Abstract
Heterozygous germline mutations in fumarate hydratase (FH) predispose to the multiple cutaneous and uterine leiomyomatosis syndrome (MCUL), which, when co-existing with renal cancer, is also known as hereditary leiomyomatosis and renal cell cancer. Twenty-seven distinct missense mutations represent 68% of FH mutations reported in MCUL. Here we show that FH missense mutations significantly occurred in fully conserved residues and in residues functioning in the FH A-site, B-site, or subunit-interacting region. Of 24 distinct missense mutations, 13 (54%) occurred in the substrate-binding A-site, 4 (17%) in the substrate-binding B-site, and 7 (29%) in the subunit-interacting region. Clustering of missense mutations suggested the presence of possible mutational hotspots. FH functional assay of lymphoblastoid cell lines from 23 individuals with heterozygous FH missense mutations showed that A-site mutants had significantly less residual activity than B-site mutants, supporting data from Escherichia coli that the A-site is the main catalytic site. Missense FH mutations predisposing to renal cancer had no unusual features, and identical mutations were found in families without renal cancer, suggesting a role for genetic or environmental factors in renal cancer development in MCUL. That all missense FH mutations associating with MCUL/hereditary leiomyomatosis and renal cell cancer showed diminished FH enzymatic activity suggests that the tumor suppressor role of fumarate hydratase may relate to its enzymatic function.
In the autosomal dominant syndrome of multiple cutaneous and uterine leiomyomatosis (MCUL, Reed syndrome, leiomyomatosis cutis et uteri, multiple leiomyomatosis; OMIM 150800), affected females develop uterine leiomyomas and affected individuals of both sexes develop cutaneous leiomyomas.1 Cutaneous leiomyomas are believed to be derived from the smooth muscle of the pilo-arrector apparatus. They generally present in the second, third, or fourth decades, typically as grouped papules or nodules on the trunk or limbs and are characteristically painful particularly in response to low temperatures or touch. Uterine leiomyomas or fibroids in MCUL are severely symptomatic with a large proportion of patients requiring symptom control by hysterectomy.2 A small proportion of families with MCUL also cluster renal cancer, either papillary renal type II cancer or renal collecting duct cancer.1,3,4,5,6 This disease variant has been referred to as hereditary leiomyomatosis and renal cancer (HLRCC, OMIM 605839). MCUL/HLRCC has been found to be caused by germline mutations in fumarate hydratase (FH) in the majority of screened cases.1,5,6 Forty-six distinct FH mutations have been reported to date in MCUL/HLRCC. Twenty-seven of these are missense mutations of 26 different residues (one residue has two reported mutations, R190H and R190L). These 27 distinct missense mutations represent 55 of 81 (68%) of the FH mutations reported in MCUL probands.1,5,6,7
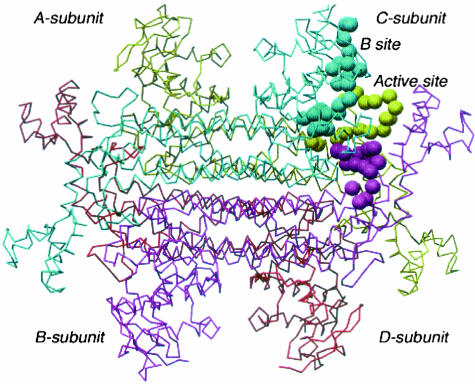
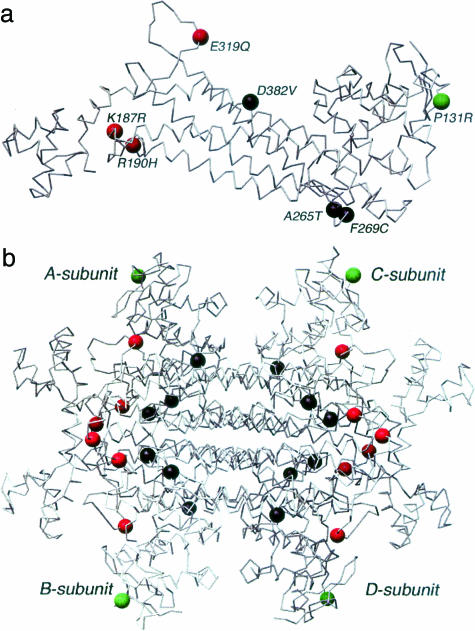
The FH locus encodes two isoforms of fumarate hydratase, cytosolic and mitochondrial, which differ only in that the latter has an initial mitochondrial signal peptide. Fumarate hydratase catalyzes the stereospecific reversible hydration of fumarate to l-malate. The mitochondrial isoform performs this reaction as part of the Krebs cycle and as such is central to aerobic respiration. The cytosolic isoform is thought to be involved in the metabolism of fumarate, which is produced in the cytosol by a number of reactions.8 FH is relatively highly evolutionarily conserved. The crystal structures of the E. coli