Abstract
Background information.
Maskin is a member of the acidic transforming coiled-coil (TACC) domain proteins found in Xenopus leavis oocytes and embryos. It is implicated in the coordination of the spindle and has been reported to mediate translational repression of cyclin B1 mRNA.
Results
We report here that maskin mRNA is translationally repressed at the level of initiation in stage 4 oocytes and becomes activated in stage 6 oocytes. The translational repression of maskin mRNA correlates with the presence of a short poly(A) tail on this mRNA in stage 4 oocytes. The 3' UTR of maskin can confer the translational regulation to a reporter mRNA, and so can the 3' UTR of human TACC3. A conserved GUCU repeat element was found to repress translation in both stage 4 and stage 6 oocytes, but deletion of this element did not abrogate repression in stage 4 oocytes. UV crosslinking experiments indicated that overlapping sets of proteins bind efficiently to both the maskin and the cyclin B1 3' UTRs. As previously reported, CPEB binds to the cyclin B1 3' UTR, but its binding to the maskin 3' UTR is minimal. By RNA affinity chromatography and mass spectrometry, we identified the embryonic deadenylation element binding protein (EDEN-BP) as one of the proteins binding to both the maskin and the cyclin B1 3' UTRs.
Conclusion
Maskin mRNA is translationally regulated by at least two repressor elements and an activation element. One of the repessor elements is the evolutionarily conserved GUCU repeat. EDEN-BP binds to both the maskin and cyclin B1 3' UTRs, indicating it may be involved in the deadenylation of these mRNAs.
Introduction
Translational control plays a major role in the regulation of oocyte development (Mendez and Richter, 2001;De Moor et al., 2005;Piccioni et al., 2005;Haccard and Jessus, 2006). Maskin is thought to repress translation of cyclin B1 and c-mos mRNA in Xenopus oocytes through its interaction with the RNA binding protein cytoplasmic polyadenylation element binding protein (CPEB) and the cap binding initiation factor eIF-4E (De Moor and Richter, 1999;Stebbins-Boaz et al., 1999;Cao and Richter, 2002;Pascreau et al., 2005). Activation of translation of these mRNAs is mediated by cytoplasmic polyadenylation, a process initiated by phosphorylation of CPEB which leads to dissociation of the deadenylase PARN and polyadenylation of the mRNA by the default activity of the cytoplasmic poly(A) polymerase Gld-2 (De Moor et al., 1999;Barnard et al., 2004;Kim and Richter, 2006). Phosphorylation of Maskin and the binding of poly(A) binding protein (PAB) to the elongated poly(A) tail itself have been implicated in this translational activation (Cao et al., 2002;Barnard et al., 2005;Pascreau et al., 2005). In addition to its role in the meiotic cell cycle, Maskin has been reported to play a role in the translational control of cyclin B1 in early embryonic cell division in Xenopus laevis (Groisman et al., 2000;Groisman et al., 2002).
Maskin is a member of the transforming acidic coiled-coil (TACC) domain family of proteins, which share a highly conserved N terminal domain of 200 amino acid residues. TACC proteins are localised at centrosomes and coordinate the formation of the mitotic spindle at least in part through their interaction with members of the XMAP/chTOG family, a group of microtubule plus end stabilising proteins (Gergely, 2002;Wiese and Zheng, 2006). Like other TACCs, Maskin is associated with centrosomes, both in embryos and tissue culture cells and has a role in the organisation of the mitotic spindle (Huang and Richter, 2004;Barnard et al., 2004;Kinoshita et al., 2005;O'Brien et al., 2005). In addition to its association with XMAP215, Maskin also binds other mitotic players such as Aurora A, importin β and NDEL1, and is found in an RNP complex with Rae1 (Blower et al., 2005;Pascreau et al., 2005;Albee et al., 2006;Mori et al., 2007).
Maskin is most closely related to mammalian TACC3, sharing N terminal homology in addition to the well-conserved C terminal coiled-coil domain (Stebbins-Boaz et al., 1999;Still et al., 2004). In addition, both maskin and TACC3 protein levels have been reported to be regulated during the mitotic cell cycle, albeit with different timing (Groisman et al., 2002;Gergely et al., 2003). However, the eIF-4E binding domain is absent from all characterised mammalian TACC3 proteins, indicating that there may be an evolutionary change in this function of the protein (De Moor et al., 2005). In addition to its function in spindle organisation in mitosis, TACC3 is localised to the nucleus in interphase, where it binds to histone acetyl transferases and has been implicated in the regulation of transcription of methylated promotors (Sadek et al., 2003;Gangisetty et al., 2004;Angrisano et al., 2006).
In this paper we show that maskin mRNA is translationally repressed in early oocytes and becomes translated in the fully grown oocyte. At the time of translational activation, the maskin mRNA gains a longer poly(A) tail. The maskin 3' UTR can transfer translation regulation to a reporter mRNA and the regulatory elements are conserved, as the 3' UTR of human TACC3 conveys the same translational control. We demonstrate that the GUCU repeat conserved in these 3' UTRs is a repressor sequence and that at least one additional translational repressor and an activator must be present in the maskin 3' UTR. Using RNA affinity chromatography and immunoprecipitation we demonstrate that the embryonic deadenylation element binding protein, EDEN-BP, associates with the maskin 3' UTR and therefore is possibly involved in the regulation of Maskin synthesis.
Results
Translational repression of Maskin in stage 4 oocytes
In order to examine the role of Maskin in the translational repression of cyclin B1 during oogenesis, we performed western and northern blots on oocytes sorted in different stages (Dumont, 1972). As can be seen in Fig. 1, cyclin B1 mRNA was maximally expressed from stage 2 onwards, while the protein was weakly expressed in stage 6 and maximally in mature oocytes, indicating that the cyclin B1 mRNA is translationally repressed in early oocytes. The maskin mRNA was similarly expressed from stage 2 onwards, and the protein was maximally expressed from stage 6 onwards (in other oocyte batches from stage 5, see Fig 2). This expression pattern indicates that maskin mRNA is also translationally repressed in early oocytes. In addition, this implies that cyclin B1 mRNA is translationally repressed in stage 2-4 oocytes independent of the presence of Maskin.
Figure 1.
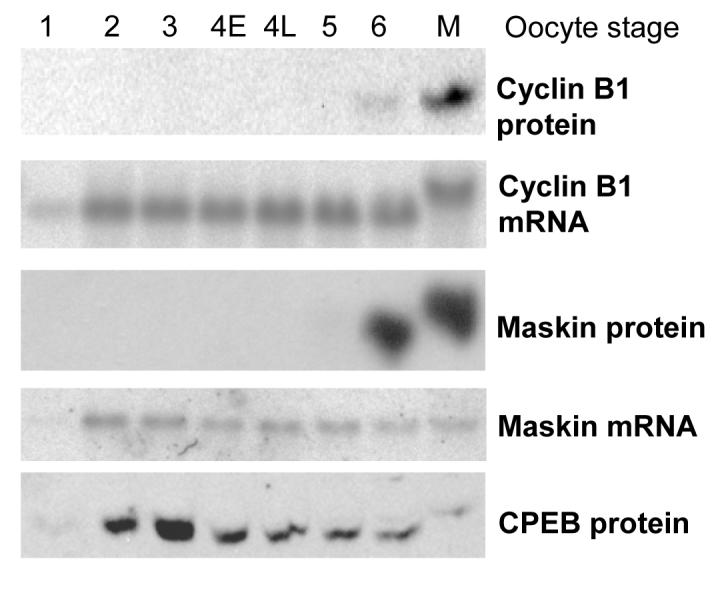
Maskin protein appears in late oogenesis, long after its mRNA
Western blots for cyclin B1, maskin and CPEB (protein panels) and Northern blots for cyclin B1 and maskin mRNA (mRNA panels). Equal numbers of oocyte equivalents from the same batch of oocytes were loaded in each lane. Oocyte stages according to Dumont. 4E: early stage 4 oocytes (0.6-0.8 mm diameter), 4L: late stage 4 oocytes (0.8-1 mm). M: mature stage 6 oocytes.
