Abstract
To establish the most sensitive and efficient strategy of clonality diagnostics via immunoglobulin and T-cell receptor gene rearrangement studies in suspected lymphoproliferative disorders, we evaluated 300 samples (from 218 patients) submitted consecutively for routine diagnostics. All samples were studied using the BIOMED-2 multiplex polymerase chain reaction (PCR) protocol. In 176 samples, Southern blot (SB) data were also available, and the two types of molecular results were compared. Results of PCR and SB analysis of both T-cell receptor and immunoglobulin loci were concordant in 85% of samples. For discordant results, PCR results were more consistent with the final diagnosis in 73% of samples. No false-negative results were obtained by PCR analysis. In contrast, SB analysis failed to detect clonality in a relatively high number of samples, mainly in cases of low tumor burden. We conclude that the novel BIOMED-2 multiplex PCR strategy is of great value in diagnosing patients with suspected B- and T-cell proliferations. Because of its higher speed, efficiency, and sensitivity, it can reliably replace SB analysis in clonality diagnostics in a routine laboratory setting. Just as with SB results, PCR results should always be interpreted in the context of clinical, immunophenotypical, and histopathological data.
In most patients with suspected lymphoproliferative disorders, discrimination between reactive and malignant cell populations can be assessed by histomorphology or cytomorphology supplemented with immunohistochemistry or flow cytometric immunophenotyping. However, in 5 to 10% of patients, diagnosis is more complicated and less straightforward. In such cases, molecular gene rearrangement studies have proved useful as an additional diagnostic tool. Molecular clonality analysis is based on the fact that, in principle, all cells of a malignancy have a common clonal origin and show clonally (identically) rearranged immunoglobulin (Ig) or T-cell receptor (TCR) genes. The diagnosis of malignant B- and T-cell proliferations is therefore supported by the finding of Ig/TCR gene clonality, whereas reactive lymphoproliferations show polyclonally rearranged Ig/TCR genes.1
Gene rearrangement analysis can be performed by Southern blot (SB)- and polymerase chain reaction (PCR)-based techniques. Despite the high reliability of SB analysis, it is increasingly replaced by PCR techniques because of the greater efficiency and sensitivity of PCR. Moreover, PCR is relatively easy, less labor intensive, and requires much less high-molecular-weight DNA. Also, SB analysis cannot be performed on paraffin-embedded tissue because the isolated DNA is often degraded. Therefore, there is a strong need to replace SB analysis with reliable PCR techniques. However, PCR studies have often suffered from false-negative results due to improper annealing of primers and/or the presence of somatic hypermutation.2
Both SB and PCR analyses of the immunoglobulin heavy chain (IGH) locus have been demonstrated to be very useful and reliable techniques in clonality assessment of suspected B-cell malignancies. However, the most useful gene target for identifying T-cell clonality is less well established. In virtually all PCR studies, only the T-cell receptor-γ (TCRG) locus was analyzed at the DNA level because of the relative structural simplicity of the gene.3,4 PCR analysis of the T-cell receptor-β (TCRB) locus as a diagnostic test has been performed mostly on cDNA using Vβ and Cβ primers. Recently, the BIOMED-2-based TCRB gene rearrangement analysis was evaluated in a large series of well-defined samples from immature and mature T-cell malignancies and was demonstrated to be a very reliable assay.5 However, these novel BIOMED-2 multiplex PCR methods for detecting B- and T-cell clonality have yet to be validated in routine diagnostic laboratory settings.
The vast majority of lymphoid malignancies encountered in the West belong to the B-cell lineage (90 to 95%). Even though B-cell clonality can be assessed by flow cytometric immunophenotyping, Ig gene rearrangement analysis is the only reliable assay for paraffin-embedded and frozen tissue biopsies. Consequently, in many routine diagnostic laboratories, TCR gene rearrangement analysis is applied to a smaller number of cases per year than Ig analysis, resulting in a lower level of experience. Because our institute is a reference center for clonality analysis in suspected T-cell proliferations, we routinely obtain many T-cell proliferation samples each year.