fumarase C and Saccharomyces cerevisiae fumarase have been elucidated.9,10 The high degree of homology of these proteins to human FH allows their use as models for predicting the effect of missense mutations on FH function. The FH protein exists as a homotetramer with two substrate-binding sites (Figure 1), designated the A-site and the B-site. The A-site is made up of residues from three of four chains in the homotetramer whereas the B-site is made up of residues from one chain. Thus any change affecting subunit-subunit interaction would be expected to affect the function of the A-site. Studies in E. coli have shown that mutating a histidine to an asparagine at the A-site results in a large decrease in enzymatic activity, but at the B-site, has little effect suggesting that the A-site may be the main catalytically active site.11 In contrast, the B-site is thought to be a substrate-binding activation site but may not be catalytically active. We have previously shown that lymphoblastoid cell lines with heterozygous mutations in FH have diminished in vitro FH activity.1,5
Figure 1.
The E. coli FumC homotetramer showing the location of one of four FumC A/B sites. The three conserved regions within the fumarate hydratase superfamily are colored cyan (corresponding to region 1 H133 to T150 in human FH), khaki (corresponding to region 2 G189 to E204 in human FH), and magenta (corresponding to region 3 G321 to E335 in human FH).
Mutations in another Krebs cycle enzyme have also been shown to cause predisposition to an inherited tumor syndrome. Germline heterozygous mutations in subunits of succinate dehydrogenase (SDH), SDHB, SDHC, and SDHD, are associated with hereditary predisposition to pheochromocytoma and paraganglioma (OMIM 168000, 115310, 605373). A major question arising from the finding of a tumor suppressor function for genes encoding Krebs cycle enzymes is whether the tumor suppressor function is related to the metabolic/bioenergetic functions of the enzymes or whether a separate as yet unknown function of these enzymes is involved.
In this study, we investigated the missense FH mutations identified in MCUL. We examined the degree of evolutionary conservation of the mutated residues. We looked for evidence of mutational hotspots and examined the likely functional effects of the mutations. We then performed FH functional assay on available lymphoblastoid cell lines to assess correlations between these features and residual FH activity. We also assessed the missense mutations associated with renal cancer for any common features.
Homozygous/compound heterozygous germline FH mutations are known to underlie an autosomal recessive inborn error of metabolism characterized by fumarate hydratase deficiency (FHD, fumarase deficiency, fumaric aciduria; OMIM 606812). Clinical features of FHD include progressive encephalopathy, developmental delay, hypotonia, cerebral malformation and atrophy, and lactic and pyruvic acidemia with death usually occurring in infancy or by the first decade.12,13,14,15 Thus this is an uncommon example of a situation in which heterozygous and homozygous mutations of a single gene give rise to very different phenotypes. The germline mutations of 11 FHD patients have been reported (Table 4).5,12,13,14,15 In all of these cases, and in some patients who have not had mutation screening, affected individuals have been shown to have low FH activity, ranging from <1 to 20% of that of controls.5,12,13,14,15 Two missense mutations, R190H and K187R, have been reported in both MCUL/HLRCC and FHD and MCUL/FHD, respectively.5 In this study, we also examined features of the missense FH mutations reported in FHD.
Table 4.
