling PA, Frederique D, Modlin RL, Porcelli SA, Furlong ST, Brenner MB. CD1-restricted microbial lipid antigen–specific recognition found in the CD8+αβ T cell pool. J Immunol. 1999;162:366–371. [PubMed] [Google Scholar]
- 20.DerSimonian H, Sugita M, Glass DN, Maier AL, Weinblatt ME, Reme T, Brenner MB. Clonal Vα 12.1+T cell expansions in the peripheral blood of rheumatoid arthritis patients. J Exp Med. 1993;177:1623–1631. doi: 10.1084/jem.177.6.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uematsu Y. A novel and rapid cloning method for the T-cell receptor variable region sequences. Immunogenetics. 1991;34:174–178. doi: 10.1007/BF00205820. [DOI] [PubMed] [Google Scholar]
- 22.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 23.Brawley JV, Concannon P. Modulation of promiscuous T cell receptor recognition by mutagenesis of CDR2 residues. J Exp Med. 1996;183:2043–2051. doi: 10.1084/jem.183.5.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 25.Jones TA, Thirup S. Using known substructures in protein model building and crystallography. EMBO (Eur Mol Biol Organ) J. 1986;5:819–822. doi: 10.1002/j.1460-2075.1986.tb04287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brunger, A.T. 1992. X-PLOR Version 3.1: A System for X-ray and NMR. Yale University Press, New Haven, CT. 382 pp.
- 27.Garcia KC, Degano M, Stanfield RL, Brunmark A, Jackson MR, Peterson PA, Teyton L, Wilson IA. An αβ T cell receptor structure at 2.5 Angstrom and its orientation in the TCR–MHC complex. Science. 1996;274:209–219. [Google Scholar]
- 28.Garboczi DN, Ghosh P, Utz U, Fan QR, Biddison WE, Wiley DC. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature. 1996;384:134–141. doi: 10.1038/384134a0. [DOI] [PubMed] [Google Scholar]
- 29.Housset D, Mazza G, Gregoire C, Piras C, Malissen B, Fontecilla-Camps JC. The three-dimensional structure of a T-cell antigen receptor Vα Vβ heterodimer reveals a novel arrangement of the Vβ domain. EMBO (Eur Mol Biol Organ) J. 1997;16:4205–4216. doi: 10.1093/emboj/16.14.4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang J, Lim K, Smolyar A, Teng M, Liu J, Tse AG, Hussey RE, Chishti Y, Thomson CT, Sweet RM, et al. Atomic structure of an αβ T cell receptor (TCR) heterodimer in complex with an anti-TCR Fab fragment derived from a mitogenic antibody. EMBO (Eur Mol Biol Organ) J. 1998;17:10–26. doi: 10.1093/emboj/17.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garcia KC, Degano M, Pease LR, Huang MD, Peterson PA, Teyton L, Wilson IA. Structural basis of plasticity in T cell receptor recognition of a self peptide MHC antigen. Science. 1998;279:1166–1172. doi: 10.1126/science.279.5354.1166. [DOI] [PubMed] [Google Scholar]
- 32.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. A program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:183–191. [Google Scholar]
- 33.Ferrin TE, Huang CC, Jarvis LE, Langridge R. The MIDAS display system. J Mol Graph. 1988;6:13–27. [Google Scholar]
- 34.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 35.Zeng DF, Dick M, Cheng LR, Amano M, Dejbakhshjones S, Huie P, Sibley R, Strober S. Subsets of transgenic T cells that recognize CD1 induce or prevent murine lupus: role of cytokines. J Exp Med. 1998;187:525–536. doi: 10.1084/jem.187.4.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Porcelli S, Yockey CE, Brenner MB, Balk SP. Analysis of T cell antigen receptor (TCR) expression by human peripheral blood CD4−8−α/β T cells demonstrates preferential use of several Vβ genes and an invariant TCR α chain. J Exp Med. 1993;178:1–16. doi: 10.1084/jem.178.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bendelac A, Rivera MN, Park SH, Roark JH. Mouse CD1-specific NK1 T cells: development, specificity, and function. Annu Rev Immunol. 1997;15:535–562. doi: 10.1146/annurev.immunol.15.1.535. [DOI] [PubMed] [Google Scholar]
- 38.Balk SP, Ebert EC, Blumenthal RL, McDermott FV, Wucherpfennig KW, Landau SB, Blumberg RS. Oligoclonal expansion and CD1 recognition by human intestinal intraepithelial lymphocytes. Science. 1991;253:1411–1415. doi: 10.1126/science.1716785. [DOI] [PubMed] [Google Scholar]