In our study, we performed a comparative prospective study of SB-PCR Ig/TCR gene rearrangements on a series of 300 specimens consecutively obtained for routine diagnostics. In general, SB analysis was performed with optimized DNA probes for the IGH locus and the TCRB locus.6,7 To determine B- or T-cell clonality by PCR analysis, we analyzed the IGH, TCRB, and TCRG genes. To this end, we used the well-defined and fully standardized set of oligonucleotide primers and PCR protocols of the BIOMED-2 Concerted Action BMH4-CT98-3936.8,9 Resulting PCR products were analyzed by both heteroduplex (HD) and GeneScan (GS) analysis to evaluate the diagnostic value of these methods.
Our results show that the BIOMED-2 PCR-based TCR gene rearrangement analysis is more sensitive in detecting T-cell clonality than SB analysis. In instances of discordance, PCR results demonstrated agreement with histopathological diagnosis more often than SB analysis. The higher sensitivity of PCR analysis over SB analysis also holds for IGH gene rearrangements. Based on these results, we discuss the most sensitive and efficient strategy of molecular clonality analysis.
Materials and Methods
Patients
From June 2001 until February 2004, 300 DNA samples from fresh or frozen tissue samples (peripheral blood [PB], n = 110; lymph node [LN], n = 68; bone marrow [BM], n = 28; skin, n = 59; bowel, n = 7; liver, n = 3; spleen, n = 1; thyroid, n = 1; vitreous fluid, n = 15; cerebrospinal fluid (CSF), n = 3; pleural fluid, n = 2; adenoid, n = 1; brain, n = 1; and maxillary sinus tissue, n = 1) were prospectively collected from a total of 218 patients with suspected malignant lymphoproliferation. Most patients were seen and followed by physicians at the Erasmus MC, University Medical Center (Rotterdam), especially at the Departments of Hematology and Dermatology. Diagnoses were based on a combination of clinical, histological, immunophenotypical, and cytomorphological data. Patients diagnosed with a malignancy were classified according to the World Health Organization classification of lymphoid neoplasms.10
DNA Isolation
High-molecular-weight DNA from fresh or frozen tissue samples was obtained by one of two methods. In the first, DNA was extracted using a phenol-chloroform extraction-based protocol, followed by ethanol precipitation and re-solubilization in Tris-EDTA buffer.1 Alternatively, DNA was isolated using the GenElute Mammalian Genomic DNA miniprep kit (Sigma-Aldrich, St. Louis, MO) according to the manufacturer’s protocol.
Southern Blot Analysis
DNA (20 μg) was digested with appropriate restriction enzymes, size fractionated in 0.7% agarose gels, and transferred to nylon filters as described elsewhere.1 The Ig and TCR gene rearrangements were detected by use of 32P random oligonucleotide-labeled probes. The IGHJ6 probe (DakoCytomation, Inc., Carpinteria, CA) was used for analyzing IGH genes in combination with BglII or BamHI/HindIII digests,6 whereas the TCRBJ1 and TCRBJ2 probes (DakoCytomation) were used for analyzing TCRB genes in combination with EcoRI, BglII, and BamHI/HindIII digests.7 In a selected number of cases (n = 11), the TCRG and TCR-δ (TCRD) gene rearrangements were studied as well. For analysis of TCRG gene rearrangements, the TCRGJ13 probe was used in combination with EcoRI and Pstl digests,11 whereas the TCRDJ1 probe was used in combination with EcoRI digests for analysis of TCRD gene rearrangements.12 In the case of clinical suspicion of natural killer cell lymphoma and Epstein-Barr virus (EBV) infection, the presence of (clonal) EBV genome was assessed using the XhoI probe in BamHI/HindIII-digested DNA.13
PCR Amplification
All amplification reactions were performed in an automated thermocycler (model ABI 9600/9700; Applied Biosystem, Foster City, CA) according to the BIOMED-2 multiplex PCR protocol.8 Each 50-μl PCR reaction included 100 ng of DNA, 10 pmol of 5′ and 3′ oligonucleotide primers, 0.2 mmol/L dNTP, 5 μl of 10× buffer II (TCRB and TCRG), or 5 μl of 10× Gold buffer (IGH and IGK), and 1 to 2 U of Ampli-Taq Gold polymerase (Applied Biosystems). The concentration of MgCl2 ranged from 1.5 mmol/L (TCRB tube C, TCRG, IGH, and IGK) to 3 mmol/L (TCRB tubes A and B). The cycling parameters were as follows: pre-activation for 7 minutes at 95°C, followed by 35 cycles of 30 seconds denaturation at 95°C, >30 seconds annealing at 60°C, and >30 seconds extension at 72°C. After the last cycle, a final extension step of at least 10 minutes at 72°C was performed.