Missense FH Mutations in FH Deficiency Patients
| Reference | Mutation | Reported activity (as percent of control activity) |
|---|---|---|
| 15 | A265T/A265T | 20 |
| 15 | D382V/D382V | <15 |
| 15 | D382V/D382V | 13 |
| 5 | P131R/IVS1–2A->G | 13 |
| 15 | F269C/435insK | 10 |
| 15 | K187R/K187R | 3.7 |
| 5 | R190H/R190H | 3 |
| 13 | E319Q/E319Q | <1 |
Materials and Methods
Missense FH mutations reported in the literature to date were examined using Clustal X PPC (1.64 b) to align human FH with the equivalent mouse, rat, Caenorhabditis elegans, Rhizopus, S. cerevisiae, and E. coli enzymes. The distribution of mutations and the degree of conservation of mutated residues were examined. Mutations were mapped onto the E. coli crystal structure 1FUO and the probable effect of the mutations examined. FH functional assay was performed on lymphoblastoid cell lines from 23 UK MCUL probands with identified heterozygous missense mutations in FH. Lymphoblastoid cell lines from 31 UK individuals without cancer were used as controls. Fumarate hydratase activity was assayed according to a modification of the method described by Hatch.16 Briefly, the assay monitors the increase in absorbance at 340 nm due to NADPH formation in a linked assay of lymphoblast sonicate, with a final reaction medium consisting of 10 mmol/L fumarate, 25 mmol/L Hepes-KOH buffer, pH7.5, *0.2 U malic enzyme/ml, 0.27 mmol/L NADP, 4 mmol/L MgCl2, and 5 mmol/L potassium phosphate. The assay was run at least twice for each sample and the final sample activity was the mean activity of these as a percentage of the mean control activity. Inter- and intra-assay variability gave a coefficient of variation of 2.6% and 5.8%, respectively.
Results
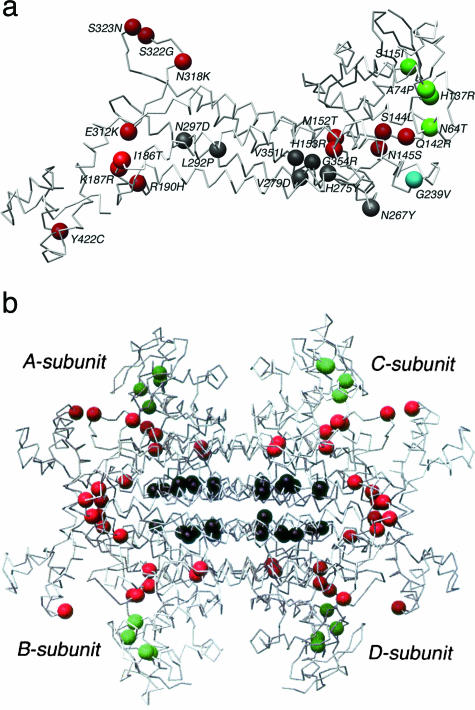
Missense mutations in FH in MCUL/HLRCC are summarized in Tables 1 and 2. There have been 27 distinct missense mutations of 26 different residues reported (one residue has two reported mutations R190H and R190L as mentioned above).1,5,6,7,17 Missense mutations in MCUL appear to cluster in certain regions in the protein (Figure 2). These include the regions: P131 to M152 (P131R, H137R and Q142R, S144L, N145S, M152T, H153R the first two mapping to the B-site and the remaining five to the A-site); I186 to R190 (delI186, I186T, K187R, R190H, and R190L, all at the A-site); A265 to N297(A265T, N267Y, F269C, H275Y, V279D, L292P, and N297D all involved in subunit-subunit interaction); and E312 to S323 (E312K, N318K, E319Q, S322G, and S323N, all at the A-site). The following residues have been the sites of more than one mutation in MCUL: Q142R, Q142X; I186T, delI186; R190H, R190L; and N318X, N318K. Eleven different US R190H families apparently did not share an ancestral haplotype suggesting that they may have arisen independently.6 However, all families in our UK study group with identical mutations, including two Spanish origin families with R190H mutations, appeared to share ancestral haplotypes suggesting the likelihood of a founder mutation in these cases.5
Table 1.
Missense FH Mutations in MCUL Patients
| Mutation | Number of probands | First report of mutation |
|---|---|---|
| N64T | 6 | 1 |
| A74P | 1 | 1 |
| S115I | 1 | 7 |
| H137R | 1 | 1 |
| Q142R | 1 | 1 |
| S144L | 1 | 6 |
| N145S | 1 | 6 |
| M152T | 1 | 6 |
| H153R | 1 | 3 |
| I186T | 1 | 5 |
| K187R | 3 | 1 |
| R190L | 1 | 6 |
| R190H | 13 | 1 |
| G239V | 1 | 1 |
| N267Y | 1 | Our UK group, previously unpublished |
| H275Y | 2 | 6 |
| V279D | 1 | 6 |
| L292P | 1 | 6 |
| N297D | 1 | 6 |
| E312K | 2 | 5 |
| N318K | 1 | 5 |
| S322G | 1 | 6 |
| S323N | 2 | 5 |
| V351L | 1 | 7 |
| G354R | 6 | 5 |
| Y422C | 2 | 6 |
| L464P | 1 | 5 |
Table 2.
