Abstract
HslVU is an ATP-dependent prokaryotic protease complex. Despite detailed crystal and molecular structure determinations of free HslV and HslU, the mechanism of ATP-dependent peptide and protein hydrolysis remained unclear, mainly because the productive complex of HslV and HslU could not be unambiguously identified from the crystal data. In the crystalline complex, the I domains of HslU interact with HslV. Observations based on electron microscopy data were interpreted in the light of the crystal structure to indicate an alternative mode of association with the intermediate domains away from HslV. By generation and analysis of two dozen HslU mutants, we find that the amidolytic and caseinolytic activities of HslVU are quite robust to mutations on both alternative docking surfaces on HslU. In contrast, HslVU activity against the maltose-binding protein-SulA fusion protein depends on the presence of the I domain and is also sensitive to mutations in the N-terminal and C-terminal domains of HslU. Mutational studies around the hexameric pore of HslU seem to show that it is involved in the recognition/translocation of maltose-binding protein-SulA but not of chromogenic small substrates and casein. ATP-binding site mutations, among other things, confirm the essential role of the “sensor arginine” (R393) and the “arginine finger” (R325) in the ATPase action of HslU and demonstrate an important role for E321. Additionally, we report a better refined structure of the HslVU complex crystallized along with resorufin-labeled casein.
Bacteria contain a number of ATP-dependent proteases, including two component systems like the ClpAP, ClpXP, and ClpQY (HslVU; ref. 1). ClpA and ClpX share a common protease, ClpP. In contrast, HslU interacts only with HslV (2). HslVU is of particular interest as a simple model system for the eukaryotic 26S proteasome (3–5). Subunits of HslV share significant sequence (6) and structural similarity (7) with the proteasomal β-subunits but form different oligomeric assemblies with D6 symmetry of HslV and D7 symmetry of the archaeal β-subcomplex. HslU, a member of the Hsp100 family of ATPases (8), exhibits both ATPase (3) and chaperone activities (9). The structure determination of HslU showed that it exists as a hexamer of subunits with three domains termed N-terminal (N), intermediate (I), and C-terminal (C) domains, respectively (10). The structure of the N and C domains classified HslU as a member of the ATPases associated with a variety of cellular activities (AAA)-ATPase superfamily of proteins as had been suggested by sequence comparison (11). HslU is therefore a useful model system for the base part of the proteasomal 19S regulatory caps, which contain six components classified as members of the AAA-ATPase family (12).
Despite abundant structural information about HslVU, the mechanism of ATP hydrolysis and protease activation is still unclear. In the x-ray structures of the HslU-ATP and HslU-5′-adenylylimidotriphosphate (AMP-PNP) complexes (10), we found an abundance of basic and acidic residues in the vicinity of the scissile anhydride bond between β- and γ-phosphates of the nucleotide. This is compatible with both base and acid catalysis of the hydrolysis reaction. To distinguish between these possibilities, we mutated residues near the nucleotide-binding site.
Additionally, and most importantly, different docking modes are observed in the crystal structure and in electron microscopy (EM) images (10, 13). In the crystal structure, HslU docks to HslV with the I domains pointing toward the HslV particle (this will be referred to as the x-ray mode of docking) (10). An attractive feature of this docking mode is that no translocation of substrate through the rather small central pore of the HslU hexamer would be required. Instead, HslU could shuttle between states of high and low affinity for HslV and deliver substrates in the process. In contrast, the EM images (13), interpreted in the light of the crystal structures of both components, indicate binding of HslU to HslV with the I domains distal to HslV (this will be referred to as the EM mode of docking).
Consistent with the weak binding of HslU to HslV, alternative configurations of the complex may be stabilized by different conditions of sample preparation (crystallization and staining, respectively). Both models present problems for functional interpretation. The crystal structure shows very weak contacts between the components and small conformational changes between free and complexed components. This is hard to reconcile with the observation that HslU stimulates the activity of HslV against small chromogenic substrates up to 100-fold. The EM model does not show how substrates could be delivered to the HslV through the narrow pore along the 6-fold axis of HslU.
To identify the functionally relevant HslVU docking mode, we have designed and generated a gallery of mutants. Here, we report the results of our extensive mutagenesis and biochemical studies. We also report a better refined structure of the HslVU complex crystallized in the presence of resorufin-labeled casein, a substrate of the HslVU protease complex.
Materials and Methods
Cloning, Expression, and Purification.
