Abstract
The three-dimensional structure of Salmonella typhimurium aspartyl dipeptidase, peptidase E, was solved crystallographically and refined to 1.2-Å resolution. The structure of this 25-kDa enzyme consists of two mixed β-sheets forming a V, flanked by six α-helices. The active site contains a Ser-His-Glu catalytic triad and is the first example of a serine peptidase/protease with a glutamate in the catalytic triad. The active site Ser is located on a strand–helix motif reminiscent of that found in α/β-hydrolases, but the polypeptide fold and the organization of the catalytic triad differ from those of the known serine proteases. This enzyme is a member of a family of serine hydrolases and appears to represent a new example of convergent evolution of peptidase activity.
Keywords: peptidase E, protease, protein crystallography, serine hydrolase
Nearly all cells are able to hydrolyze proteins to free amino acids. Most of the enzymes that carry out the initial steps of protein degradation produce peptides as products, and the action of enzymes specialized in the hydrolysis of small peptides is necessary to complete the degradation process (1). To ensure the rapid hydrolysis of the great variety of peptides produced by protein degradation, most cells contain a number of different peptidases. Some of these enzymes display a broad specificity, hydrolyzing peptides of various lengths and amino acid compositions. Others have a narrow specificity hydrolyzing only a restricted subset of peptides. One such enzyme, aspartyl dipeptidase (peptidase E), is uniquely able to hydrolyze only Asp-X dipeptides (where X is any amino acid), and one tripeptide, Asp-Gly-Gly (2). This enzyme, originally found in bacteria but now known also to be present in the eukaryotes Xenopus laevis and Drosophila melanogaster, differs in specificity from all other known peptidases. In bacteria, the enzyme is believed to play a role in the degradation of peptides generated by intracellular protein breakdown or imported into the cell as nutrient sources. In X. laevis, the enzyme's production is developmentally regulated in response to thyroid hormone, and it is thought to play a role in apoptosis during tail resorption (3).
The primary sequence of aspartyl dipeptidase does not allow it to be classified in any known peptidase family (4, 5). Recently sequence comparison and site-directed mutagenesis data have been obtained, suggesting that aspartyl dipeptidase is a serine hydrolase (6). Because the enzyme appears to be unrelated by sequence to any other known serine hydrolase, it seemed possible that aspartyl dipeptidase might belong to a new family of serine hydrolases. Serine proteases or peptidases that possess a catalytic triad can be classified into five groups: the trypsin-like proteases (7), the subtilisin-like proteases (8), the serine carboxypeptidases (9), ClpP protease (10), and cytomegalovirus protease (11–13). The catalytic triad consists of a serine, a histidine, and a third residue, which is an aspartic acid in all of these enzymes except cytomegalovirus protease, where it is a histidine. However, the three-dimensional fold of the polypeptide chain and the order in which the catalytic triad residues appear in the sequence are characteristic for each group. Some lipases and esterases (14–16) are structurally similar to the serine carboxypeptidases, and enzymes with this fold are generally referred to as α/β-hydrolases (17). This paper reports a high resolution structure of Salmonella typhimurium aspartyl dipeptidase. The structure reveals that this enzyme is indeed the prototype of a new serine peptidase family with a unique fold and an unusual Ser-His-Glu catalytic triad.
Materials and Methods
Aspartyl dipeptidase was purified from S. typhimurium strains TN5213 and methionine auxotroph TN5517 (6), by using gel filtration and anion exchange chromatography (2, 4). Crystals grown in PEG 35000 (4) were flash frozen at 100 K after the addition of 40% glycerol to the mother liquor; they belong to space group C2 with a = 92.1 Å, b = 42.8 Å, c = 62.9 Å, and β = 106.5°, with one molecule per asymmetric unit. Data on selenomethionine protein crystals were collected at four different wavelengths λ1–λ4 at the National Synchrotron Light Source beam station X4A, and the native data were collected at beam station X12C (Table 1). Data were processed and reduced by denzo and scalepack (18). The three selenium atom positions, phases, and the initial electron density maps were calculated with the cns program package (19). The structure was initially refined by using xplor (20), followed by cns refinement. Finally, the model was refined with anisotropic B-factors, by using shelx (21). The crystallographic free R (22) was monitored by using 5% of the data. The graphics program o (23) was used to build the molecule and display the 2|Fo|-|Fc| and |Fo|-|Fc| electron density maps. The bound substrate model was created by manually docking an Asp-Ala dipeptide into the active site, followed by xplor energy minimization with fixed enzyme coordinates.