Features of Reported Distinct Missense FH Mutations in MCUL
| Mutated residue | Evolutionary conservation of residue | Homologous residue in E. coli fumarase C | Likely function of mutated residue based on studies in E. coli | Mean FH activity of heterozygote as percentage of control activity |
|---|---|---|---|---|
| N64T | Fully | N59 | B-site | 47.2 |
| A74P | Weakly | A70 | B-site | 53 |
| S115I | Weakly | A111 | B-site | |
| H137R | Weakly | D133 | B-site | 51.5 |
| Q142R | Fully | Q138 | A-site | 46.1 |
| S144L | Fully | S140 | A-site | |
| N145S | Fully | N141 | A-site | |
| M152T | Fully | M148 | A-site | |
| H153R | Fully | H149 | A-site | |
| I186T | Strongly | I181 | A-site | 22.8 |
| K187R | Fully | K183 | A-site | 28.2 |
| R190H R190L | Fully | R186 | A-site | 26.9 (R190H) |
| G239V | Fully | G235 | Unknown | 39.1 |
| N267Y | Fully | N263 | Subunit-subunit interaction | |
| H275Y | Unconserved (fully conserved except in E. coli) | C271 | Subunit-subunit interaction | |
| V279D | Fully | V275 | Subunit-subunit interaction | |
| L292P | Strongly | L288 | Subunit-subunit interaction | |
| N297D | Strongly | N293 | Subunit-subunit interaction | |
| E312K | Fully | E308 | A-site | 17.4 |
| N318K | Fully | N314 | A-site | |
| S322G | Fully | S318 | A-site | |
| S323N | Fully | S319 | A-site | |
| V351L | Strongly | I347 | Subunit-subunit interaction | |
| G354R | Weakly | G350 | Subunit-subunit interaction | 33.9 |
| Y422C | Fully | Y418 | A-site | |
| L464P | Strongly | V460 | Not resolved in E. coli structure | 16.9 |
Figure 2.
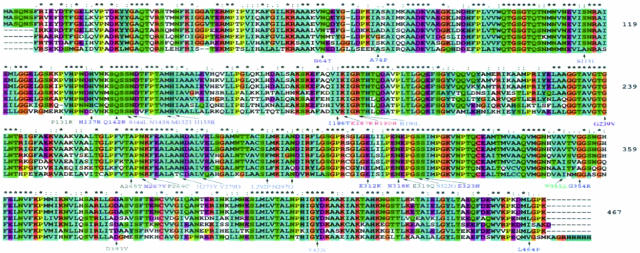
Clustal alignment of FH across seven species (from top, rat, mouse, human, C. elegans, Rhizopus, S. cerevisiae, E. coli) using the default Clustal setting based on biochemical properties of residues as follows: *, positions that have a single, fully conserved residue; :, indicates that one of the following strong groups is fully conserved: STA NEQK NHQK NDEQ QHRK MILV MILF HY FYW; ., indicates that one of the following weaker groups is fully conserved: CSA ATV SAG STNK STPA SGND SNDEQK NDEQHK NEQHRK FVLIH FYM. We have referred to these in the text as follows: *, fully conserved residues, :, strongly conserved residues, ., weakly conserved residues. All other residues are considered to be unconserved. Missense mutations are shown. Mutations reported in FH deficiency are black, mutations reported in MCUL are blue, mutations reported in both MCUL and FHD are red.
The clustal alignment demonstrated the relatively high degree of evolutionary conservation of FH (Figure 2). Three particular regions of amino acid sequence identity have been previously noted. These correspond to region 1 H133 to T150, region 2 G189 to E204, and region 3 G321 to E335 in human FH.9 These have been shown to be important in formation of the active A-site and B-site and have some overlap with the regions that may represent mutational hotspots (Figure 2). The clustal alignment across seven species showed that, of distinct missense mutations of FH in MCUL, 16 of 26 (60%) were of fully conserved, 5 of 26 (20%) of strongly conserved, 4 of 26 (16%) of weakly conserved, and 1 of 26 (4%) of nonconserved residues (Table 2, Figure 1). The proportion of mutations at fully conserved versus nonconserved residues was significantly greater (P = 0.04, Fisher’s exact test) than the proportion of such residues in the native FH protein (47.3% fully conserved, 18.2% strongly conserved, 9.0% weakly conserved, and 25.5% nonconserved, respectively).