The isolation of wild-type HslV (7) and HslU (10) has been described. Mutants were generated by using standard PCR techniques and ligated into pET12b or pET22b. These constructs were then transformed into BL21(DE3) or BL21(DE3)[pLysS] cells for expression. Wild-type HslV and HslU and the mutant enzymes were purified as previously described (7, 10). Each mutagenesis was confirmed by DNA sequencing as well as electron spray ionization mass spectroscopic (ESI-MS) analyses. Maltose-binding protein and SulA fusion protein (MBP-SulA) was overexpressed and purified as described previously (14). The state of oligomerization was estimated by gel filtration and comparison with the elution profile of the wild-type protein in the absence of exogenous nucleotide.
Assays.
ATPase activity was measured as described earlier (15), by determining the amount of inorganic phosphate formed on ATP hydrolysis and detected at 660 nm as a complex with malachite green and ammonium molybdate.
Peptide hydrolysis was assayed by using the chromogenic peptide, carbobenzoxy-Gly-Gly-Leu-7-amido-4-methylcoumarin (Z-Gly-Gly-Leu-AMC; Bachem) as a substrate as reported earlier (3) with 1 μg of HslV and 2.5 μg of HslU.
Resorufin-labeled casein (Roche Molecular Biochemicals) (16) or FITC-labeled casein (17) was used as a model protein substrate. The proteolytic activity of HslV (10 μg) in the presence of HslU (25 μg) against resorufin-casein was assayed following the manufacturer's instructions. The degradation of FITC-casein was measured by using HPLC. Enzyme samples (15 μg of HslV; 37.5 μg of HslU) in buffer U (50 mM Tris⋅HCl, pH 7.5/5 mM MgCl2) were incubated for 45 min with 2 mM adenosine 5′-O-(3-thiotriphosphate; ATP-γS) and 1 mM FITC-casein. The reaction was stopped by the addition of calpain inhibitor I (acetyl-Leu-Leu-norleucinal) to a final concentration of 1 mM.
The MBP-SulA assay was carried out as reported earlier (14, 18) with minor modifications.
Crystallization and Refinement.
HslV-HslU complexed with resorufin-casein was crystallized as follows: 2 μl of an HslV-HslU mixture prepared as previously described (10) was mixed with 0.3 μl of 4 mg/ml resorufin-labeled casein and 2.3 μl of reservoir buffer, that contained 100 mM Mes, pH 6.5, and 1.95 M sodium acetate. Purple-colored crystals were grown in hanging drops by vapor diffusion at room temperature. They are isomorphous to the complex crystals described previously (10).
Diffraction data were collected on a charge-coupled device detector at the BW6 beamline of the Deutsche Elektronen Synchroton Centre, Hamburg (Table 1). Data processing and subsequent refinements were carried out as previously described (10). Solvent molecules were added by using model-phased difference Fourier maps by using cns (19).
Table 1.
Data collection and refinement statistics
| Data collection statistics | |
| Space group | P6322 |
| Cell parameters, Å | a = b = 172.02, c = 276.57 |
| Resolution range, Å | 25-2.8 |
| Unique/total reflections | 59,863/706,383 |
| Redundancy | 11.8 |
| Completeness, %* | 99.8 (99.8) |
| Rmerge† (%) | 12.0 |
| Refinement and model statistics | |
| R factor/Rfree‡ | 0.254/0.304 |
| Resolution range, Å | 15-2.8 |
| Modeled residues | HslV; (1–174) × 4 |
| HslU; (1–139, 151–165, 218–443), (1–129, 221–443) | |
| Number of ligands | 2 (AMP-PNP) |
| Number of solvent molecules | 293 |
| Rmsd bond lengths, Å | 0.014 |
| Rmsd bond angles, ° | 1.65 |
| Rmsd NCS protein, Å | HslV (172 matching Cα); 0.26 (free), 0.24 (complexed) |
| HslU (333 matching Cα); 0.27 | |
Rmsd, root-mean-square deviation.
Values are for the reflections in the highest resolution shell (2.8–2.87 Å).
Rmerge = ΣhΣi|I(h, i) − 〈I(h)〉|/ΣhΣiI(h, i), where I(h, i) is the intensity of the ith measurement of reflection h, and 〈I(h)〉 is the average value over multiple measurements.
R = Σ∥Fo| − |Fc∥/Σ|Fo|, where Rfree is calculated for the 10% test set of reflections.
Results
Mutations of ATP-Binding Sites of HslU.