Table 1.
Crystallographic data
| Data collection and phasing statistics | |||||
|---|---|---|---|---|---|
| Data set | λ1 | λ2 (edge) | λ3 (peak) | λ4 | Native |
| Anomalous statistics | Yes | Yes | Yes | Yes | No |
| Wavelength, Å | 0.9879 | 0.9793 | 0.9788 | 0.9668 | 1.000 |
| Resolution range, Å | 20.0–1.8 | 20.0–1.8 | 20.0–1.8 | 20.0–1.8 | 20.0–1.2 |
| Completeness (final shell)* | 97.4 (94.3) | 97.3 (94.7) | 97.3 (94.7) | 97.4 (94.4) | 93.6 (76.1) |
| Total no. of reflections | 147 797 | 146 841 | 147 191 | 147 914 | 137 483 |
| Unique no. of reflections* | 41 624 | 41 511 | 41 541 | 41 612 | 68 270 |
| Rsym (final shell), % | 2.8 (8.7) | 2.8 (7.2) | 2.8 (7.7) | 2.8 (8.2) | 4.7 (27.4) |
| I/σ(I) | 30.1 (12.9) | 32.9 (16.4) | 32.4 (16.3) | 30.4 (15.7) | 14.0 (2.6) |
| Phasing power, disp./anom.† | 1.7/2.3 | 1.6/4.1 | 1.1/4.8 | /3.5 | |
| RCullis, disp./anom.† | 0.66/0.58 | 0.64/0.39 | 0.74/0.34 | /0.43 | |
| f′ obs/f" obs | −4.3/0.6 | −6.7/3.9 | −5.3/3.9 | −3.8/2.5 | |
| Refinement statistics (based on all data, 1684 protein atoms and 193 solvent atoms) |
| B factors | Resolution range, Å | R, % | Rfree, % | rms
deviation
|
Bave Main/side/protein/solvent, Å | |
|---|---|---|---|---|---|---|
| Bonds, Å | Angles | |||||
| Isotropic | 20.0–1.2 | 21.8 | 23.0 | 0.013 | 1.6° | 11.0/13.7/12.3/21.5 |
| Anisotropic | 10.0–1.2 | 15.8 | 19.1 | 0.012 | 0.031 Å | 13.3/18.0/15.6/28.0 |
A Friedel pair is considered as two unique reflections for the anomalously processed data sets but as a single unique reflection for the nonanomalously processed data set.
40,865 structure factors were phased with a figure of merit of 0.79.
Results and Discussion
Overall Structure.
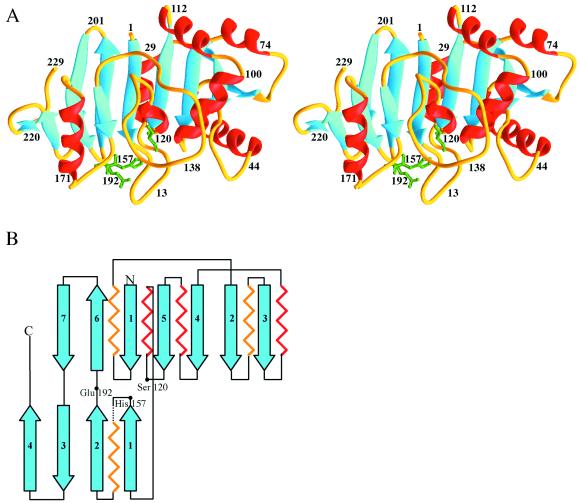
The structure of aspartyl dipeptidase was solved to 1.8 Å through multiple wavelength anomalous dispersion methodology, by using a selenomethionine derivative and four different wavelengths. The structure was subsequently refined with a 1.2-Å native data set (Table 1). With the exception of residues 161–169, for which no electron density was found, the peptide could be unambiguously traced from the N terminus to the C terminus. In the refined native structure, some disorder is evident among residues 42–50, which is presumably why the R value is relatively high (21.8% with isotropic B-factors and 15.8% with anisotropic B-factors). Aspartyl dipeptidase is composed of two β-sheets forming a V and contains a Ser-His-Glu catalytic triad situated at the base where the two mixed β-sheets meet (Fig. 1). The active site, Ser120, belongs to the larger of the two β-sheets, His157 belongs to the smaller, and the third catalytic triad residue, Glu192, is found at