Figure 2.
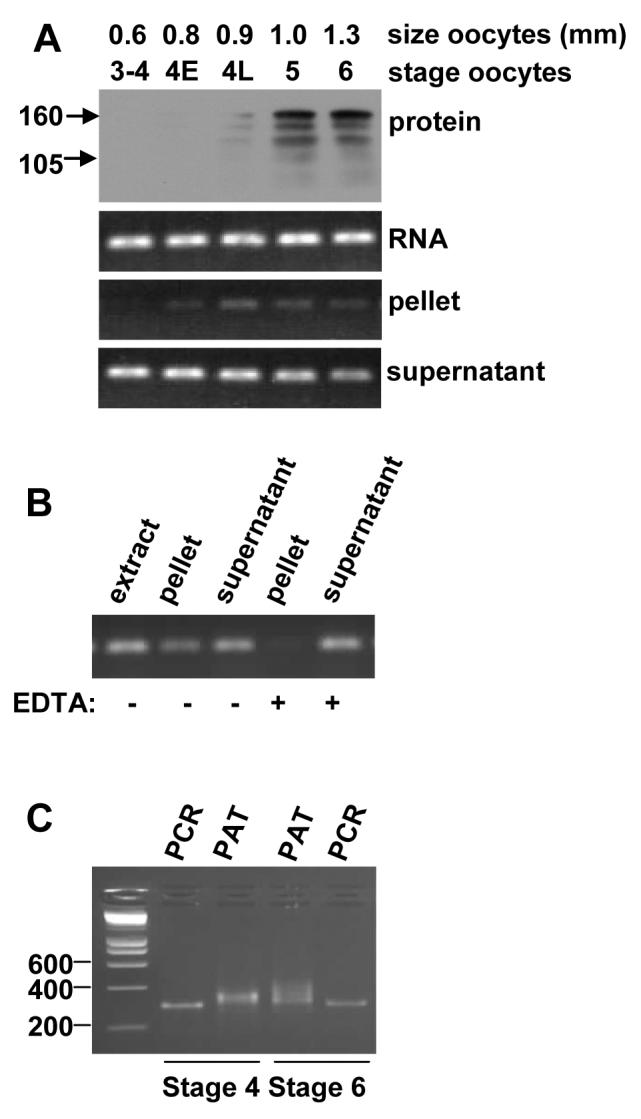
Maskin mRNA in early stage 4 oocytes is not bound to polyribosomes and has a short poly(A) tail.
(A) protein: Western for maskin on stage 4-6 oocytes. RNA: RT-PCR for maskin mRNA on total RNA from stage 4-6 oocytes. Pellet: RT-PCR on RNA from the polyribosomal pellet (translated mRNA). Supernatant: RT-PCR on mRNA from the free mRNP (untranslated mRNA). Marker sizes in KDa are indicated on the left of the panel. (B) Stage 6 oocyte extracts were treated with EDTA to release the ribosomes before pelleting (EDTA) or not, the RNA isolated from the fractions was amplified as in A. (C) Poly(A) test on stage 4 and stage 6 oocyte RNA. PCR: PCR using a fixed forward primer and a reverse primer at the end of the 3' UTR, thus amplifying the 3' UTR excluding the poly(A) tail. PAT: PCR using a fixed forward primer and the adaptor primer wich amplifies the same region including the poly(A) tail. Marker sizes are indicated in numbers of base pairs.
Since the maskin mRNA is present long before the protein can be detected, we suspected that this mRNA is also translationally repressed in small oocytes. However, because the protein is detected in full grown, but immature, oocytes, it appears that it is translationally activated before the cyclin B1 mRNA. To examine the translation state of the maskin mRNA during oocyte development, we separated polyribosomal and free mRNP by centrifugation over sucrose cushions and examined the localisation of maskin mRNA (Fig. 2A). The expression of Maskin protein was examined by western blot on extracts from the same batch of oocytes. As expected, the maskin mRNA appeared in the pellet (polyribosomes) just before the Maskin protein became detectable (in this batch in late stage 4). Dissociation of the ribosomes with EDTA resulted in the maskin mRNA being depleted from the polyribosomal fraction, confirming that a portion of the mRNA is in polyribosomes in stage 6 oocytes (Fig. 2B). This result shows that the maskin mRNA is translationally repressed at an initiation step in early oocytes and becomes translated between late stage 4 and stage 6.
The poly(A) tail length of the maskin mRNA is regulated
Because many mRNAs in the oocyte and early embryo are translationally regulated by cytoplasmic polyadenylation and/or deadenylation, we examined the length of the poly(A) tail of maskin mRNA in early stage 4 and stage 6 oocytes using a PCR based poly(A) test (PAT). As figure 2C shows, the poly(A) tail of the maskin mRNA was approximately 50 nucleotides in stage 4 oocytes and 100 nucleotides in stage 6 oocytes. This indicates that the maskin mRNA is potentially activated by cytoplasmic polyadenylation or repressed by deadenylation.
The 3' UTR of maskin mRNA contains translational control elements
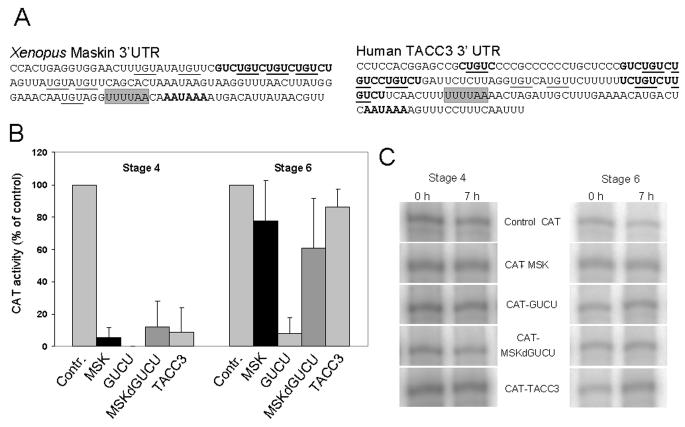
Translational control elements functioning in oocytes are commonly located in the 3' untranslated region (De Moor et al., 2005). To identify potential translational control sequences, we compared the 3' UTRs of the mRNAs for Maskin and its human homologue TACC3 (Fig 3A). No consensus CPEs were apparent, although both mRNAs contained short stretches of U's followed by A's that could represent weak CPEB binding sites. Strikingly, both mRNAs contained multiple repeats of GUCU. Such repeats can also be found in the mouse, bovine and porcine TACC3 mRNAs. This indicates that the maskin 3' UTR is likely to have an evolutionarily conserved function. To assess the function of the maskin and TACC3 3' UTRs we injected CAT reporter RNAs with these sequences into stage 4 and stage 6 oocytes. The maskin (MSK) and TACC3 3' UTRs strongly repressed the expression from the reporter mRNA in early stage 4 oocytes, and not in stage 6 oocytes (Fig 3B). This expression pattern is entirely consistent with the translation characteristics of the endogenous maskin mRNA demonstrated in Fig. 2A and indicates that the maskin 3' UTR contains translational control elements that have been conserved with human TACC3.