For amplification of IGH rearrangements, we used six framework FR1-VH primers, seven FR2-VH primers, seven FR3-VH primers, and one carboxyfluorescein-labeled JH consensus primer in three multiplex combinations (IGH multiplex tubes A, B, and C). DNA from the precursor B-cell line NALM-6 was used as a positive control. In the case of inconclusive IGH gene results, the samples were further analyzed using multiplex PCR reactions for the Ig-κ (IGK) genes. For amplification of the IGK locus, we used seven Vκ primers, two FAM-labeled Jκ primers, one intron RSS primer, and one FAM-labeled Kde primer in two combinations (IGK multiplex tubes A and B). For amplification of TCRB rearrangements, Vβ family and FAM-labeled Jβ primers were used in two different combinations (TCRB multiplex tubes A and B) and in one combination containing Dβ and Jβ primers (TCRB multiplex tube C). DNA obtained from the following immature T-cell lines was used as a positive control: RPMI-8402 (tube A), CML-T1 (tube B), and Jurkat (tube C). For amplification of TCRG genes, we used Vγ and FAM-labeled Jγ primers in two multiplex combinations (tubes A and B).8 Positive controls consisted of DNA from immature T-cell lines MOLT 3, RPMI-8402 (tube A), and Jurkat (tube B). In a few cases, TCRD rearrangements were analyzed using a single multiplex tube containing Vδ, Jδ, and Dδ primers. All BIOMED-2 multiplex PCR tubes were obtained from InVivoScribe Technologies (Carlsbad, CA; www.invivoscribe.com). All cell lines are available at the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (Braunschweig, Germany). Features of these cell lines have been summarized in detail elsewhere.14
After Ig/TCR amplification, 10 μl of PCR products was loaded on 1% agarose gels to check whether PCR products had been formed. Subsequently, PCR products were further analyzed by heteroduplex and GeneScan analysis (see below) to assess whether the obtained PCR products were derived from monoclonal or polyclonal cell populations.
Heteroduplex and GeneScan Analyses
The PCR products for HD analysis were denatured at 94°C for 5 minutes and subsequently renatured at 4°C for 60 minutes to induce duplex formation.15 Afterwards, the duplexes were immediately loaded on 6% nondenaturing polyacrylamide gels in 0.5× Tris-boric acid-EDTA buffer, run at ambient temperature, and visualized by ethidium bromide staining. A 100-bp DNA ladder (Promega Corporation, Madison, WI) was used as size marker.
GeneScan analysis was performed using an automated ABI PRISM 377 fluorescent sequencer (Applied Biosystems) for the majority of PCR products, whereas the remaining samples were analyzed using an ABI 3100 Genetic Analyzer (Applied Biosystems). When using the first detection method (ABI 377), 2 μl of 10× diluted PCR products were mixed with 2.0 μl of formamide, 0.5 μl of 6-carboxytetramethylrhodamine-labeled internal standard (Genescan 500-TAMRA; Applied Biosystems), and 0.5 μl of loading buffer (blue dextran). After denaturation at 95°C for 2 minutes and cooling, 3 μl of the mixture was size-separated on a high-resolution polyacrylamide gel and analyzed. The size and profile of the PCR products was determined using GeneScan Analysis software v. 2.1 (Applied Biosystems).16,17
For the second method (ABI 3100), 1 μl of a 5× dilution of PCR products was added to 10 μl of a MilliQ:rhodamine-labeled internal standard (GeneScan-500 ROX; Applied Biosystems) mixture (40:1). After denaturation at 95°C for 2 minutes and cooling, the samples were size-separated and detected. The size and profile of the PCR products were determined using GeneScan Analysis software v. 3.7.1. (Applied Biosystems). GeneScan analysis results in a Gaussian distribution of multiple peaks, representing many different PCR products in case of reactive lymphoproliferations but gives a single peak in case of monoclonal lymphoproliferation. Oligoclonality is defined as multiple peaks in a polyclonal background.