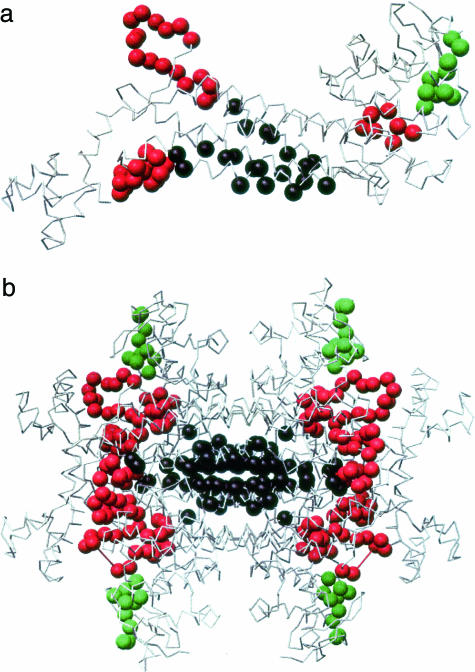
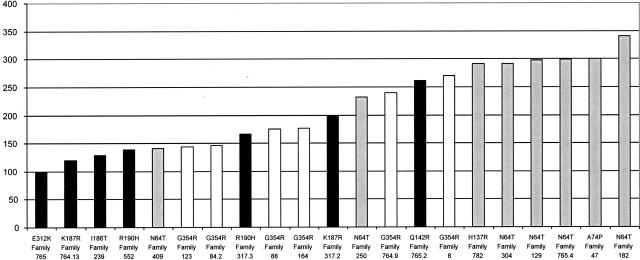
The proportion of residues functioning in the A-site, B-site, and in homotetramer formation in the FH monomer is 56 of 467 (12%) amino acids, 10 of 467 (2%) amino acids, and 22 of 467 (5%) amino acids with the remaining 379 residues (81%) having no known direct role in these functions (Figure 3). Mapping of the 26 residues with reported missense mutations in MCUL onto the homologous E. coli structure showed that one mutation (L464P) was of a residue that has not been mapped in the E. coli structure (Table 2). Of 25 missense mutations, 1 was of unknown function (G239V) (4%), 13 were predicted to affect the A-site (52%), 4 the B-site (16%), and 7 (28%) subunit-subunit interaction in the FH homotetramer (Table 2, Figure 4). Thus MCUL missense mutations are significantly likely to be of residues that have a known role (P < 0.001, Fisher’s exact test). The missense mutation of a residue of unknown function (G239V) was not found in control populations and has diminished FH activity in vitro of 39.1% suggesting that it is a true mutation and that it may affect FH enzymatic function by an unknown mechanism. Of note, there are no features to suggest that this mutation affects splicing. FH functional assay on 23 UK lymphoblastoid cell lines with missense FH mutations showed diminished in vitro FH activity in all cases. There was no significant association between residual FH activity and degree of evolutionary conservation of the mutated residue (data not shown). A-site mutants were shown to have significantly less residual activity (mean, 159 nmol/mg/minute; range, 99 to 261 nmol/mg/minute) than B-site mutants (mean, 274 nmol/mg/minute; range, 141 to 341 nmol/mg/minute) (Mann-Whitney test, P = 0.008) (Figure 5). A mutation, G354R, which, by virtue of its location on the interface between subunits would be predicted to impair homotetramerization, had an effect intermediate to that of A-site mutations and B-site mutations (mean, 192 nmol/mg/minute; range, 144 to 270 nmol/mg/minute). As a caveat, we have previously shown that heterozygous missense FH mutants have lower residual activity in vitro than deletion/truncating mutants, possibly due to a dominant-negative action of the missense mutant, preventing fully functional tetramers from forming in all but 1 of 16 of molecules.5 This therefore complicates the assessment of the effects of the missense mutations because it is not currently possible to separate the dominant-negative effects from the other effects of missense mutations.
Figure 3.
The E. coli FumC (a) monomer (b) homotetramer with B site (green), active site (red), and subunit interface (black) amino acids mapped. Coordinates from 1FUO A-subunit. Illustration prepared using Chimera.