Figure 3.
The maskin 3' UTR controls translation by multiple elements
(A) The maskin and TACC3 3' UTRs are depicted with the GUCU repeats and the poly(A) signal (AAUAAA) in bold, the potential weak CPEB binding site shaded and the UGU trinucleotides underlined. (B) CAT activity in injected oocytes. CAT reporter mRNAs were injected into stage 4 and stage 6 oocytes and incubated for 7 hours. CAT enzyme activity is expressed as percentage of the activity obtained for the CAT control mRNA. Control: control CAT mRNA, MSK: CAT mRNA with the full maskin 3' UTR, GUCU: CAT mRNA with the GUCU repeat in the 3' UTR, MSKdGUCU: CAT mRNA with the maskin 3' UTR lacking the GUCU repeat, TACC3: CAT mRNA with the full TACC3 3' UTR. Error bars indicate 1 standard deviation. (C) mRNA levels at the start and end of the incubation period as determined by RNase protection.
To assess the function of the GUCU repeats, we constructed mRNAs with the repeat alone (GUCU) or the maskin 3' UTR lacking the repeat (MSKdGUCU). As can be seen in Fig 3B, both the GUCU repeat and the MSKdGUCU construct repressed translation in stage 4 oocytes. In stage 6, however, the MSKdGUCU construct behaved like the full length maskin 3' UTR, while the GUCU repeat on its own still repressed. These data indicate that the GUCU repeat is a conserved translational repressor element and that the maskin 3' UTR contains sequences that are necessary to overcome this repression in full grown oocytes. Because the MSKdGUCU construct represses translation in stage 4 oocytes, an additional translation repression element must also be present in the maskin 3' UTR.
To demonstrate that the maskin 3' UTR and its sequences are not destabilising the mRNAs to which they are attached, we performed RNase protection assays for the CAT mRNA. As can bee seen in figure 3C, no mRNA degradation was detected over the 7 hour time course of our experiments.
The maskin mRNA binds multiple proteins including EDEN-BP
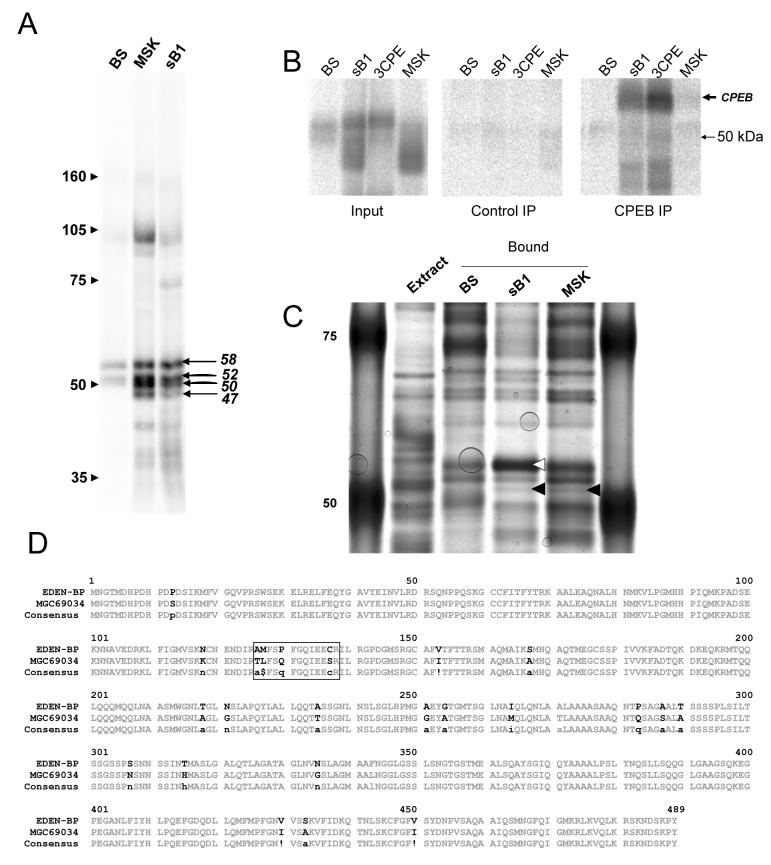
To investigate which proteins might be involved in the translational regulation by the maskin 3' UTR, we performed UV crosslinking experiments using the maskin (MSK) and cyclin B1 (sB1) 3' UTRs and a control vector transcript (BS). Figure 4A shows a representative experiment. We consistently found a similar set of proteins of 60 to 47 kDa crosslinking to both the cyclin B1 and maskin 3' UTRs. At least four bands of 58, 52, 50 and 47 kDa were present in this region. In addition to these highly reproducible bands, we also regularly observed weaker crosslinking to the maskin 3' UTR of proteins of 75, 42, 37 and 36 kDa.
Figure 4.
Multiple proteins including EDEN-BP bind to the maskin and cyclin B1 3' UTRs
(A) UV crosslinking assay with stage 4 extract on radioactive vector (BS), maskin 3' UTR (MSK) and cyclin B1 (sB1) 3' UTR transcripts. (B) UV crosslinking as above (UV) followed by immunoprecipitation with αCPEB antibody. (C) RNA affinity purification with RNAs as above, separated on SDS-PAGE and stained with silver stain. Triangles indicate CPEB (white) and EDEN-BP (black), as identified by mass spectrometry. The circles are bubbles trapped under the gel during photography. (D) Comparison of EDEN-BP and MGC69034. The peptides identified by mass spectrometry are boxed.
The 58 kDa protein could potentially be CPEB, since the cyclin B1 3' UTR is known to bind this 62 kDa protein. We therefore performed a crosslink-immunoprecipitation to investigate the binding of CPEB to the maskin 3' UTR. As can be seen in Fig 4B, CPEB was readily labelled by UV crosslinking to the cyclin B1 3' UTR, but only a very weak signal was detected for the maskin 3' UTR. These data indicate that CPEB may bind weakly to the maskin 3' UTR, but the 58 kDa band seen in the crosslinking assays is likely to be another protein.
To identify the proteins binding to the maskin 3' UTR, we developed an RNA affinity chromatography method in which a short biotinylated antisense RNA is annealed to the common 5' polylinker region of our 3' UTR transcripts. This hybrid RNA was then incubated with oocyte extract and recovered on streptavidin paramagnetic particles. After several washes, the bound protein was recovered using gel loading buffer and separated on an SDS PAGE gel. As can be seen in Fig 4C, CPEB was recovered specifically and efficiently when using the cyclin B1 3' UTR in this method. In addition, a specific band of approximately 52 kDa was observed binding to both the cyclin B1 and the maskin 3' UTR. Tandem mass spectrometry of this protein binding to the maskin 3' UTR yielded two related peptide sequences, AMFSPFGQIEECR and TLFSQFGQIEESR. This identified the protein as a mixture of EDEN-BP (AAC41243) and a closely related protein, the MGC69034 protein (AAH70706), as depicted in Fig 4D. The high homology between the two proteins indicates that MGC69034 is probably generated from the second EDEN-BP gene in the partially tetraploid Xenopus laevis.