a short crossover segment between the two β-sheets. The order of the strands of the larger, eight-stranded β-sheet is 32451(−6)7, and that of the smaller, four-stranded β-sheet, 12(−3)4. The connectivities are 3×, 1×, −2×, −1×, −2, −1 and −1×, −1, −1, respectively (25). The larger of the two sheets is relatively flat, with the exception of the highly twisted strand 3 at the edge. The highest score (6.4) for structural similarity obtained by the program dali (26) was assigned to catalase, which has a fold unrelated to aspartyl dipeptidase. Thus, aspartyl dipeptidase appears to be a member of a novel family of serine peptidases unrelated to the trypsin-like proteases (7), subtilisin-like proteases (8), serine carboxypeptidase (9), cytomegalovirus protease (11–13), or ClpP protease (10). Although the active site aspartate is replaced by a glutamate in some of the esterases or lipases and by a histidine in cytomegalovirus protease, aspartyl dipeptidase is the first peptide bond-hydrolyzing enzyme with a glutamate in its catalytic triad.
Figure 1.
Structure of aspartyl dipeptidase. (A) Ribbon diagram (35). Helices are shown in red and β-strands in blue. The side chains of the catalytic triad residues are shown in green. (B) Topological representation.
Strand–Helix Motif and Catalytic Triad.
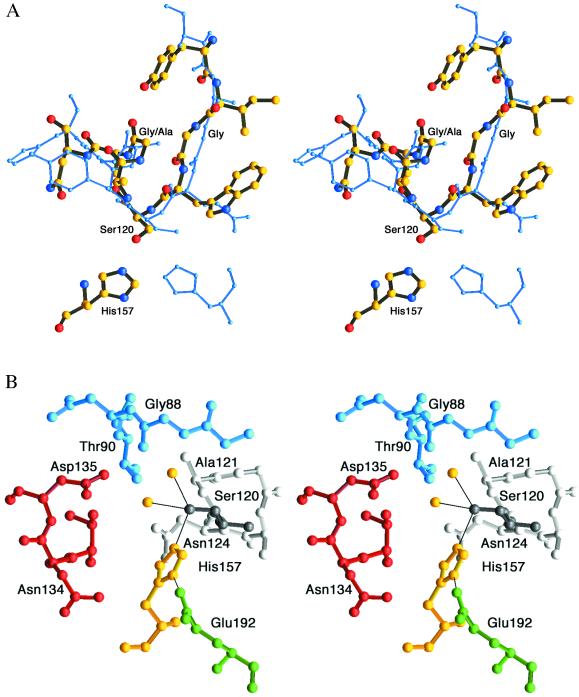
Despite its different fold, the active site serine of aspartyl dipeptidase is found at a strand–helix motif reminiscent of that found in α/β-hydrolases, e.g., serine carboxypeptidase (9), proline iminopeptidase (27), and acetylcholinesterase (15), although the helix in aspartyl dipeptidase makes a single turn only (Fig. 2A). Similar to these proteins, the active site serine is positioned in a less favorable part of the Ramachandran plot (Φ = 57°, ψ = −122°). This positioning is a consequence of the secondary structure elements that are joined by the active site serine. The Φ value reflects its position at the end of a β-strand and is characteristic of β-hairpin turns. At the same time this residue is part of an α-helix, and the hydrogen bond between its carbonyl oxygen atom and the main-chain nitrogen atom of Asn124 depends on the ψ value. The sharp turn of the strand–helix junction also explains why aspartyl dipeptidase conforms to the GXSXG sequence motif similar to that of the α/β-hydrolases. The two glycines in this motif are found in favored regions of the Ramachandran plot, but side chains at these positions would sterically exclude each other. Serine carboxypeptidase is able to host an alanine in the second but not the first of the Gly/Ala positions because its strand–helix motif adopts a broader turn.
Figure 2.