Figure 4.
The E. coli FumC (a) monomer (b) homotetramer with MCUL mutations indicated in terms of function of the mutated residue with B-site (green), active A-site (red), and subunit interface (black) amino acids mapped. Mutated residues of unknown function are shown in cyan. Coordinates from 1FUO A-subunit. Illustration prepared using Chimera.
Figure 5.
FH activity of lymphoblastoid lines from MCUL/HLRCC probands with heterozygote missense FH mutation with the likely function of the mutated residue shown as follows: black, A-site; white, subunit-subunit interactions; and gray, B-site. The y axis is FH activity in nmol/g/minute.
The missense mutations that predispose to renal cancer in MCUL (N318K, R190H, and H275Y) did not appear to differ greatly from other MCUL/HLRCC missense mutations not associated with renal cancer, nor, apart from all being mutations of fully conserved residues, did they appear to share any common functions or features (Table 3). The R190H mutation had also been identified in several other MCUL families without renal cancer. These included 10 US families reported by Toro and colleagues6 and two families from our UK study group who were of Spanish origin and had a shared ancestral haplotype.1,5 The H275Y mutation had also been reported in one MCUL/HLRCC family without renal cancer.7
Table 3.
Missense FH Mutations Predisposing to Renal Cancer
| Ethnic origin | Reference | FH missense mutation | Histology of renal cancer | Evolutionary conservation of residue | Likely function of mutated residue based on studies in E. coli |
|---|---|---|---|---|---|
| Finnish | 3 | H153R | PRCCII | Fully | A-site |
| USA | 6 | R190H | PRCCII | Fully | A-site |
| USA | 6 | H275Y | PRCCII | Fully | Subunit-subunit interaction |
| UK | 5 | N318K | CDC | Fully | A-site |
Missense mutations reported in FHD are summarized in Tables 4 and 5. They appear to cluster in some of the same regions as MCUL mutations (Figures 2 and 3). The missense mutations reported in FHD affected the A-site in three of seven cases, B-site in one of seven cases, and subunit-subunit interaction in three of seven cases. Six of seven reported missense mutations in FHD were of fully conserved residues while one (H275Y) was of an unconserved residue although further examination showed that this residue was fully conserved except for in E. coli. We did not further analyze the missense FH mutations occurring in FHD for a number of reasons. Firstly, the number of cases was so small that meaningful analysis or comparison with MCUL was difficult. Secondly, we did not have enough cell lines available for assay within our own laboratory and the FH activities reported in the literature would not have been directly comparable as many were performed in different laboratories. Thirdly, missense mutations in FHD often occurred in conjunction with other, nonidentical mutations, that is, as compound heterozygotes, making analysis of the effects of each individual mutation difficult (Figures 1 and 6, Table 5).
Table 5.
Features of Reported Distinct Missense FH Mutations in FH Deficiency
| Mutated residue | Evolutionary conservation of residue | Homologous residue in E. coli fumarase C | Likely function of mutated residue based on studies in E. coli |
|---|---|---|---|
| P131R | Unconserved | K127 | B-site |
| K187R | Fully | K183 | A-site |
| R190H | Fully | R186 | A-site |
| A265T | Fully | A261 | Subunit-subunit interaction |
| F269C | Fully | F265 | Subunit-subunit interaction |
| E319Q | Fully | E315 | A-site |
| D382V | Fully | D378 | Subunit-subunit interaction |
Figure 6.
The E. coli FumC (a) monomer (b) homotetramer with FHD mutations indicated in terms of function of the mutated residue with B-site (green), active A-site (red), and subunit interface (black) amino acids mapped Coordinates from 1FUO A-subunit. Illustration prepared using Chimera.
Discussion
Analysis of reported missense FH mutations results in several interesting findings. Firstly, the distribution of missense mutations within the FH protein suggests the existence of mutational hotspots. These are significantly likely to be of fully conserved residues and have known functions, for example, they form part of the A-site, the B-site, or affect subunit-subunit interaction.