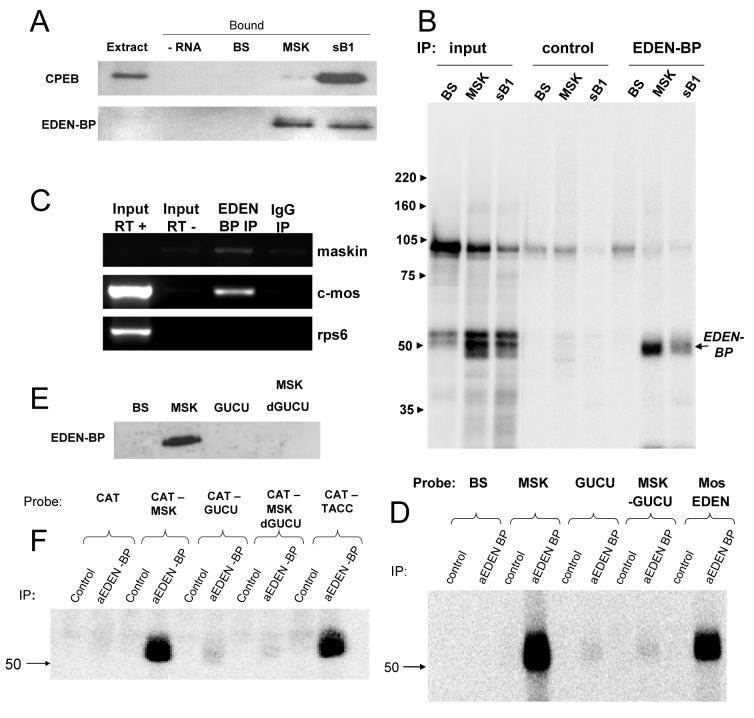
In order to confirm the interaction of EDEN-BP with the maskin and cyclin B1 3' UTRs, we performed western blots on RNA affinity purification eluates. As can be observed in Fig 5A, CPEB bound strongly to the cyclin B1 3' UTR and weakly to the maskin 3' UTR, confirming the results in Fig 4B. EDEN-BP was found to be greatly enriched on the beads holding cyclin B1 and maskin 3' UTRs, but not on the control RNA. In addition, immunoprecipitation of crosslinked maskin and cyclin B1 3' UTRs with an EDEN-BP antibody confirmed the specific interaction with both these RNAs (Fig 5B). To confirm that EDEN-BP is associated with endogenous maskin mRNA, we performed immunoprecipitation followed by RT-PCR (Fig. 5 C). As expected, the maskin and c-mos mRNAs, but not the ribosomal protein S6 mRNA (rpS6), were specifically precipitated with the EDEN-BP antibody.
Figure 5.
EDEN-BP binds to the full length maskin 3' UTR only
(A) Western for CPEB and EDEN-BP on RNA affinity purification eluates from beads bearing control (BS), maskin 3' UTR (MSK) and cyclin B1 3' UTR (sB1) RNAs. (B) UV crosslinking and immunoprecipitation with a control rabbit IgG and the EDEN-BP antibody using radioactive forms of the RNAs listed for (A). (C) RT-PCR for maskin, c-mos and ribosomal protein S6 (rsp6) on RNA isolated from immunoprecipitates obtained with control (IgG) and EDEN-BP antibody. RT+/RT−: reverse transcriptase added or omitted. (D) UV crosslinking followed by immunoprecipitation as in (B) using the control (BS), full maskin 3' UTR (MSK), the GUCU repeat alone (GUCU), the maskin 3' UTR lacking the GUCU repeat (MSKdGUCU) or the EDEN from the c-Mos 3' UTR (Mos EDEN). (E) Western for EDEN-BP on RNA affinity purification eluates from RNAs as described in (D). (F) UV crosslinking and IP with capped and polyadenylated CAT mRNAs as used in Fig. 3.
To determine what part of the maskin 3' UTR binds EDEN-BP, we performed crosslink-immunoprecipitations with the GUCU repeat and the maskin 3' UTR lacking the GUCU repeat (MSKdGUCU). Surprisingly, neither of these RNAs was crosslinked efficiently to EDEN-BP (Fig 5D). To ascertain that this result was not due to a peculiarity of the crosslinking assays, we performed RNA affinity chromatography with the maskin 3' UTR mutants. As can be seen in Fig 5E, neither the GUCU repeat nor the MSKdGUCU construct retained EDEN-BP on the columns. An alternative explanation for the lack of EDEN-BP binding to the GUCU repeat or MSKdGUCU is that the short transcripts used for the crosslinking are not folding correctly, while the same sequences included in the CAT mRNAs used in Fig.3 could fold correctly and bind EDEN-BP. We therefore did crosslink-immunoprecipitation experiments with the full capped and polyadenylated CAT mRNAs. As can be seen in Fig. 5F, the result was identical to that obtained with the shorter RNAs. In addition, we found that the TACC3 3' UTR also binds EDEN-BP, indicating that the binding site for EDEN-BP is conserved. However, the lack of overlap between the EDEN-BP binding sites and the translational control sequences indicates that the EDEN-BP binding sequence in the maskin 3' UTR is not equivalent to either of the repression elements or the activation element.
Conclusion
In this paper we demonstrate that maskin mRNA is translationally repressed by its 3' UTR in early stage 4 oocytes. The translational activation of the maskin mRNA coincides with elongation of its poly(A) tail. Mapping of the elements in the 3' UTR revealed that it contains two repression elements, one of which is the conserved GUCU repeat. As the GUCU repeat alone still represses translation in stage 6 oocytes, sequences elsewhere in the maskin 3' UTR must mediate translational activation. UV crosslinking experiments indicate that protein complexes of similar compositions bind the cyclin B1 and maskin 3' UTRs and we have demonstrated that one of the binding proteins they have in common is EDEN-BP.
The maskin 3' UTR regulates translation
Our studies show that the maskin 3' UTR represses translation in stage 4 oocytes, but not in stage 6. The localisation of the maskin mRNA in the subpolysomal fraction in stage 4 oocytes indicates that the translational repression is at the level of initiation. Because the human TACC3 3' UTR mediates the same regulation in Xenopus oocytes, it appears that these regulatory sequences have been preserved in the human mRNA and are recognised by frog proteins. This makes the TACC3 mRNA a likely candidate for translational regulation as well. We have demonstrated that the poly(A) tail of maskin mRNA is longer in stage 6 than in stage 4 oocytes, this indicates that the mRNA may be regulated by cytoplasmic polyadenylation or deadenylation. However, the fact that repression occurs on reporter mRNAs with short (30A) poly(A) tails, typical of the poly(A) tail length found in repressed maternal mRNAs, implies that deadenylation is unlikely to be the cause of the repression mediated by the maskin 3' UTR.