(A) Strand–helix motif. Superimposition of aspartyl dipeptidase (thick multicolored model) and serine carboxypeptidase (blue) illustrates similarities of the strand–helix motifs and the different histidine positions (35). The Cα atoms of the nine residues surrounding the active site Ser were superimposed and have a rms deviation of 0.9 Å. (B) View of the active site with Ser120 (dark gray) and the helix following it with Ala121-Asn124 (light gray). The active site Ser is coordinated to His157 and two water molecules (yellow). Glu192 is shown in green, Thr133-Asp135 in red, and Gly87-Thr90 in blue.
The active site histidine is presented from an orientation, vis-à-vis the active site serine, that is different from that of serine carboxypeptidase and proline iminopeptidase (9, 27). This orientation is best illustrated by superimposing the active site serine segments of aspartyl dipeptidase and serine carboxypeptidase (Fig. 2A). The handedness (17), i.e., the orientations and positions of the serine and histidine main chains relative the imidazole plane, are similar to those of trypsin and subtilisin and quite different from the serine carboxypeptidase active site constellation.
The hydrogen bonds of the catalytic triad in aspartyl dipeptidase display a very favorable geometry (Fig. 2B). The Oγ of the active site Ser120 is coordinated to His157 (2.7 Å) and to two water molecules (3.0 and 3.4 Å, respectively). Glu192 hydrogen bonds to His157 (2.7 Å), but surprisingly it is not hydrogen bonded to any other protein atom. In contrast to other serine proteases, the carboxylate hydrogen bond adopts an anti rather than the more common syn conformation (28). The angles between the two hydrogen bonds and the O—C bonds of the Ser (107°) and Glu (132°) do not deviate much from the values expected for tetrahedral and trigonal coordination. Both the Ser120 Oγ and the Glu192 Oɛ2 atoms are essentially within the imidazole ring plane (3° and 6°, respectively), although the imidazole and carboxylate planes make an angle of 65° and are far from coplanar. The relatively strong hydrogen bonds leave little room for interactions between the His and the substrate P1 side chain. Thus, from a structural point of view, a substrate-assisted mechanism (29) seems unlikely. Glu192 is conserved among all of the available aspartyl dipeptidase sequences (6), and to test its role in hydrolysis we constructed an E192A mutant. The mutant is approximately 1% as active as the wild type. (Using Asp-Leu as the substrate, we determined the specific activity for the E192A mutant to be 0.021 nmol min−1 μg−1 compared with a specific activity of 2.13 nmol min−1 μg−1 for the wild type.) The residual activity of the E192A mutant is greater than that of trypsin Asp102 mutants (30) but less than that of cytomegalovirus protease mutants affecting His157, the third member of the catalytic triad in this enzyme (13). Thus the presence of a hydrogen bond between Glu192 and His157 increases the catalytic efficiency of aspartyl dipeptidase by approximately 100-fold.
Active Site Pocket.
The active site (Fig. 2B) is fully exposed to the solvent. Thus, discrimination against larger peptides and proteins appears to be achieved by the chemical properties of the active site itself rather than by restricted access, as in the case of prolyl oligopeptidase (31). There is no positively charged residue in the active site that could ion-pair with the negative charge of the substrate aspartate side chain. Two positively charged and conserved residues, Arg171 and Arg174, are essential for catalysis (6) but are both too far away from the active site to be able to interact directly with the substrate. Arg171 hydrogen bonds to the main-chain oxygen of the active site His157. In addition, it probably influences the orientation of the Asn134 side chain in such a way as to direct the obligate hydrogen bond donor nitrogen, rather than the acceptor oxygen, toward the active site, which has to accommodate an aspartate side chain. The role of the more remotely located Arg174 is more difficult to explain, but its positive charge might enhance the electrostatic effect of Arg171. The two arginines are located on an α-helix and are relatively close to each other (Fig. 1a). Another essential residue, the conserved Asp135 (6), is situated in the active site and might interact with the N-terminal amino group of the substrate. A similar role has been indicated for two conserved glutamates in the active site of proline iminopeptidase (27).