It is possible that some single nucleotide variants, for example, those affecting unconserved residues or amino acids not involved in the A-site, B-site, or subunit-subunit interaction may present with a mild or absent MCUL phenotype and are therefore not represented in our MCUL study group. In support of this, we have recently identified a P131R (unconserved residue) variant carrying mother of an FH deficiency patient with no obvious MCUL phenotype.11 The finding of lower residual FH activity in cell lines with missense mutations of the A-site compared to the B-site supports the suggestion from studies in E. coli that in FH, the A-site is of greater importance in catalytic activity than the B-site.11 The significant effect of the mutation affecting subunit-subunit interaction on enzymatic activity was not unexpected given the requirement of homotetramerization for formation of the A-site.
Renal cancers in MCUL are an uncommon association but are aggressive and of early onset. That missense mutations associated with renal carcinoma did not appear to have any unusual features compared to other MCUL mutations, and that identical FH missense mutations were present in families with and without renal cancers suggests that apart from the underlying FH mutation, genetic or environmental modifying factors play a role in predisposition to renal cancer in MCUL.
The pathway by which tumorigenesis occurs in MCUL is unknown. As mentioned, heterozygous germline mutations of another Krebs cycle enzyme, succinate dehydrogenase, have been shown to predispose to a familial neoplastic syndrome, features of which include paragangliomas and pheochromocytomas (OMIM 168000, 115310, 605373). Thus the reduction of activity of the Krebs cycle may be important in tumorigenesis. This study supports the likely role of diminished enzyme function as the mechanism of tumorigenesis because almost all of the missense mutations reported in MCUL are of residues that would be predicted to disrupt A-site or B-site or formation of the A-site by homotetramerization and because all of the MCUL missense mutations that have been tested cause diminished FH activity.
Acknowledgments
We thank the patients, their families, and their referring clinicians for their involvement in this study.
Footnotes
Supported by Cancer Research UK (clinical research fellowship to N.A.A.).
References
- Tomlinson IPM, Alam NA, Rowan AJ, Barclay E, Jaeger EE, Kelsell D, Leigh I, Gorman P, Lamlum H, Rahman S, Roylance RR, Olpin S, Bevan S, Barker K, Hearle N, Houlston RS, Kiuru M, Lehtonen R, Karhu A, Vilkki S, Laiho P, Eklund C, Vierimaa O, Aittomaki K, Hietala M, Sistonen P, Paetau A, Salovaara R, Herva R, Launonen V, Aaltonen LA. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat Genet. 2002;30:406–410. doi: 10.1038/ng849. [DOI] [PubMed] [Google Scholar]
- Alam NA, Barclay E, Rowan AJ, Tyrer JP, Calonje E, Manek S, Kelsell D, Leigh I, Olpin S, Tomlinson IP. Clinical features of multiple cutaneous and uterine leiomyomatosis: an underdiagnosed tumor syndrome. Arch Dermatol. 2005;141:199–206. doi: 10.1001/archderm.141.2.199. [DOI] [PubMed] [Google Scholar]
- Kiuru M, Launonen V, Hietala M, Aittomaki K, Vierimaa O, Salovaara R, Arola J, Pukkala E, Sistonen P, Herva R, Aaltonen LA. Familial cutaneous leiomyomatosis is a two-hit condition associated with renal cell cancer of characteristic histopathology. Am J Pathol. 2001;159:825–829. doi: 10.1016/S0002-9440(10)61757-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Launonen V, Vierimaa O, Kiuru M, Isola J, Roth S, Pukkala E, Sistonen P, Herva R, Aaltonen LA. Inherited susceptibility to uterine leiomyomas and renal cell cancer. Proc Natl Acad Sci USA. 2001;98:3387–3392. doi: 10.1073/pnas.051633798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam NA, Rowan AJ, Wortham NC, Pollard PJ, Mitchell M, Tyrer JP, Barclay E, Calonje E, Manek S, Adams SJ, Bowers PW, Burrows NP, Charles-Holmes R, Cook LJ, Daly BM, Ford GP, Fuller LC, Hadfield-Jones SE, Hardwick N, Highet AS, Keefe M, MacDonald-Hull SP, Potts ED, Crone M, Wilkinson S, Camacho-Martinez F, Jablonska S, Ratnavel R, MacDonald A, Mann RJ, Grice K, Guillet G, Lewis-Jones MS, McGrath H, Seukeran DC, Morrison PJ, Fleming S, Rahman S, Kelsell D, Leigh I, Olpin S, Tomlinson IP. Genetic and functional analyses of FH mutations in multiple cutaneous and uterine leiomyomatosis, hereditary leiomyomatosis and renal cancer, and fumarate hydratase deficiency. Hum Mol Genet. 