Results
We consecutively investigated 300 samples (from 218 patients) that were submitted for routine diagnostics. In all patients a clinical diagnosis of malignant lymphoproliferative disease was initially considered. All samples were studied by multiplex PCR-based HD and GS analysis of Ig/TCR gene rearrangements, whereas SB analysis could be performed on only 176 out of the 300 samples (Table 1). Some samples were analyzed for both Ig and TCR clonality. Overall, 258 samples were analyzed by PCR for T-cell clonality; of these samples, 150 were analyzed by TCRB SB. PCR IGH gene rearrangement analysis was performed in 87 samples, and SB analysis was performed in 48 of these samples.
Table 1.
Summary of Performed SB- and PCR-Based Ig/TCR Analysis
| Clonality analysis | Samples (patients)
|
|
|---|---|---|
| Multiplex PCR | SB | |
| TCR only | 214 (155) | 28 (24) |
| Ig only | 39 (30) | 3 (3) |
| Ig and TCR | 47 (33) | 145 (119) |
| Total | 300 (218) | 176 (146) |
Because of our position as a reference center and our extensive expertise on TCR gene rearrangement analysis, the focus of our study lies on T-cell clonality analysis. SB and PCR analyses were, in principle, performed once. In practice this means that Ig clonality was evaluated with the IGHJ6 probe and in three different FR IGH multiplex PCR reactions. In limited cases, additional SB and PCR analysis of the IGK gene was performed. TCR clonality was analyzed with two TCRB SB probes as well as with TCRB and TCRG multiplex PCR protocols. Few samples were studied by SB analysis for TCRG/TCRD gene rearrangements or for EBV genome and PCR analysis of TCRD gene rearrangements. In case of doubtful or discrepant results, assays were repeated.
High Level of Concordance between SB and PCR Ig/TCR Gene Rearrangement Studies
Of 300 samples, 176 were analyzed simultaneously by multiplex PCR and SB analysis (Table 1). Molecular data were concordant for TCR analysis in 127 of 150 samples (85%), whereas Ig gene rearrangement analysis was concordant in 41 of 48 samples (85%). Table 2 shows the proportion of concordant SB-PCR results in polyclonal and clonal cases. The finding of a monoclonal T- or B-cell population both by SB and PCR clonality analysis strongly suggests a clonal (probably malignant) lymphoproliferation. Polyclonal gene rearrangements in both assays are indicative of a reactive (benign) lymphoproliferation. Nevertheless, it should be emphasized that the results of molecular clonality studies should always be interpreted in the context of clinical, histological, and immunophenotypical data. In 30 samples derived from 27 patients, discordances were found between multiplex PCR and SB analysis. In all of these samples, monoclonality was demonstrated by PCR analysis, whereas SB did not show evidence for clonal Ig/TCR gene rearrangements.
Table 2.
Concordance between SB- and PCR-Based Ig/TCR Analysis in Polyclonal and Clonal Cases
| SB-PCR concordance | Gene rearrangement target
|
|
|---|---|---|
| Ig | TCR | |
| Total concordance | 41/48 (85%) | 127/150 (85%) |
| Polyclonal cases | 25/41 (61%) | 83/127 (65%) |
| Clonal cases | 16/41 (39%) | 44/127 (35%) |
Evaluation of TCR Clonality as Demonstrated by PCR Analysis with Nonclonal Results Observed by SB Analysis
In 23 samples derived from 21 patients, multiplex PCR analysis demonstrated clonal TCRB/TCRG gene rearrangements, which were not detected by SB analysis. In 17 samples, the molecular findings correlated with the histopathological diagnosis of malignant T-cell proliferations (74%). (For an example of histologically proven ALCL, see Figure 1.) In the remaining samples there was no clear indication for a T-cell malignancy at the time of diagnosis (Table 3).
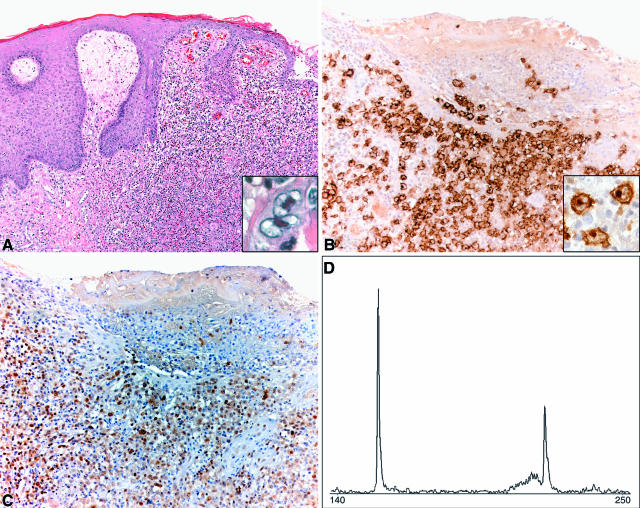
Figure 1.