The maskin 3' UTR contains multiple conserved regulatory elements
Our mapping of the translational control elements in the maskin 3' UTR indicates that it contains multiple elements, at least two repression elements and an activation element. In view of the poly(A) tail length increase of the maskin mRNA in stage 6 oocytes, the activation element could very well be a cytoplasmic polyadenylation element. It seems unlikely that the weak binding of CPEB is involved in this process, as CPEB mediated polyadenylation is inactive in immature stage 6 oocytes. The GUCU repeat is clearly one of the conserved translational repression elements. Because both the TACC3 and the maskin 3' UTR mediate translational activation in stage 6 oocytes, the unidentified activation element is also likely to be amongst the conserved features. In addition to the functional elements characterised by functional mapping, an EDEN-BP binding site is also conserved between the maskin and TACC3 3' UTRs. TACC3 protein is present in full grown mammalian oocytes and the mRNA is expressed in all oocyte stages, but unfortunately the protein expression in growing oocytes has not been examined (Hao et al., 2002). In addition, TACC3 mRNA and the EDEN-BP homologue CUG-BP1 are co-expressed in a large number of tissues and cell types, indicating that regulation of TACC3 by CUG-BP1 may occur elsewhere (Sadek et al., 2003). These data indicate that TACC3 and Maskin are subject to a highly conserved and complex post-transcriptional control.
The maskin 3' UTR binds multiple RNA binding proteins, including EDEN-BP
Our crosslinking experiments indicate that the maskin and cyclin B1 mRNAs bind a similar set of proteins, including prominent bands of 58, 52, 50 and 47 kDa. By immunoprecipitation, CPEB was found to crosslink only weakly to the maskin 3' UTR and thus probably is not the 58 kDa protein detected in extracts. Nevertheless, we confirmed the weak binding of CPEB to the maskin 3' UTR using RNA affinity chromatography and western blotting. By affinity chromatography and mass spectrometry as well as by UV crosslinking, we identified EDEN-BP as another protein binding to both the cyclin B1 and the maskin 3' UTR.
EDEN-BP is a 52 kDa protein that mediates embryonic deadenylation of mRNAs bearing its binding sequence, the embryonic deadenylation element or EDEN (Paillard et al., 1998). EDEN-BP is necessary for normal somitic segmentation in the Xenopus embryo (Gautier-Court et al., 2004). The mammalian homologue of EDEN-BP, CUG-BP1, can substitute for EDEN-BP in deadenylation assays and is thought to be involved in a large variety of physiological processes, including the causes of muscular dystophy (Barreau et al., 2006;de Haro et al., 2006). EDEN-BP contains three RNA recognition motifs (RRMs). Binding of EDEN-BP to classical EDEN sequences only requires the two N terminal RRMs and some adjacent sequence that mediates dimerisation or oligomerisation (Bonnet-Corven et al., 2002;Cosson et al., 2006;Barreau et al., 2006). Recently, it has been shown that CUG-BP1 co-immunoprecipitates with poly(A) ribonuclease (PARN) (Moraes et al., 2006). A similar complex containing EDEN-BP and PARN in the embryo could explain its role in deadenylation.
The EDEN sequence has been described as rich in uridine-purine dinucleotide repeats. A synthetic sequence of six contiguous UGUA motifs can confer deadenylation (Audic et al., 1998;Paillard and Osborne, 2003;Osborne et al., 2005). Recently, a SELEX screen for sequences binding to CUG-BP1 defined high affinity binding sites as regions of approximately 36 nucleotides with a high UG dinucleotide content and at least four UGU trinucleotides (Marquis et al., 2006). These elements mediated efficient deadenylation in Xenopus embryos, indicating they are genuine EDEN sequences. Using this definition, the maskin 3' UTR contains an EDEN that is comprised of 41 nucleotides that include the GUCU repeat and its flanking sequence (7 UGU) and the cyclin B1 3' UTR has an EDEN of 45 nucleotides upstream of and overlapping the first CPE (6 UGU). The maskin 3' UTR lacking the GUCU repeat still contains 4 UGU sequences, but the GC richness of the inserted sequence may be disturbing the EDEN-BP binding. Similarly, the GUCU repeat construct contains 4 UGU sequences, but these are grouped very closely together and therefore may not all be functional in EDEN-BP binding.
Because the binding of EDEN-BP to the maskin 3' UTR constructs does not correlate with any of the observed regulatory functions of this sequence, the significance of this interaction is not clear. The fact that both cyclin B1 and maskin mRNA have short poly(A) tails in their untranslated state and bind EDEN-BP in their 3' UTRs suggests that EDEN-BP could be responsible for deadenylation after these mRNAs are exported from the nucleus. However, EDEN sequences do not mediate deadenylation in stage 6 oocytes or unfertilised eggs (Legagneux et al., 1995), consequently such shortening of the poly(A) tails would be likely to happen in an earlier stage of oogenesis. Recently, deadenylation of the cyclin B1 3' UTR in stage 6 oocytes was reported to be mediated by the association of CPEB with a complex containing poly(A) ribonuclease (PARN) (Kim et al., 2006). In contrast, the maskin 3' UTR does not bind CPEB efficiently, therefore its deadenylation is unlikely to be mediated by this protein. This makes EDEN-BP the only currently available candidate to mediate deadenylation of the maskin mRNA in small oocytes.
Maskin protein expression is regulated in the cell cycle in early embryos. It appears likely that this regulation is a combination of translational control and protein degradation, as it is for cyclin B1 (Groisman et al., 2000;Groisman et al., 2002). It is therefore possible that the association of EDEN-BP with the maskin 3' UTR has little or no function in the oocyte, but exerts its effect during embryogenesis in the translational control of maskin mRNA during cell division, when EDEN mediated deadenylation is known to be active.
The three translational control elements we defined in the maskin 3' UTR by deletion analysis of reporter constructs do seem not to overlap with the binding site of EDEN-BP in this RNA. This indicates that the EDEN-BP binding site is likely to represent a fourth element. Combined with the detection by UV crosslinking of 4 to 7 specific proteins, this relatively short 3' UTR appears to be packed with regulatory complexes. It is as yet unclear if these elements are adjacent or overlapping, whether all complexes bind simultaneously and which complexes actually form on the endogenous mRNA. It is therefore crucial that the composition of maskin and cyclin B1 mRNP complexes is studied in further detail.
Methods
Plasmid construction
The pT3CAT vector for in vitro transcription of CAT mRNAs have been described previously (De Moor et al., 1999). BamH1 and EcoR1 sites were appended to the maskin 3' UTR by PCR on a cloned maskin cDNA (Genbank AF200212) with the primers MSK-3F5 (CGCGGATCCACTGAGGTGGAACTTTG) and MSK-3R3 (CCGGAATTCAACGTTATAATGTCATTTTATTTG). This fragment was inserted in a Bluescript II SK-vector, generating pT3-MSK. This was cut with BamH1 and the CAT containing BamH1 fragment from pT3CAT was inserted to create pT3CAT-MSK. The pT3CAT-TACC3 plasmid was created in a similar way using IMAGE clone AW5149489 with primers HTACC3-1S (CGGATCCTCCACGGAGCCGCTG) and HTACC3-1A (GCGAATTCAAATTGAAAGGAAACTTTTATTGAG) for the PCR. For the creation of CMdGUCU a modified pGEM vector was digested with Smal and BstZI. The annealed oligos MSK-3F2 (GGCCGCCACTGAGGTGGAACTTTGTATATGTTCCC) and MSK-3R2 (GGGAACATATACAAAGTTCCACCTCAGTGGC), representing the 5' end of the maskin 3' UTR, were inserted into these sites. The 3' end of the maskin 3' UTR was amplified by PCR using T3CAT-MSK as template and oligos MSK-3F3 (AACTGCAGTTATGTATGTTCAGCAC) and M13R (GGAAACAGCTATGACCATG). The resulting PCR product was digested with Pst1 and Xbal and ligated into the plasmid with the 5' end of the maskin 3' UTR digested with the same enzymes. The resulting plasmid was a used as a template in a PCR with oligos MSK-3F5 and MSK-3R3. The product was ligated into pBluescript IISK- cut with BamH1 and EcoR1. The resulting plasmid was then digested with BamH1 to insert the CAT coding region as described for pT3CAT-MSK. To make the pT3CAT-GUCU construct the oligos AGCTGTCTGTCTGTCTGTCTGCCCGGGCTGCA and GCCCGGGCAGACAGACAGACAGACAGCT were annealed and cloned into the Sma1 and Pst1 sites of pT3CAT.