2003;12:1241–1252. doi: 10.1093/hmg/ddg148. [DOI] [PubMed] [Google Scholar]
- Toro JR, Nickerson ML, Wei MH, Warren MB, Glenn GM, Turner ML, Stewart L, Duray P, Tourre O, Sharma N, Choyke P, Stratton P, Merino M, Walther MM, Linehan WM, Schmidt LS, Zbar B. Mutations in the fumarate hydratase gene cause hereditary leiomyomatosis and renal cell cancer in families in North America. Am J Hum Genet. 2003;73:95–106. doi: 10.1086/376435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Mir A, Glaser B, Chuang GS, Horev L, Waldman A, Engler DE, Gordon D, Spelman LJ, Hatzibougias I, Green J, Christiano AM, Zlotogorski A. Germline fumarate hydratase mutations in families with multiple cutaneous and uterine leiomyomata. J Invest Dermatol. 2003;121:741–744. doi: 10.1046/j.1523-1747.2003.12499.x. [DOI] [PubMed] [Google Scholar]
- Pines O, Even-Ram S, Elnathan N, Battat E, Aharonov O, Gibson D, Goldberg I. The cytosolic pathway of L-malic acid synthesis in Saccharomyces cerevisiae: the role of fumarase. Appl Microbiol Biotechnol. 1996;46:393–399. doi: 10.1007/BF00166235. [DOI] [PubMed] [Google Scholar]
- Weaver TM, Levitt DG, Donnelly MI, Stevens PP, Banaszak LJ. The multisubunit active site of fumarase C from Escherichia coli. Nat Struct Biol. 1995;2:654–662. doi: 10.1038/nsb0895-654. [DOI] [PubMed] [Google Scholar]
- Weaver T, Lees M, Zaitsev V, Zaitseva I, Duke E, Lindley P, McSweeny S, Svensson A, Keruchenko J, Keruchenko I, Gladilin K, Banaszak L. Crystal structures of native and recombinant yeast fumarase. J Mol Biol. 1998;280:431–442. doi: 10.1006/jmbi.1998.1862. [DOI] [PubMed] [Google Scholar]
- Weaver T, Lees M, Banaszak L. Mutations of fumarase that distinguish between the active site and a nearby dicarboxylic acid binding site. Protein Sci. 1997;6:834–842. doi: 10.1002/pro.5560060410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellera C, Uziel G, Rimoldi M, Zeviani M, Laverda A, Carrara F, DiDonato S. Fumarase deficiency is an autosomal recessive encephalopathy affecting both the mitochondrial and the cytosolic enzymes. Neurology. 1990;40:495–499. doi: 10.1212/wnl.40.3_part_1.495. [DOI] [PubMed] [Google Scholar]
- Bourgeron T, Chretien D, Poggi-Bach J, Doonan S, Rabier D, Letouze P, Munnich A, Rotig A, Landrieu P, Rustin P. Mutation of the fumarase gene in two siblings with progressive encephalopathy and fumarase deficiency. J Clin Invest. 1994;93:2514–2518. doi: 10.1172/JCI117261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustin P, Bourgeron T, Parfait B, Chretien D, Munnich A, Rotig A. Inborn errors of the Krebs cycle: a group of unusual mitochondrial diseases in human. Biochim Biophys Acta. 1997;1361:185–197. doi: 10.1016/s0925-4439(97)00035-5. [DOI] [PubMed] [Google Scholar]
- Coughlin EM, Christensen E, Kunz PL, Krishnamoorthy KS, Walker V, Dennis NR, Chalmers RA, Elpeleg ON, Whelan D, Pollitt RJ, Ramesh V, Mandell R, Shih VE. Molecular analysis and prenatal diagnosis of human fumarase deficiency. Mol Genet Metab. 1998;63:254–262. doi: 10.1006/mgme.1998.2684. [DOI] [PubMed] [Google Scholar]
- Hatch MD. A simple spectrophotometric assay for fumarate hydratase in crude tissue extracts. Anal Biochem. 1978;85:271–275. doi: 10.1016/0003-2697(78)90299-3. [DOI] [PubMed] [Google Scholar]
- Kiuru M, Launonen V. Hereditary leiomyomatosis and renal cell cancer (HLRCC). Curr Mol Med. 2004;4:869–875. doi: 10.2174/1566524043359638. [DOI] [PubMed] [Google Scholar]