Superimposition of the active site His of trypsin, subtilisin, serine carboxypeptidase, and aspartyl dipeptidase suggests that the oxyanion hole is formed by the electrostatic action of the main-chain nitrogen atoms of Gly88 and Ala121. If this supposition is correct, the main-chain nitrogen atom of the residue following the active site Ser is involved in stabilizing the tetrahedral intermediate both in serine carboxypeptidase and aspartyl dipeptidase, although the strand–helix motifs are oppositely oriented with respect to the imidazole ring plane in the two enzymes. This arrangement suggests that the strand–helix motif, with its hydrogen bond between the carbonyl oxygen of Ser120 and the main-chain nitrogen of Asn124, has evolved to maximize the electrophilicity of the oxyanion hole through the dipole moment of the helix.
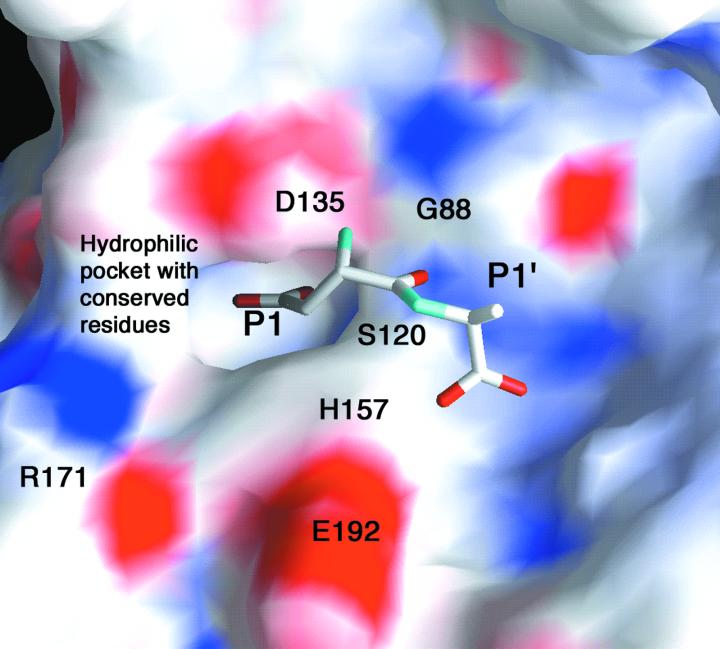
An Asp-Ala dipeptide was docked into the active site of aspartyl dipeptidase, as shown in Fig. 3. This model is based on the supposed location of the oxyanion hole, the suggested interaction between Asp135 and the substrate amino group, the location of conserved residues, and the fact that the aspartyl dipeptidase active site contains a pocket with obligate (main chain nitrogen and side chain of Asn134) and possible (Thr90, Asn124, Thr133) hydrogen bond donors into which the substrate aspartate side chain has been placed. The substrate amino group is also suggested to interact with the carbonyl oxygen of Gly88. Both the amino group and the aspartate carboxylate group are close to experimentally determined water positions. Discrimination against asparaginyl dipeptides is probably because of hydrogen bond interactions rather than ion pairing, because there is no positively charged residue in the active site pocket. Docking the peptide into the active site is restricted by the distance between enzyme serine and the substrate carbonyl group, suggesting either that tetrahedral intermediate formation and binding are simultaneous processes or that minor conformational changes take place on binding. The side chain of the P1′ residue is facing the solvent, consistent with the observed lack of specificity with respect to this residue (2). Removal of the P1′ side chain would perhaps permit a rotation around the N—Cα bond of this residue, orienting the C-terminal carboxylate group toward the exterior. This hypothesis would explain the enzyme's activity toward Asp-Gly-Gly, the only known tripeptide substrate.
Figure 3.
The active site of aspartyl dipeptidase (36). Positive charge is shown in blue and negative charge in red. An Asp-Ala dipeptide has been modeled into the active site. The amino group of the substrate interacts with Asp135, and the P1 side chain is found in a hydrophilic pocket with residues that are conserved throughout the sequences of Salmonella typhimurium, Escherichia coli, Haemophilus influenzae, Actinobacillus actinomycetemcomitans, Shewanella putrefaciens, Pasteurella multocida, Xenopus laevis, and Drosophila melanogaster (6). The P1′ side chain is oriented toward the solvent. The substrate can move further into the active site only at the cost of very close contacts between its carbonyl atom and the active site serine hydroxyl group.