Skin localization of ALCL in case 4. A: Histology of ulcerating skin tumor. Skin biopsy specimen demonstrates localization of dermal infiltrate of large cells with irregular nuclei (HE; magnification, ×100). Note the large hallmark cells (inset) (HE; magnification, ×600). B: The large neoplastic cells are strongly positive for CD30 (magnification, ×200). Note the strongly stained membrane and the dot-like staining in the Golgi complex area (inset) (magnification, ×600). C: The neoplastic cells also stain for CD4 (magnification, ×200). D: PCR-based GeneScan analysis. Bi-allelic monoclonal TCRG gene rearrangements could be identified in skin DNA.
Table 3.
Discordances between SB and PCR Results in Suspect T-Cell Proliferations
| Case no. | Sample no. | Final clinicohistological diagnosis* | SB
|
PCR procedure
|
||||
|---|---|---|---|---|---|---|---|---|
| TCRB | TCRG | TCRD | TCRB | TCRG | TCRD | |||
| 1 | 2001-146 (skin) | LyP | G | ND | ND | C | C | ND |
| 2 | 2001-148 (LN) | AILT | G | ND | ND | C | C | ND |
| 3 | 2001-152 (PB) | T-LGL | G | G | ND | C | C | ND |
| 4 | 2001-162 (skin) | ALCL | G | G | ND | C | C | ND |
| 5 | 2002-003 (LN) | Peripheral T-NHL, NOS | G | ND | ND | C | P | P |
| 6 | 2002-022 (PB) | MF | G | ND | ND | C | C | ND |
| 7 | 2002-033 (LN) | CD30+ CTCL | G | ND | ND | C | C | ND |
| 8 | 2002-047 (PB) | T-LGL | G | ND | ND | C | C | ND |
| 9 | 2002-050 (LN) | ALCL | G | ND | G | C | P | C |
| 10 | 2002-113 (BM) | T-ALL | G | ND | G | C | P | C |
| 11 | 2002-144 (BM) | MPD-U | G | G | G | C | C | P |
| 12 | 2003-022 (LN) | AILT | G | G | G | C | C | ND |
| 13 | 2003-172 (BM) | T-LBL | G | G | G | C | C | C |
| 14 | 2003-236 (BM) | HES | G | ND | ND | C | P | ND |
| 2003-296 (PB) | HES | G | ND | ND | C | C | ND | |
| 15 | 2003-245 (skin) | MF | G | G | G | C | P | ND |
| 16 | 2003-270 (PB) | SS | G | ND | ND | C | P | ND |
| 2004-007 (LN) | SS | G | ND | ND | C | P | ND | |
| 17 | 2004-001 (skin) | SS | G | ND | ND | C | P | ND |
| 18† | 2004-004 (PB) | Reactive | G | ND | ND | C | C | ND |
| 19 | 2004-005 (skin) | CD30+ CTCL | G | ND | ND | C | P | ND |
| 20 | 2004-011 (BM) | Reactive | G | ND | ND | C | P | ND |
| 21 | 2004-020 (LN) | Reactive | G | ND | ND | C | C | ND |
R, rearranged; G, germline; C, clonal; P, polyclonal; ND, not done.
AILT, angioimmunoblastic T-cell lymphoma; ALCL, anaplastic large-cell lymphoma; CTCL, cutaneous T-cell lymphoma; HES, hypereosinophilic syndrome; LyP, lymphomatoid papulosis; MF, mycosis fungoides; MPD-U, myeloproliferative disease, unclassifiable; PTLD, post-transplant lymphoproliferative disorder; SS, Sézary syndrome; T-ALL, T-cell acute lymphoblastic leukemia; T-LBL, T-cell lymphoblastic lymphoma; T-LGL, T-cell large granular lymphocyte leukemia; T-NHL NOS, T-cell non-Hodgkin’s lymphoma, not otherwise specified.
Case 18 was diagnosed with a CD30+ CTCL 10 years before.