In the HslU-ATP and HslU-AMP-PNP complexes (10), the nucleotide is bound between adjacent subunits at the hinge region of the N and C domains. We found the ɛ-amino groups of K63 (part of the Walker A motif) and K80 close to the γ-phosphate of ATP (see figure 4 in ref. 10). The latter's involvement in catalysis would not be expected from sequence comparisons between members of the AAA-ATPases, because it is not conserved (11). Mutation of this residue to threonine showed that amidolytic and caseinolytic activities were reduced by factors of 4 and 2, respectively. Surprisingly, the ATPase activity and the rate of degradation of the MBP-SulA fusion protein were unchanged (Table 2).
Table 2.
Summary of HslU mutants
| Peptide | Casein | SulA | ATPase | Hexamer | Comments | |
|---|---|---|---|---|---|---|
| Wild type | ++++ | ++++ | ++++ | ++++ | ++++ | |
| ATP-binding site mutants | ||||||
| K80T | + | ++ | ++++ | ++++ | ++ | |
| E286Q | ++ | ++ | ++ | ++++ | +++ | ATP-bound (structure)* |
| E321Q | − | − | − | − | ++++ | |
| R325E | − | − | − | − | − | ATP-bound (structure)* |
| R393A | − | − | − | − | ++ | ATP-bound (CD)* |
| I domain mutants | ||||||
| Δ137–150 | ++++ | ++++ | ++++ | ++++ | ++++ | 2 Gly linker |
| Δ175–209 | ++++ | ++++ | − | ++++ | ++++ | 2 Gly linker |
| Δ111–239 | +++ | +++ | − | +++ | ++++ | 3 Gly linker |
| Contact region in the EM mode | ||||||
| E266Q | ++++ | ++++ | ++++ | ++++ | ++++ | |
| E266Q/E385K | ++++ | ++++ | ++++ | ++++ | ++++ | |
| I312W | +++++ | +++++ | +++++ | +++++ | ++++ | |
| Ins(264, 265) | ++++ | ++++ | − | ++++ | ++++ | 5 Gly insertion |
| Ins(311, 312) | − | − | − | + | ++ | 5 Gly insertion |
| Ins(387, 388) | ++++ | +++ | − | +++++ | − | 5 Gly insertion† |
| Ins(435, 436) | NA | NA | NA | NA | NA | Insoluble, 5 Gly insertion |
| Δ432–443 | NA | NA | NA | NA | NA | Insoluble, C-term deletion |
| E436A/D437A | +++ | ND | ND | ND | ++++ | |
| E436K/D437K | +++ | ++++ | − | ++++ | ++++ | Charge reversal |
| Hexamer pore mutants | ||||||
| E88Q | ++ | − | − | ++++ | ++++ | |
| E88Q/E266Q | ++ | − | − | ++++ | ++++ | |
| Y91G | +++ | +++ | − | ++++ | ++++ | |
| V92G | ++++ | ++++ | − | ++++ | +++ | |
| G93A | + | + | − | ++ | ++++ | |
| E95W | ++++ | ++++ | + | ++++ | +++ | |
| Δ88–92 | − | − | − | − | ++ | 3 Gly linker |
| Δ89–92 | ++ | ++ | − | ++ | +++ | 1 Gly linker |
The values are relative to the wild-type HslU activity or hexamer content as a 100% (++++, 80–100%; +++, 60–80%; ++, 40–60%; +, 20–40%; −, <20%); NA, not applicable; ND, not determined.
Unpublished results.
† Hexamer in the presence of exogenous nucleotide.
A prominent and conserved residue R325 from the adjacent subunits, homologous to R315 in FtsH, is essential for activity in FtsH (20). The R325E mutant exhibited a complete loss of all protease and ATPase activities, but the crystal structure of the mutant complex was nearly identical to the native complex (data not shown). In particular, the mutant is still able to bind nucleotide under crystallization conditions.
The adjacent subunit also contributes two acidic residues, E286 and E321, to the active site. In the HslVU structure, E321 is closer to the γ-phosphate, but E286 and not E321 appears to be conserved among AAA-ATPases (11). The replacement of E286 with glutamine had no effect on ATPase activity, but the activities against peptide, casein, and MBP-SulA substrates were reduced. The analogous mutation in E321 had a more drastic effect: E321Q was inactive in all assays, including the ATPase assay.
R393 of the C domain, a conserved residue of the AAA-ATPases (11), is part of the GΦRXΦ (Φ hydrophobic) signature sequence V of the Clp family (8). This “sensor arginine” is likely to mediate the change in the relative orientation of the N and C domains of HslU on nucleotide binding and release. Mutation of this residue to alanine abolished both the ATPase and protease activities.
Activity of HslVU Under Crystallization Conditions and Refined Model of HslV-HslU.