Aspartyl dipeptidase represents a new class of serine hydrolases, with a unique structure, overall polypeptide fold, and sequence arrangement of the catalytic residues. The location of the active site Ser in a strand–helix motif is similar to that of the serine carboxypeptidases, but the geometry of the catalytic triad and its orientation relative to the strand–helix motif are different. It is therefore not likely that aspartyl dipeptidase and the serine carboxypeptidases are evolutionarily related. We suggest that the superficial resemblance of aspartyl dipeptidase to the other proteases, i.e., the presence of a hydrogen-bonded Ser-His-carboxylic acid catalytic triad and the strand–helix location of the active site serine are the results of convergent evolution.
Interestingly, the GXSAG (residues 119–123), GGXT (residues 87–90), and PH (residues 156–157) motifs can be found in ORFs with little or no overall sequence similarity to aspartyl dipeptidase. Examples of such sequences are Bacillus subtilis YgaJ (32), Leishmania major CAB71275 (33), and Deinococcus radiodurans D75442 (34). Hence the residues directly involved in the catalytic mechanism, i.e., the active site Ser (Ser120), the active site His (His157), and the two residues that form the oxyanion hole (Gly88 and Ala121) are present in these proteins. The other residues defining the P1 pocket in aspartyl dipeptidase cannot be found in these sequences, suggesting that they represent enzymes with the same fold and same basic mechanism as aspartyl dipeptidase, but with different substrate specificities. It appears, therefore, that the aspartyl dipeptidases are all members of a larger family of hydrolytic enzymes present in both prokaryotes and eukaryotes.
Acknowledgments
We thank T. Knox for site-directed mutagenesis and for continuous assistance, H. Robinson and Y.-G. Gao for x-ray room assistance, C. Ogata (NSLS X4A) and R. Sweet (NSLS X12C) for beamline assistance, M. Churchill for sharing beam time, R. Lassy for making the aspartyl dipeptidase clones and for stimulating discussions, and Professor A. Liljas for having initiated the project. This work was supported by National Institute for Allergy and Infectious Diseases Grant AI10333 (to C.G.M.) and National Institute of General Medical Sciences Grant GN41612 (to A.H.-J.W.).
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes 1FY2 and 1FYE).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.260376797.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.260376797
References
- 1.Miller C G. In: Protein Degradation and Proteolytic Modification. Neidhardt F C, Curtis R III, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Vol. 1. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 938–954. [Google Scholar]
- 2.Carter T H, Miller C G. J Bacteriol. 1984;159:453–459. doi: 10.1128/jb.159.2.453-459.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown D D, Wang Z, Furlow J D, Kanamori A, Schwartzman R A, Remo B F, Pinder A. Proc Natl Acad Sci USA. 1996;93:1924–1929. doi: 10.1073/pnas.93.5.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conlin C A, Håkansson K, Liljas A, Miller C G. J Bacteriol. 1994;176:166–172. doi: 10.1128/jb.176.1.166-172.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrett A J, Rawlings N D, Woessner J F. Handbook of Proteolytic Enzymes. San Diego: Academic; 1998. [Google Scholar]
- 6.Lassy R A, Miller C G. J Bacteriol. 2000;182:2536–2543. doi: 10.1128/jb.182.9.2536-2543.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matthews B W, Sigler P B, Henderson R, Blow D M. Nature (London) 1967;214:652–656. doi: 10.1038/214652a0. [DOI] [PubMed] [Google Scholar]
- 8.Wright C S, Alden R A, Kraut J. Nature (London) 1969;221:235–242. doi: 10.1038/221235a0. [DOI] [PubMed] [Google Scholar]