PCR analysis of the TCR locus identified clonal gene rearrangements in six samples derived from five patients in whom no diagnosis of T-cell leukemia or lymphoma could be made. Case 11 was diagnosed as myeloproliferative disease, unclassifiable. Case 14, diagnosed as idiopathic hypereosinophilic syndrome (HES), showed identical clonal TCR rearrangements in the BM and PB samples by PCR analysis. Immunophenotyping of the PB sample demonstrated a small T-cell population (0.3% of leukocytes) with an aberrant phenotype (CD3−/CD4+/CD5+). It has been shown that in some HES patients, abnormal monoclonal T cells can be found in PB.18,19,20,21 However, in the literature so far, no clonal T-cell populations identical to the one in PB have been detected in BM. Further research should reveal the meaning of the clonal T-cell population in BM. Because patients diagnosed with idiopathic HES and associated clonal T-cell populations are at risk of developing T-cell lymphoma,19,22,23 careful follow-up is required in this case. Case 18 was admitted to our hospital in 1994 and was diagnosed with CD30+ cutaneous T-cell lymphoma. Although the patient was clinically healthy without any signs of relapse, the PB sample demonstrated monoclonal TCRB rearrangements in 2004. The PCR analysis on the paraffin-embedded skin tissue sample from 1994 demonstrated weak clonal TCRB rearrangements, identical to the ones found in the PB sample 10 years after presentation. We strongly recommend follow-up of this patient as well. A skin relapse of an ALCL was diagnosed in case 20. Staging resulted in the detection of weak clonal TCRB gene rearrangements in the BM sample, not identical to the ones found in the skin. Flow cytometry and histopathology did not demonstrate localization of malignant lymphoma. Finally, case 21 was diagnosed as reactive lymphadenopathy after extensive immunohistochemical and flow cytometric analysis of the LN sample.
Evaluation of Ig Clonality as Demonstrated by PCR Analysis with Nonclonal Results Observed by SB Analysis
In seven samples derived from six patients, multiplex PCR analysis demonstrated clonal B-cell populations that were not detected by SB analysis (Table 4). IGH monoclonality was detected in the LN sample of case 27, who suffered from human immunodeficiency virus and EBV infection. The histopathological diagnosis of B-post-transplantation lymphoproliferative disorder-associated disease was made from the LN sample (Figure 2). This polymorphic B-cell lymphoproliferative disorder also occurs in immunodeficient states outside the posttransplant setting and comprises 5% of human immunodeficiency virus-associated lymphomas. Most cases have clonally rearranged IGH and IGK genes with the TCRB gene in the germline configuration as found by SB analysis.24 In addition, TCR oligoclonality was detected in the sample by PCR analysis (Figure 2). Although SB analysis of the IGH locus did not provide evidence for malignancy, the presence of a clonal EBV genome in the LN sample was identified by SB analysis (data not shown). In four of the other six samples, the IGH PCR results agreed with biopsy histology and immunophenotypical analysis. All together, five of seven (71%) B-cell proliferations showed concordant PCR and histology results.
Table 4.
Discordances between SB and PCR Results in Suspect B-Cell Proliferations
| Case no. | Sample no. | Final clinico-histological diagnosis* | SB
|
PCR procedure
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| IGH | IGK | EBV | FR1 | FR2 | FR3 | IGK | |||
| 22 | 2001-125 (skin) | Dermatitis | G | ND | ND | C | C | C | ND |
| 23 | 2001-170 (PB) | MM | G | ND | ND | C | C | C | ND |
| 24 | 2002-011 (BM) | Reactive | G | ND | ND | C | C | P | ND |
| 25 | 2002-096A (LN) | DLBCL | G | ND | ND | C | C | C | ND |
| 2002-096B (BM) | DLBCL | G | ND | ND | C | C | C | ND | |
| 26 | 2002-122 (skin) | DLBCL | G | G | ND | C | C | P | C |
| 27† | 2003-242 (LN) | B-PTLD | G | G | R | C | C | C | ND |
G, germline; R, rearranged; C, clonal; P, polyclonal; ND, not done.
DLBCL, diffuse large B-cell lymphoma; MM, multiple myeloma.
Case no. 27 was diagnosed with a PTLD-associated disease (HIV and EBV positive) and demonstrated oligoclonal TCR rearrangements in the LN sample by PCR analysis (Figure 2).