To examine the influence of the crystallization conditions on enzymatic activity, we crystallized the complex in the presence of resorufin-casein and investigated the effect of this component in the crystallization mix. HslVU is expected to be less active under our crystallization conditions than in the presence of Mg2+, ATP, and low salt (10). We found that crystals of the same space group and lattice constants could be grown with an excess of magnesium and without exogenous ATP. They were not pursued further because they diffracted less well. Additionally, HslV-HslU complex crystals grown in the presence of resorufin-labeled casein were purple in color because of trapped fully/partially digested colored substrate. The presence of casein fragments in the washed crystals was confirmed by SDS/PAGE analysis (data not shown). Further support for the activity of the enzyme came from caseinolytic assays performed under crystallization conditions. Also, crystals of the complex grown in the presence of resorufin-casein by using an HslV mutant that hydrolyzes casein very slowly had varying amounts of fully/partially digested colored substrate trapped in them (data not shown). Crystals that appeared earlier were almost colorless, whereas those that appeared later were colored to varying extents. It is obvious that active enzyme species exist under the conditions of crystallization.
HslVU crystals grown in the presence of resorufin-labeled casein diffracted to the same resolution (2.8 Å) as the native HslVU complex, but the quality of data was better (Table 1). Although the resulting structure is almost identical to that of the native HslVU complex, the electron density maps corresponding to the I domain were somewhat better defined. On the basis of these maps, we could model additional residues in this domain, one of the six N-terminal histidine tags in the HslU molecule and two C-terminal residues in HslV, which were not visible in the previously reported structure (10). In addition, water molecules could be added to the model by using these maps. Electron density corresponding to casein or its digested fragments could not be observed, probably because of random positions of the digested fragments in the crystal.
Deletions in the I domain of HslU.
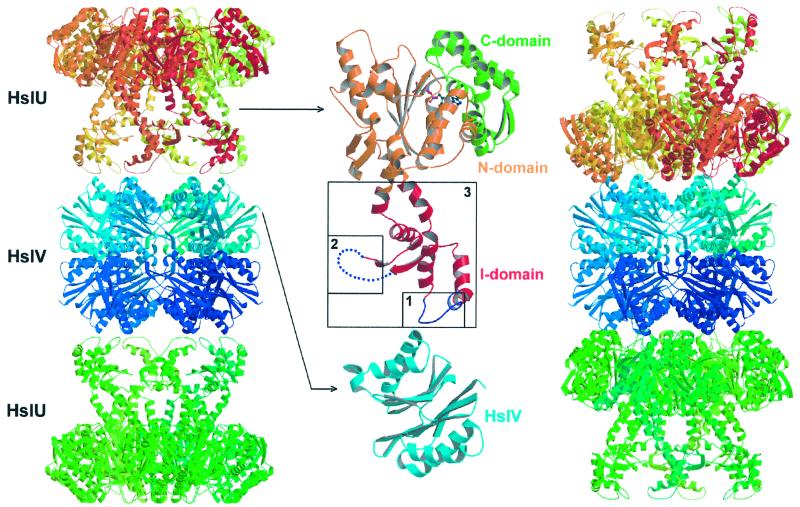
The I domain protrudes from the N domain with residue M110 and returns to it with residue A243 (ref. 10; Fig. 1 Left). It has a loosely folded coiled helical structure, and large parts are disordered in our earlier structures (10) and in the current refined model. In the crystal structures of the complex, this domain is in contact with HslV.
Figure 1.
Two possible docking modes of the HslVU complex and sites of deletion in the I domain of HslU. Ribbon diagram of the HslVU crystal structure (Left) and modeled complex structure based on EM images and considerations of steric complementarity (Right). Model of the disordered part in the I domain is taken from free (HslU)6-(AMP-PNP)3 structure (Protein Data Bank ID code 1DO2). Closeup view of deleted regions in the I domain (Middle). Numbers 1, 2, and 3 correspond to the Δ137–150, Δ175–209, and Δ111–239 deletion mutants, respectively. The N, C, and I domains are shown in orange, green, and red, respectively. This figure was drawn by using MOLSCRIPT (26) and rendered by using RASTER3D (27).
To investigate the role of the I domain and especially its involvement in the docking between HslU and HslV, we designed three different deletion mutants (Δ137–150, Δ175–209, Δ111–239) in this region of HslU (Table 2, and Fig. 1 Middle).
In the Δ137–150 mutant, we deleted residues that appear to mediate the interaction between HslU and HslV in the crystalline complex. As these and the adjacent residues in HslV are more disordered in the structure of the complex than in the structures of free HslV (7) and of HslU-AMP-PNP, we had suggested that an order–disorder transition could play a role in the mechanism of protease activation. The Δ137–150 mutant exhibits wild-type-like activities against small chromogenic peptides, casein, and MBP-SulA, ruling out an essential role of this segment in complex formation.