To make the template for the biotinylated antisense RNA used in the RNA affinity chromatography, we made pROD by digesting Bluescript SK- with Xba1, filling in the site with Klenow DNA polymerase, re-digesting with EcoR5 and religating the plasmid. The lineup of the EDEN sequences was performed with MultiAlin (Corpet et al., 1998).
RNA synthesis
Templates for in vitro transcription of mRNAs with short (30A) poly (A) tails were generated by PCR as described previously (De Moor et al., 1999). For the synthesis of capped probes and competitor RNAs, the previously described vectors pT3-sB1 (pscyclin B1), pT3-GM-CSF, pT3-CPE1, pT3-3CPE1 as well as pT3-MSK (see above) and pT3-GUCU (GUCU oligo described above in pBluescript II SK-) were linearised with EcoR5. pBluescript II SK – was used for control transcripts. c-Mos EDEN RNA was transcribed from pGbORF-mosEDEN linearised with BamH1 (Paillard et al., 1998). RNA for injection and competitions was transcribed with the appropriate Ambion message machine kit , purified over a spun column of Sephadex G50M and concentrated by ethanol precipitation.
Oocyte preparation and injection, CAT assays
Young adult female Xenopus laevis were primed with 50 IU of pregnant mare serum gonadotropin. 2-10 days later, the ovaries were collected after humane killing. Ovaries and oocytes were kept in NOM (50% Leibovitz L-15 medium with L-glutamine, 15 mM HEPES pH 7.5, 50 U/ml of each streptomycin, penicillin and nystatin, gentamycin 0.1 mg/ml) (Kloc and Etkin, 1999). Ovaries were treated with collagenase and dispase in NOM (Kuge and Richter, 1995) and sorted under a microscope according to Dumont (Dumont, 1972) or by sieving (extracts for affinity chromatography). For stage 4 oocyte injections, early stage 4 oocytes of 0.6-0.7 mm were used. For injections of reporter mRNAs 0.1 to 0.6 fmole of capped RNA with a trace label of α-32P-UTP was injected, and the oocytes incubated for 7 hours at room temperature. The oocytes were then collected in batches of 8-10, the medium was removed and the sample frozen on dry ice and stored at −80°C. CAT essays were performed as described previously, except that the incubation time was 2 hours (De Moor et al., 1999).
Polysome pellets
Polysomal and free messenger ribonucleoproteins (mRNPs) were separated by layering the extract of 20 oocytes on a sucrose cushion and subjecting it to ultracentrifugation as described (Wormington, 1991). RNA was isolated from total extract and the supernatant and pellet using guanidinium thiocyanate (Chomczynski and Sacchi, 1987). Levels of maskin mRNA in the different fractions was analysed by RT-PCR using an oligo-dT primer for the reverse transcription and oligos MS6 (5'-CGTAAGAATGAGGAGGCACTG-3') and MA1 (5'-CTTCCATTCCAGTACTTCTAAATCC-3') for the PCR.
PAT assay
The PAT assay was peformed as described by Sallés et al. (1999). The RT reaction was performed using 200 ng total RNA from stage 4 or stage 6 oocytes and 20 pmol PATdTanchor primer (5'-GCGAGCTCCGCGGCCGCG-T12-3'). For control reactions the gene specific primer PAT-MSK-R1 (5'-GCGAGCTCCGCGGCCGCGAACGTTATAATGTCATTTTATTTG-3") was used. For the PCR reaction the primers PATanchor (5'-GCGAGCTCCGCGGCCGCG-3') and PAT-MSK-F1 (5'-GCGACAGTCTGCAGGCAACGC-3') were used.
RNase protection assays
Probes for RNase Protection Assays (RPA) on CAT mRNAs were synthesised using pBSK-CAT plasmid linearised with NcoI using T7RNA polymerase (Promega). RNase Protection Assays were performed according to the protocol provided with the Direct Protect ™ Lysate RPA kit (Ambion) with minor alterations. Hybridisation was overnight at 45°C. RNase digestion was for 1 hour at 37°C. Precipitation of the RNAs was aided by the addition of 10 μg of yeast tRNA (Ambion). Protected fragments were separated at 6% acrylamide/urea TBE gel and detected by exposure to a phosphor imaging screen.
UV crosslinking assays and gel retardation
UV crosslinking and gel retardation reactions were in 100 mM KC1, 2mM MgC12, 5 mM dithiothreitol, 0.2 mg/ml heparin, 20 mM HEPES, pH 7. 0.5-2 ×106 cpm of radiolabelled RNA was heat-denatured and incubated with 1-2 oocyte equivalents in 10 μl at room temperature for 10 minutes. For UV crosslinking, the samples were placed in a 96 well plate on ice and exposed to UV for 15 minutes, followed by RNase A digestion and SDS-PAGE electrophoresis as described previously (De Moor et al., 1999). For gel retardation sucrose was added to 8% and the reactions were loaded on a 4% acrylamide gel in 0.5x TBE. For CPEB immunoprecipitation, the optimal crosslinking time for CPEB, 5 minutes, and a heparin concentration of 0.5 mg/ml was used. For both UV crosslinking and gel retardation the gels were dried and exposed to a phosphor imaging screen.
RNA affinity chromatography
For RNA affinity chromatography, a nucleotide mix containing 16 biotin UTP and all four standard ribonucleotides was added to a MegaShortScript T7 (Ambion) mixture to a concentration of 200 nM of each nucleotide. This mix was used to transcribe pROD digested with restriction enzyme Pvu2. 20 μg of this antisense RNA was hybridized to an equal amount of mMessage mMachine (Ambion) RNA transcripts from various templates (pT3-sB1 and pT3-MSK digested with EcoR5 and Bluescript digested with BssH2) in RNase protection hybridization buffer (80% formamide, 400 mM NaC1, 2 mM EDTA, 80mM PIPES, pH6.8) at 50°C for 14 hours or more. This mixture was diluted with 10 volumes of RNA binding buffer (100 mM KC1, 2mM MgC12, 2 mM dithiothreitol, 20 mM HEPES, pH 7.5, 0.02 mg/ml heparin, 0.02 mg/ml BSA, 1% Tween20) and bound to 0.1 ml of washed streptavidin paramagnetic bead suspension (Promega) and washed extensively with RNA binding buffer. 1000 oocytes (stage 2-4), were homogenized in 5 ml of RNA binding buffer with protease inhibitors, and centrifugated for 10 minutes at 10,000 g. The supernatant was distributed among the three sets of beads and incubated at room temperature. The beads were washed extensively with RNA binding buffer and eluted with 7.7 M urea, 2.2 M thiourea, 2% CHAPS, 50 mM DTT. For mass spectrometry, the gels were stained with silver stain, the bands cut out, digested with trypsin and treated with iodoacetamide. Mass spectrometry was performed on a Micromass Q-Tof at the Biopolymer Synthesis and Analysis Unit, University of Nottingham.