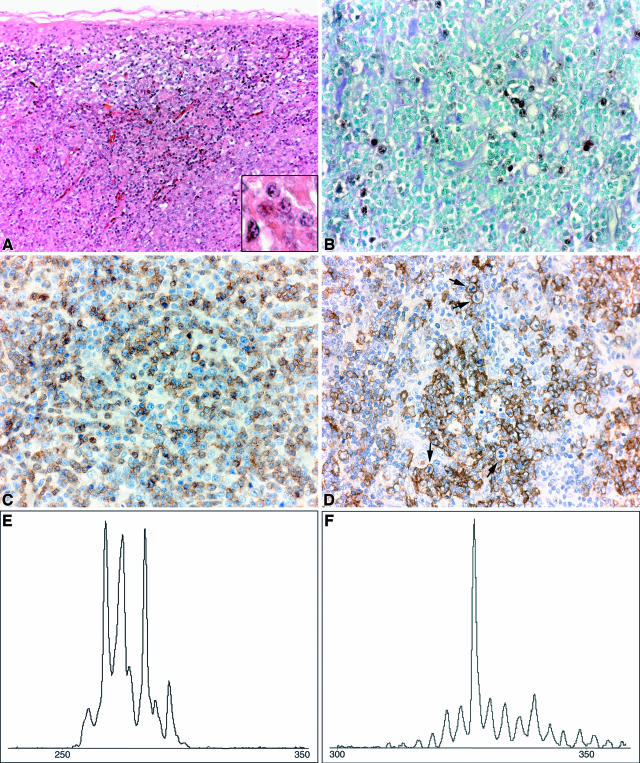
Figure 2.
Polymorphic B-cell PTLD-associated disease in case 27. A: Histology of LN. There was no recognizable lymph node architecture (HE; magnification, ×200). There were numerous blasts (inset) (HE; magnification, ×600) intermingled with an infiltrate of smaller lymphocytes. B: EBER-ISH showed scattered positively stained nuclei, indicating the presence of EBV RNA (magnification, ×400). C: CD3 staining of the predominantly small T cells. Occasionally, larger cells stained positively (magnification, ×400). D: CD20 stained the majority of the large blasts. Note the numerous mitotic figures (arrows) (magnification, ×400). E and F: PCR-based GeneScan analysis of LN DNA. Oligoclonal TCRB gene rearrangements could be identified (E), whereas a monoclonal IGH gene rearrangement was detected (F).
The two remaining samples were diagnosed as reactive lesions after extensive histopathological evaluation. In the skin sample of case 22, a weak monoclonal gene rearrangement was detected by PCR analysis. On histopathological examination, the lymphocytic infiltrate almost exclusively contained T cells. This case was discussed in the Dutch Cutaneous Lymphoma Working Group, and a diagnosis of dermatitis was made. The small amount of polyclonal B cells in the cutaneous infiltrate might explain the finding of (oligo)clonal IGH gene rearrangements.25,26 Although a weak clonal IGH gene rearrangement was detected in the BM sample of case 24 by PCR analysis, immunophenotyping only detected a reactive plasma cell population, and no abnormal B cells were seen by cytomorphological evaluation.
Discussion
We evaluated a group of 218 patients with an initial suspicion of lymphoproliferative disorders to estimate whether the newly designed primers and PCR protocols of the BIOMED-2 Concerted Action method could reliably replace SB analysis in a routine diagnostic setting. A total of 300 DNA samples was analyzed by two PCR-based strategies (HD and GS analysis), whereas a series of 176 DNA samples were analyzed by both SB and PCR analyses. The fact that SB analysis could not be performed in 124 samples further stresses the need for reliable PCR assays. When the SB and PCR approaches were compared and related to the final diagnosis, PCR and SB results of both T- and B-cell clonality analyses were concordant in 85% of samples.
In those samples in which the SB-PCR results were discordant, the PCR results appeared to correlate with the histopathological diagnosis in 22 of 30 cases (73%). Although PCR analyses showed unconfirmed clonal results in 27% of cases in comparison with the SB analysis, the number of false-negative SB results in cases of true malignancy was unexpectedly high. Further research and follow-up should estimate the meaning of small clonal T- or B-cell populations in nonmalignant cases as determined by PCR-based techniques. Most recently, the BIOMED-2-based primers and protocols were studied in a series of ∼100 well-defined samples of reactive lymphoproliferations, and (oligo)clonality was detected in 10% of cases (A.W. Langerak, unpublished data).