- 9.Liao D I, Breddam K, Sweet R M, Bullock T, Remington S J. Biochemistry. 1992;31:9796–9812. doi: 10.1021/bi00155a037. [DOI] [PubMed] [Google Scholar]
- 10.Wang J, Hartling J A, Flanagan J M. Cell. 1997;91:447–456. doi: 10.1016/s0092-8674(00)80431-6. [DOI] [PubMed] [Google Scholar]
- 11.Shieh H S, Kurumbail R G, Stevens A M, Stegeman R A, Sturman E J, Pak J Y, Wittwer A J, Palmier M O, Wiegand R C, Holwerda B C, Stallings W C. Nature (London) 1996;383:279–282. doi: 10.1038/383279a0. [DOI] [PubMed] [Google Scholar]
- 12.Qiu X, Culp J S, DiLella A G, Hellmig B, Hoog S S, Janson C A, Smith W W, Abdel-Meguid S S. Nature (London) 1996;383:275–279. doi: 10.1038/383275a0. [DOI] [PubMed] [Google Scholar]
- 13.Tong L, Qian C, Massariol M J, Bonneau P R, Cordingley M G, Lagace L. Nature (London) 1996;383:272–275. doi: 10.1038/383272a0. [DOI] [PubMed] [Google Scholar]
- 14.Schrag J D, Li Y G, Wu S, Cygler M. Nature (London) 1991;351:761–764. doi: 10.1038/351761a0. [DOI] [PubMed] [Google Scholar]
- 15.Sussman J L, Harel M, Frolow F, Oefner C, Goldman A, Toker L, Silman I. Science. 1991;253:872–879. doi: 10.1126/science.1678899. [DOI] [PubMed] [Google Scholar]
- 16.Winkler F K, D'Arcy A, Hunziker W. Nature (London) 1990;343:771–774. doi: 10.1038/343771a0. [DOI] [PubMed] [Google Scholar]
- 17.Ollis D L, Cheah E, Cygler M, Dijkstra B, Frolow F, Franken S M, Harel M, Remington S J, Silman I, Schrag J, et al. Protein Eng. 1992;5:197–211. doi: 10.1093/protein/5.3.197. [DOI] [PubMed] [Google Scholar]
- 18.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 19.Brünger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 20.Brünger A T, Kuriyan K, Karplus M. Science. 1987;235:458–460. doi: 10.1126/science.235.4787.458. [DOI] [PubMed] [Google Scholar]
- 21.Sheldrick G M, Schneider T R. Methods Enzymol. 1997;277:319–343. [PubMed] [Google Scholar]
- 22.Brünger A T. Nature (London) 1992;355:472–474. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 23.Jones T A, Zou J-Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 24.Bernstein F C, Koetzle T F, Williams G J, Meyer E E, Jr, Brice M D, Rodgers J R, Kennard O, Shimanouchi T, Tasumi M. J Mol Biol. 1977;112:535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- 25.Richardson J S. Adv Protein Chem. 1981;34:167–339. doi: 10.1016/s0065-3233(08)60520-3. [DOI] [PubMed] [Google Scholar]
- 26.Holm L, Sander C. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 27.Medrano F J, Alonso J, Garcia J L, Romero A, Bode W, Gomis-Ruth F X. EMBO J. 1998;17:1–9. doi: 10.1093/emboj/17.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ippolito J A, Alexander R S, Christianson D W. J Mol Biol. 1990;215:457–471. doi: 10.1016/s0022-2836(05)80364-x. [DOI] [PubMed] [Google Scholar]
- 29.Dall'Acqua W, Halin C, Rodrigues M L, Carter P. Protein Eng. 1999;12:981–987. doi: 10.1093/protein/12.11.981. [DOI] [PubMed] [Google Scholar]
- 30.Craik C S, Roczniak S, Largman C, Rutter W J. Science. 1987;237:909–913. doi: 10.1126/science.3303334. [DOI] [PubMed] [Google Scholar]
- 31.Fülöp V, Bocskei Z, Polgar L. Cell. 1998;94:161–170. doi: 10.1016/s0092-8674(00)81416-6. [DOI] [PubMed] [Google Scholar]
- 32.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, et al. Nature (London) 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 33.Ivens A C, Lewis S M, Bagherzadeh A, Zhang L, Chan H M, Smith D F. Genome Res. 1998;8:135–145. doi: 10.1101/gr.8.2.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.White O, Eisen J A, Heidelberg J F, Hickey E K, Peterson J D, Dodson R J, Haft D H, Gwinn M L, Nelson W C, Richardson D L, et al. Science. 1999;286:1571–1577. doi: 10.1126/science.286.5444.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carson M. J Appl Crystallogr. 1991;24:958–961. [Google Scholar]
- 36.Nicholls A, Honig B J. J Comput Chem. 1991;12:435–445. [Google Scholar]