In the Δ175–209 mutant, we deleted a segment that is disordered in all three HslU crystal structures (10). This deletion mutant exhibits the same behavior as the wild-type protein toward Z-Gly-Gly-Leu-AMC and casein. However, unlike in the case of the wild-type enzyme, no degradation of the MBP-SulA fusion protein was observed.
The Δ111–239 mutant, although lacking the entire I domain, was still able to stimulate peptide and casein hydrolysis. However, this mutant did not degrade the MBP-SulA fusion protein.
Mutations of Putative Contact Regions in HslU in the EM-Docking Mode.
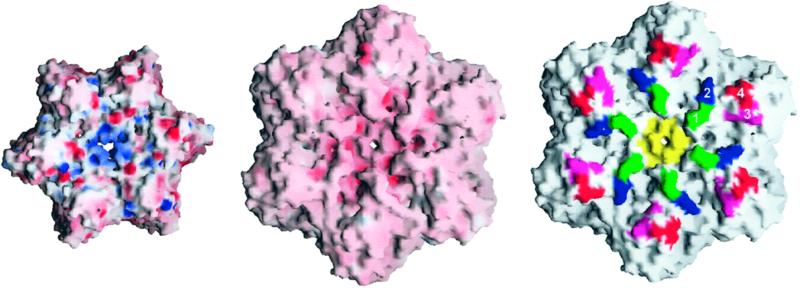
Next, we attempted to disrupt the surface of HslU that would be in contact with HslV in the EM mode of docking. In the absence of a high-resolution structure of the HslVU complex in the EM-mode of docking, we selected residues based primarily on the prominent positions on the surface of HslU and their contribution to the electrostatic potential (Fig. 1 Right). As charges on HslU and HslV appear to complement each other in this docking mode (Fig. 2 Left and Center), we first converted charged residues in HslU to neutral or oppositely charged residues (D436K/E437K, D436A/E437A, E266Q, and E266Q/E385K). The removal of negative charges near the carboxy terminus (D436K/E437K, D436A/E437A) leads a loss of activity against only MBP-SulA. The other mutants exhibit wild-type-like activities against the small peptide, casein, and MBP-SulA (Table 2).
Figure 2.
Representation of the electrostatic potential surfaces of HslV (Left) and HslU (Center) involved in the EM mode of docking. Negatively charged regions are in red, and positively charged regions are in blue. Sites of mutations in the HslU (Right). Numbers 1 (green) and 3 (pink) mark sites of pentaglycine insertions after residues 264 and 387 as well as changes of surface charges (E266Q; E266Q/E385K), 2 (blue) marks the site of introduction of a bulky side chain (I312W), and 4 (red) marks the site of a charge reversal (E436K/D437K). The hexamer pore is colored in yellow. This figure was drawn by using GRASP (28).
To generate more drastic effects, we introduced pentaglycine linkers at various positions on the surface of HslU (Table 2, and Fig. 2 Right). These insertion mutants are Ins(265,266; pentaglycine residues between 265 and 266), Ins(311,312; pentaglycine residues between 311 and 312), Ins(387,388; pentaglycine residues between 387 and 388), and Ins(436,437; pentaglycine residues between 436 and 437). Despite these profound steric alterations of the surface of HslU, the Ins(265,266) and Ins(387,388) mutants behaved like the wild-type protein toward peptide and casein but did not degrade the MBP-SulA fusion protein (Table 2). The Ins(311,312) mutant had much lower activities in all assays, including the ATPase assay, compared with wild-type enzyme. Thus, we mutated isoleucine 312 to a bulky residue, tryptophan. This mutant exhibited at least wild-type-like activities against all of the three substrates used in our assays (Table 2).
The carboxy terminus of HslU, which is conserved among the HslU family, is buried inside the C domain with its charge balanced by R394 and R329 of the adjacent subunit. It seems to be very important for the folding or oligomerization of HslU. When we added some residues [Ins(436,437)] in this region or deleted the C-terminal residues (Δ432–443), the mutants were expressed in insoluble forms (Table 2).
Mutations of Hexamer Pore of HslU.