Antibodies, western blotting and immunoprecipitation
Western blotting for maskin, cyclin B1, EDEN-BP and CPEB were performed as described (Hake and Richter, 1994;Paillard et al., 1998;De Moor et al., 1999;Stebbins-Boaz et al., 1999). Immunoprecipitation of proteins labelled by UV crosslinking was performed with protein A agarose in RIPA buffer (1xPBS, 0.5% Igepal, 0.5% sodium deoxycholate, 0.1% SDS). The immunoprecipitation-RT-PCR was performed as described by Kim and Richter (2006). For each sample 100 stage 4 oocytes were used. RTs were performed using specific primers (maskin: 5′-GCCAGCTCTAGAAACGTTATAATGTCATTTTATTTG-3′, c-mos: 5′-CCATTCACACTTCTGATTG-3′, rps6: 5′-TGACTGGATTCAGATTTG-3′). PCRs were performed using the following primers: maskin 5′-ATTGGAGGATTGTACCCAAGC-3′ and 5′-CTTCCATTCCAGTACTTCTAAATCC-3′, c-mos 5′-AGGGAGCATCACTCCTTGTC-3′ and 5′-CGGAACATAGGATAGTGCAC-3′, rps6 5′-GTTGTTCGCATCAGCGGTG-3′ and 5′-CAATACGTCTGCGCTTATGC-3′. All PCR products were analysed by agarose gel electrophoresis.
Acknowledgements
We thank Joel Richter (University of Massachusetts, Worcester, USA) for the gift of CPEB and maskin polyclonal antibodies, Luc Paillard (Rennes, France) for the EDEN-BP antibody and the pGbORF-mosEDEN plasmid, Andrew Counsell for some of the early optimisations of the crosslinking experiments and Kevin Bailey of the Biopolymer Synthesis and Analysis Unit, University of Nottingham, for mass spectrometry. This work was supported by grants from the Wellcome Trust and the BBSRC.
References
- Albee AJ, Tao W, Wiese C. Phosphorylation of maskin by Aurora-A is regulated by RanGTP and importin beta. J. Biol. Chem. 2006;281:38293–38301. doi: 10.1074/jbc.M607203200. [DOI] [PubMed] [Google Scholar]
- Angrisano T, Lembo F, Pero R, Natale F, Fusco A, Avvedimento VE, Bruni CB, Chiariotti L. TACC3 mediates the association of MBD2 with histone acetyltransferases and relieves transcriptional repression of methylated promoters. Nucleic Acids Res. 2006;34:364–372. doi: 10.1093/nar/gkj400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audic Y, Omilli F, Osborne HB. Embryo deadenylation element-dependent deadenylation is enhanced by a cis element containing AUU repeats. Mol. Cell Biol. 1998;18:6879–6884. doi: 10.1128/mcb.18.12.6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard DC, Cao Q, Richter JD. Differential phosphorylation controls Maskin association with eukaryotic translation initiation factor 4E and localization on the mitotic apparatus. Mol. Cell Biol. 2005;25:7605–7615. doi: 10.1128/MCB.25.17.7605-7615.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard DC, Ryan K, Manley JL, Richter JD. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell. 2004;119:641–651. doi: 10.1016/j.cell.2004.10.029. [DOI] [PubMed] [Google Scholar]
- Barreau C, Paillard L, Mereau A, Osborne HB. Mammalian CELF/Bruno-like RNA-binding proteins: molecular characteristics and biological functions. Biochimie. 2006;88:515–525. doi: 10.1016/j.biochi.2005.10.011. [DOI] [PubMed] [Google Scholar]
- Blower MD, Nachury M, Heald R, Weis K. A Rae1-containing ribonucleoprotein complex is required for mitotic spindle assembly. Cell. 2005;121:223–234. doi: 10.1016/j.cell.2005.02.016. [DOI] [PubMed] [Google Scholar]
- Bonnet-Corven S, Audic Y, Omilli F, Osborne HB. An analysis of the sequence requirements of EDEN-BP for specific RNA binding. Nucleic Acids Res. 2002;30:4667–4674. doi: 10.1093/nar/gkf586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Q, Richter JD. Dissolution of the maskin-eIF4E complex by cytoplasmic polyadenylation and poly(A)-binding protein controls cyclin B1 mRNA translation and oocyte maturation. EMBO J. 2002;21:3852–3862. doi: 10.1093/emboj/cdf353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Corpet F, Gouzy J, Kahn D. The ProDom database of protein domain families. Nucleic Acids Res. 1998;26:323–326. doi: 10.1093/nar/26.1.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosson B, Gautier-Court, Maniey D, it-Ahmed O, Lesimple M, Osborne HB, Paillard L. Oligomerization of EDEN-BP is required for specific mRNA deadenylation and binding. Biol. Cell. 2006;98:653–665. doi: 10.1042/BC20060054. [DOI] [PubMed] [Google Scholar]
- de Haro M, Al-Ramahi I, De GB, Ukani L, Rosa A, Faustino NA, Ashizawa T, Cooper TA, Botas J. MBNL1 and CUGBP1 modify expanded CUG-induced toxicity in a Drosophila model of myotonic dystrophy type 1. Hum. Mol. Genet. 2006;15:2138–2145. doi: 10.1093/hmg/ddl137. [DOI] [PubMed] [Google Scholar]
- De Moor CH, Meijer HA, Lissenden S. Mechanisms of translational control by the 3′ UTR in development and differentiation. Semin. Cell Dev. Biol. 2005;16:49–58. doi: 10.1016/j.semcdb.2004.11.007. [DOI] [PubMed] [Google Scholar]
- De Moor CH, Richter JD. Cytoplasmic polyadenylation elements mediate masking and unmasking of cyclin B1 mRNA. EMBO J. 1999;18:2294–2303. doi: 10.1093/emboj/18.8.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont JN. Oogenesis in Xenopus laevis (Daudin). I. Stages of oocyte development in laboratory maintained animals. J. Morphol. 1972;136:153–180. doi: 10.1002/jmor.1051360203. [DOI] [PubMed] [Google Scholar]
- Gangisetty O, Lauffart B, Sondarva GV, Chelsea DM, Still IH. The transforming acidic coiled coil proteins interact with nuclear histone acetyltransferases. Oncogene. 2004;23:2559–2563. doi: 10.1038/sj.onc.1207424. [DOI] [PubMed] [Google Scholar]
- Gautier-Court, Le CC, Barreau C, Audic Y, Graindorge A, Maniey D, Osborne HB, Paillard L. EDEN-BP-dependent post-transcriptional regulation of gene expression in Xenopus somitic segmentation. Development. 2004;131:6107–6117. doi: 10.1242/dev.01528. [DOI] [PubMed] [Google Scholar]
- Gergely F. Centrosomal TACCtics. BioEssays. 2002;24:915–925. doi: 10.1002/bies.10162. [DOI] [PubMed] [Google Scholar]
- Gergely F, Draviam VM, Raff JW. The ch-TOG/XMAP215 protein is essential for spindle pole organization in human somatic cells. Genes Dev. 2003;17:336–341. doi: 10.1101/gad.245603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman I, Huang YS, Mendez R, Cao Q, Theurkauf W, Richter JD. CPEB, maskin, and cyclin B1 mRNA at the mitotic apparatus: implications for local translational control of cell division. Cell. 2000;103:435–447. doi: 10.1016/s0092-8674(00)00135-5. [DOI] [PubMed] [Google Scholar]