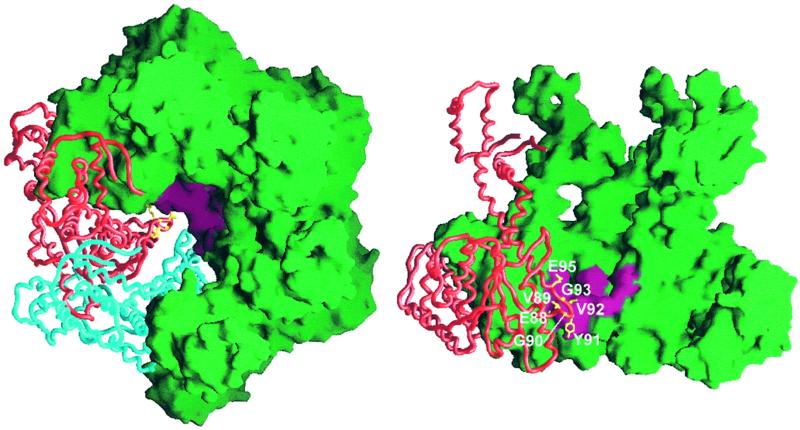
Despite similar architectural arrangements of HslVU and the core particle of the proteasome, there is a large functional difference. Although HslV is activated by HslU, the 20S particle consisting of the α and β rings is activated by the 17-subunit regulatory particle of the 26S proteasome. Structural features of the 20S proteasome have suggested (21, 22) and mutational studies have shown (23) that the unfolded substrate is threaded through the gate formed by the α subunits. We therefore mutated the hexameric pore formed by HslU to study its role.
The residues numbered 87–95, which form the hexamer pore of HslU (Fig. 3), are highly conserved in all known HslU sequences. This loop was ordered only in the trigonal complex but disordered in the other two HslU structures (10). Y91 is central in the pore (Fig. 3) and was mutated to glycine. The Y91G mutant exhibited almost similar peptidase, protease (assayed against Z-Gly-Gly-Leu-AMC and FITC-casein) and ATPase activities as the wild-type protein but did not degrade MBP-SulA. The V92G mutant was similar to Y91G. Two deletion mutants, Δ88–92 and Δ89–92, showed no activity and much reduced activity in all assays, respectively (Table 2). E88 seems critical, as its substitution with a neutral residue in the E88Q and E266Q/E88Q mutants leads to reduced peptidase activity. The activities against casein and SulA were much reduced, although the ATPase activity was similar to that of the wild-type enzyme.
Figure 3.
Sites of mutations in the hexamer pore. Side-chain atoms (yellow) are shown only in one subunit for clarity. Mutation sites in the hexamer pore are colored in pink. Top view of HslU (Left). Side view of the central pore of HslU hexamer (Right). Two subunits from the ring nearest to the reader are removed to expose the interior. This figure was drawn by using GRASP (28).
In contrast to these mutants, which were designed largely to widen the pore, we attempted to block the pore by introducing a bulky tryptophan side chain at position 95 (E95W) and in another instance (G93A) by reducing its flexibility. The E95W mutant exhibited wild-type-like peptidase, caseinolytic, and ATPase activities, but its activity against MBP-SulA was much lower (Table 2). The G93A mutant, on the other hand, exhibited much reduced activities in all assays including ATPase activity (Table 2).
Discussion
In the presence of the nonhydrolyzable ATP-γS, wild-type HslVU degrades the chromogenic peptide and casein, but not MBP-SulA (18). On the basis of these results, it has been suggested that only ATP binding but not its hydrolysis is a prerequisite for peptidase and caseinolytic activities, whereas ATP binding and hydrolysis are necessary for MBP-SulA degradation.
As expected, we find that the “arginine finger” R325 and the “sensor arginine” R393 are required for all HslVU activities. The crystal structure of the R325E mutant and preliminary circular dichroism spectroscopic data of the R393A mutant indicate that both proteins can still bind nucleotide. Failure of the R325E and R393A mutants to degrade peptide can be attributed to weaker intersubunit contacts. R325E fully and R393A partially dissociates in gel filtration experiments in the presence of salt.
In addition, we discovered an essential role for E321, a residue that is close to the nucleotide in our structure but is not conserved among AAA-family proteins (11). Lack of any hydrolytic activity in this mutant cannot be attributed to deficiencies in oligomerization. Further work is required to determine whether the protein retains affinity for nucleotide.
The involvement of the other residues is less clear. Mutations of K80 and E286 have differential effects on the different activities, probably reflecting a more indirect role of these residues in catalysis. We consistently find that mutations in the vicinity of the nucleotide-binding site of HslU tend to have a more drastic effect on peptide and casein hydrolysis than on MBP-SulA degradation (Table 2). This is in stark contrast with our mutagenesis results on the putative docking surfaces that affect mostly MBP-SulA degradation and have little effect on peptide and casein hydrolysis.
Deletion of the whole I domain abolished degradation of MBP-SulA, but caseinolytic and peptidase activities were unaffected. Deletion of the residues that mediate HslV–HslU interactions in the crystal structure had no effect on any of the HslVU activities. In contrast, deletion of the residues that are invisible in the crystal structure had the same effect as a deletion of the full I domain. These results suggest that the I domain and, in particular, residues 175–209, are involved in MBP-SulA recognition and/or unfolding. We had proposed earlier that the I domain might be involved in substrate recognition and binding, although the defined parts of the I domain of HslU show no detectable structural similarity with the substrate-binding domains of other chaperones (24). A structural comparison of the I domain with proteins in the DALI database (25) indicated a notable structural similarity (Z score >2.0) to a domain of arginyl–tRNA synthetase, high mobility group I protein, and RNA polymerase primary σ-factor fragment. The significance of this result is not clear because, unlike in those proteins, we could not detect any affinity between HslU and DNA in gel shift assays.
To test the relevance of the EM-docking mode for the degradation of the various HslVU substrates, we initially mutated charged residues on the surface of HslU. With the exception of the charge reversal at the carboxy terminus, all these mutants behaved like wild type. The same was true when we introduced a bulky residue. To our surprise, even the introduction of 30 glycine residues (5 per monomer) into the putative docking surface does not interfere with peptidase and caseinolytic activities.
The hexameric pore of HslU occurs in a dish-shaped depression of HslU (Fig. 3). It is unlikely that residues in the pore are involved in direct HslV–HslU interaction, also because the docking surface of HslV has a concave shape (Fig. 1). As a general trend, we find that mutations in the hexameric pore have little effect on the ATPase, peptidase, and caseinolytic activities. In contrast, they all have drastic effects on MBP-SulA degradation. We conclude that for this substrate, but not necessarily for casein, this region is involved in substrate recognition and/or translocation.
In summary, we conclude that peptidase and caseinolytic activities are surprisingly robust to changes in both putative docking surfaces of HslU. For these substrates, a precise complex between HslV and HslU may not be required, with HslU acting as a substrate concentrator and delivery system to HslV. In contrast, requirements for structural integrity of HslU are much more stringent for the degradation of MBP-SulA, most likely because of the extra unfolding step.
Our mutagenesis data suggest that different HslV-HslU complexes may be involved in the degradation of different substrates. We speculate that the EM mode of docking may be relevant for MBP-SulA degradation, whereas the x-ray mode of docking seems to play a more important role for peptidase and caseinolytic activities.
Acknowledgments
We thank M. Boicu for DNA sequencing and S. Körner for mass spectroscopic analysis, H. Bartunik and G. Bourenkov for help with data collection, P. Zwickl for discussions, and A. Higashitani (National Institute of Genetics, Japan) for a generous gift of the MBP-SulA fusion protein clone. H.K.S. and R.R are supported by postdoctoral fellowships from The Korea Science and Engineering and The Alexander von Humboldt Foundations, respectively. The financial support of the Deutsche Forschungsgemeinschaft and of the Human Frontier Science Program is also acknowledged.
Abbreviations
- N domain
N-terminal domain
- I domain
intermediate domain
- C domain
C-terminal domain
- AAA
ATPases associated with a variety of cellular activities
- AMP-PNP
5′-adenylylimidotriphosphate
- MBP-SulA
maltose-binding protein and SulA fusion protein
- EM
electron microscopy
Note Added in Proof.
While this paper was in press, small angle x-ray scattering data and a crystal structure of HslVU complex from Haemophilus influenzae were reported (29).