- Groisman I, Jung MY, Sarkissian M, Cao Q, Richter JD. Translational control of the embryonic cell cycle. Cell. 2002;109:473–483. doi: 10.1016/s0092-8674(02)00733-x. [DOI] [PubMed] [Google Scholar]
- Haccard O, Jessus C. Oocyte maturation, Mos and cyclins--a matter of synthesis: two functionally redundant ways to induce meiotic maturation. Cell Cycle. 2006;5:1152–1159. doi: 10.4161/cc.5.11.2800. [DOI] [PubMed] [Google Scholar]
- Hake LE, Richter JD. CPEB is a specificity factor that mediates cytoplasmic polyadenylation during Xenopus oocyte maturation. Cell. 1994;79:617–627. doi: 10.1016/0092-8674(94)90547-9. [DOI] [PubMed] [Google Scholar]
- Hao Z, Stoler MH, Sen B, Shore A, Westbrook A, Flickinger CJ, Herr JC, Coonrod SA. TACC3 expression and localization in the murine egg and ovary. Mol. Reprod. Dev. 2002;63:291–299. doi: 10.1002/mrd.90012. [DOI] [PubMed] [Google Scholar]
- Huang YS, Richter JD. Regulation of local mRNA translation. Curr. Opin. Cell Biol. 2004;16:308–313. doi: 10.1016/j.ceb.2004.03.002. [DOI] [PubMed] [Google Scholar]
- Kim JH, Richter JD. Opposing polymerase-deadenylase activities regulate cytoplasmic polyadenylation. Mol. Cell. 2006;24:173–183. doi: 10.1016/j.molcel.2006.08.016. [DOI] [PubMed] [Google Scholar]
- Kinoshita K, Noetzel TL, Pelletier L, Mechtler K, Drechsel DN, Schwager A, Lee M, Raff JW, Hyman AA. Aurora A phosphorylation of TACC3/maskin is required for centrosome-dependent microtubule assembly in mitosis. J. Cell Biol. 2005;170:1047–1055. doi: 10.1083/jcb.200503023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloc M, Etkin LD. Analysis of localized RNAs in Xenopus oocytes. In: Richter JD, editor. A Comparative Methods Approach to the Study of Oocytes and Embryos. Oxford University Press; New York: 1999. pp. 256–278. [Google Scholar]
- Kuge H, Richter JD. Cytoplasmic 3′ poly(A) addition induces 5′ cap ribose methylation: Implications for translational control of maternal mRNA. EMBO J. 1995;14:6301–6310. doi: 10.1002/j.1460-2075.1995.tb00320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legagneux V, Omilli F, Osborne HB. Substrate-specific regulation of RNA deadenylation in Xenopus embryo and activated egg extracts. RNA. 1995;1:1001–1008. [PMC free article] [PubMed] [Google Scholar]
- Marquis J, Paillard L, Audic Y, Cosson B, Danos O, Le BC, Osborne HB. CUG-BP1/CELF1 requires UGU-rich sequences for high affinity binding. Biochem. J. 2006;400:291–301. doi: 10.1042/BJ20060490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez R, Richter JD. Translational control by CPEB: a means to the end. Nat. Rev. Mol. Cell Biol. 2001;2:521–529. doi: 10.1038/35080081. [DOI] [PubMed] [Google Scholar]
- Moraes KC, Wilusz CJ, Wilusz J. CUG-BP binds to RNA substrates and recruits PARN deadenylase. RNA. 2006;12:1810–1816. doi: 10.1261/rna.59606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori D, Yano Y, Toyo-Oka K, Yoshida N, Yamada M, Muramatsu M, Zhang D, Saya H, Toyoshima YY, Kinoshita K, Wynshaw-Boris A, Hirotsune S. NDEL1 Phosphorylation by Aurora-A Kinase is Essential for Centrosomal Maturation, Separation and TACC3 Recruitment. Mol. Cell Biol. 2007;27:352–367. doi: 10.1128/MCB.00878-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien LL, Albee AJ, Liu L, Tao W, Dobrzyn P, Lizarraga SB, Wiese C. The Xenopus TACC homologue, maskin, functions in mitotic spindle assembly. Mol. Biol. Cell. 2005;16:2836–2847. doi: 10.1091/mbc.E04-10-0926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne HB, Gautier-Court, Graindorge A, Barreau C, Audic Y, Thuret R, Pollet N, Paillard L. Post-transcriptional regulation in Xenopus embryos: role and targets of EDEN-BP. Biochem. Soc. Trans. 2005;33:1541–1543. doi: 10.1042/BST0331541. [DOI] [PubMed] [Google Scholar]
- Paillard L, Omilli F, Legagneux V, Bassez T, Maniey D, Osborne HB. EDEN and EDEN-BP, a cis element and an associated factor that mediate sequence-specific mRNA deadenylation in Xenopus embryos. EMBO J. 1998;17:278–287. doi: 10.1093/emboj/17.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paillard L, Osborne HB. East of EDEN was a poly(A) tail. Biol. Cell. 2003;95:211–219. doi: 10.1016/s0248-4900(03)00038-8. [DOI] [PubMed] [Google Scholar]
- Pascreau G, Delcros JG, Cremet JY, Prigent C, Arlot-Bonnemains Y. Phosphorylation of maskin by Aurora-A participates in the control of sequential protein synthesis during Xenopus laevis oocyte maturation. J. Biol. Chem. 2005;280:13415–13423. doi: 10.1074/jbc.M410584200. [DOI] [PubMed] [Google Scholar]
- Piccioni F, Zappavigna V, Verrotti AC. Translational regulation during oogenesis and early development: the cap-poly(A) tail relationship. C. R. Biol. 2005;328:863–881. doi: 10.1016/j.crvi.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Sadek CM, Pelto-Huikko M, Tujague M, Steffensen KR, Wennerholm M, Gustafsson JA. TACC3 expression is tightly regulated during early differentiation. Gene Expr. Patterns. 2003;3:203–211. doi: 10.1016/s1567-133x(02)00066-2. [DOI] [PubMed] [Google Scholar]
- Stebbins-Boaz B, Cao Q, De Moor CH, Mendez R, Richter JD. Maskin is a CPEB associated factor that transiently interacts with eIF-4E. Mol. Cell. 1999;4:1017–1027. doi: 10.1016/s1097-2765(00)80230-0. [DOI] [PubMed] [Google Scholar]
- Still IH, Vettaikkorumakankauv AK, DiMatteo A, Liang P. Structure-function evolution of the Transforming acidic coiled coil genes revealed by analysis of phylogenetically diverse organisms. BMC. Evol. Biol. 2004;4:16. doi: 10.1186/1471-2148-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese C, Zheng Y. Microtubule nucleation: {gamma}-tubulin and beyond. J. Cell Sci. 2006;119:4143–4153. doi: 10.1242/jcs.03226. [DOI] [PubMed] [Google Scholar]
- Wormington M. Preparation of synthetic mRNAs and analyses of translational efficiency in microinjected Xenopus oocytes. In: Kay BK, Peng HB, editors. Xenopus laevis: Practical Uses in Cell and Molecular Biology. Academic Press, Inc.; San Diego: 1991. pp. 167–183. [DOI] [PubMed] [Google Scholar]