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1e94).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.250491797.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.250491797
References
- 1.Gottesman S, Maurizi M R, Wickner S. Cell. 1997;91:435–438. doi: 10.1016/s0092-8674(00)80428-6. [DOI] [PubMed] [Google Scholar]
- 2.Missiakas D, Schwager F, Betton J M, Georgopoulos C, Raina S. EMBO J. 1996;15:6899–6909. [PMC free article] [PubMed] [Google Scholar]
- 3.Rohrwild M, Coux O, Huang H-C, Moerschell R P, Yoo S J, Seol J H, Chung C H, Goldberg A L. Proc Natl Acad Sci USA. 1996;93:5808–5813. doi: 10.1073/pnas.93.12.5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoo S J, Seol J H, Shin D H, Rohrwild M, Kang M S, Tanaka K, Goldberg A L, Chung C H. J Biol Chem. 1996;271:14035–14040. doi: 10.1074/jbc.271.24.14035. [DOI] [PubMed] [Google Scholar]
- 5.Bochtler M, Ditzel L, Groll M, Hartmann C, Huber R. Annu Rev Biophys Biomol Struct. 1999;28:295–317. doi: 10.1146/annurev.biophys.28.1.295. [DOI] [PubMed] [Google Scholar]
- 6.Chuang S E, Burland V, Plunkett G, III, Daniels D L, Blattner F R. Gene. 1993;134:1–6. doi: 10.1016/0378-1119(93)90167-2. [DOI] [PubMed] [Google Scholar]
- 7.Bochtler M, Ditzel L, Groll M, Huber R. Proc Natl Acad Sci USA. 1997;94:6070–6074. doi: 10.1073/pnas.94.12.6070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schirmer E C, Glover J R, Singer M A, Lindquist S. Trends Biochem Sci. 1996;21:289–296. [PubMed] [Google Scholar]
- 9.Seong I S, Oh J Y, Lee J W, Tanaka K, Chung C H. FEBS Lett. 2000;477:224–229. doi: 10.1016/s0014-5793(00)01808-1. [DOI] [PubMed] [Google Scholar]
- 10.Bochtler M, Hartmann C, Song H K, Bourenkov G P, Bartunik H D, Huber R. Nature (London) 2000;403:800–805. doi: 10.1038/35001629. [DOI] [PubMed] [Google Scholar]
- 11.Neuwald A F, Aravind L, Spouge J L, Koonin E V. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- 12.Glickman M H, Rubin D M, Coux O, Wefes I, Pfeifer G, Cjeka Z, Baumeister W, Fried V A, Finley D. Cell. 1998;94:615–623. doi: 10.1016/s0092-8674(00)81603-7. [DOI] [PubMed] [Google Scholar]
- 13.Rohrwild M, Pfeifer G, Santarius U, Müller S A, Huang H-C, Engel A, Baumeister W, Goldberg A L. Nat Struct Biol. 1997;4:133–139. doi: 10.1038/nsb0297-133. [DOI] [PubMed] [Google Scholar]
- 14.Higashitani A, Ishii Y, Kato Y, Horiuchi K. Mol Gen Genet. 1997;254:351–357. doi: 10.1007/s004380050426. [DOI] [PubMed] [Google Scholar]
- 15.Lanzetta P A, Alvarez L J, Reinach P S, Candia O A. Anal Biochem. 1979;100:95–97. doi: 10.1016/0003-2697(79)90115-5. [DOI] [PubMed] [Google Scholar]
- 16.Matsuka Y V, Pillai S, Gubba S, Musser J M, Olmsted S B. Infect Immun. 1999;67:4326–4333. doi: 10.1128/iai.67.9.4326-4333.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Twining S S. Anal Biochem. 1984;143:30–34. doi: 10.1016/0003-2697(84)90553-0. [DOI] [PubMed] [Google Scholar]
- 18.Seong I S, Oh J Y, Yoo S J, Seol J H, Chung C H. FEBS Lett. 1999;456:211–214. doi: 10.1016/s0014-5793(99)00935-7. [DOI] [PubMed] [Google Scholar]
- 19.Brünger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 20.Karata K, Inagawa T, Wilkinson A J, Tatsuta T, Ogura T. J Biol Chem. 1999;274:26225–26232. doi: 10.1074/jbc.274.37.26225. [DOI] [PubMed] [Google Scholar]
- 21.Löwe J, Stock D, Jap B, Zwickl P, Baumeister W, Huber R. Science. 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- 22.Groll M, Ditzel L, Löwe J, Stock D, Bochtler M, Bartunik H D, Huber R. Nature (London) 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 23.Groll M, Bajorek M, Köhler A, Moroder L, Rubin D M, Huber R, Glickman M H, Finley D. Nat Struct Biol. 2000;7:1062–1067. doi: 10.1038/80992. [DOI] [PubMed] [Google Scholar]
- 24.Saibil H. Curr Opin Struct Biol. 2000;10:251–258. doi: 10.1016/s0959-440x(00)00074-9. [DOI] [PubMed] [Google Scholar]
- 25.Holm L, Sander C. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 26.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 27.Merritt E A, Bacon D J. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 28.Nicholls A, Bharadwaj R, Honig B. Biophys J. 1993;64:A166. [Google Scholar]
- 29.Sousa M C, Trame C B, Tsuruta H, Wilbanks S M, Reddy V S, McKay D B. Cell. 2000;103:633–643. doi: 10.1016/s0092-8674(00)00166-5. [DOI] [PubMed] [Google Scholar]