Based on our research and published reports, we strongly recommend careful follow-up of nonmalignant cases with clonal lymphoproliferations, because Ig/TCR clonality might be an early sign of an underlying hematological malignancy. Frequent clinical and histological follow-up on PB, BM, or LN samples is required. This might result in diagnosing patients in early stage disease, thus favoring prognosis and survival. The detection of identical T- or B-cell clones in a tissue sample and in PB is of great importance in staging lymphoid malignancies and is of prognostic relevance.27,28,29,30 It should, however, be noted that the finding of (oligo)clonality might not always be clinically significant. (Oligo)clonal T-cell populations can be detected in PB of the elderly,31,32 in patients diagnosed with autoimmune diseases, and in patients with viral infection.33 In cutaneous T-cell lymphoma, the detection of an identical monoclonal TCR gene rearrangement in skin and extracutaneous tissues has been proven to be an independent prognostic marker.34,35 In addition, detection of the same clonal gene rearrangements in multiple skin biopsy specimens at the time of diagnosis may provide prognostic information related to disease progression.36 Therefore, if possible, we recommend studying multiple (parallel) samples from multiple suspect localizations at initial disease presentation. In addition, follow-up samples should be evaluated when disease progression is suspected.
The detection of a clonal B- or T-cell population can facilitate diagnosis in patients in whom clinical, histopathological, and immunophenotypical findings are consistent but not entirely typical of a malignant lymphoproliferation. The importance of accurate molecular clonality diagnostics is supported by the fact that the current diagnostic criteria for patients with SS and T-LGL leukemia include the presence of a clonal TCR gene rearrangement in PB.37,38
The BIOMED-2 multiplex PCR approach is a rapid and reliable procedure that is far more sensitive than SB analysis in detecting clonality in suspect lymphoproliferations. The final clinico-histopathological diagnosis correlates well with PCR results in a higher number of patients in comparison with SB results. Therefore, it seems that the “gold standard” of SB analysis can now be reliably replaced in a routine laboratory setting. Essentially, this not only holds true for Ig clonality analysis but also for TCR clonality assessment, as demonstrated in this study. There was no difference in clonality detection rate between TCRB and TCRG gene rearrangement analysis, whereas the combined analysis clearly showed additional value. We detected the highest number of clonal IGH gene rearrangements when using multiplex combination A and B (FR1 + FR2 primers) (Table 5), but the combined information from all three FR PCR reactions resulted in a higher number of clonal cases. Although SB IGH gene rearrangement analysis remains a reliable and helpful additional test, the PCR approach is strongly preferred. The finding of polyclonal IGH gene rearrangements by PCR analysis in case of strong suspicion of a B-cell malignancy would indicate performing additional PCR-based IGK analysis. In two cases of our series, analysis of the IGK locus was of additional value. We detected clonal IGK gene rearrangements in a case of follicular lymphoma and a case of cutaneous B-cell lymphoma, whereas polyclonal gene rearrangements were found on IGH analysis. In case of T-cell clonality diagnostics, our TCRB/TCRG protocols are superior to SB analysis of the TCRB locus. The latter, however, may still be a valuable test under certain conditions.
Table 5.
Clonality Detection Rate of Different Targets in PCR-Based Ig/TCR Clonality Assessment
|
IGH
|
TCR
|
||||||
|---|---|---|---|---|---|---|---|
| FR1 | FR2 | FR3 | FR1+2+3 | TCRB | TCRG | TCRB + TCRG | |
| Clonality (%) | 94 | 88 | 55 | 100 | 87 | 86 | 100 |
| Per target | (31/33) | (29/33) | (18/33) | (33/33) | (103/119) | (102/119) | (119/119) |
In conclusion, both HD and GS are reliable techniques with completely concordant results. Because of its enhanced speed, accuracy, and interpretation, GS analysis is slightly favored over HD analysis. Based on our current results and extensive experience with TCR gene studies in general, we now propose a flowchart that demonstrates the most efficient and sensitive strategy in detecting T-cell clonality (Figure 3). This strategy holds true for fresh or frozen tissue samples and may be applicable to paraffin-embedded tissue samples, provided that DNA quality is such that PCR products of ∼300 bp can be amplified.
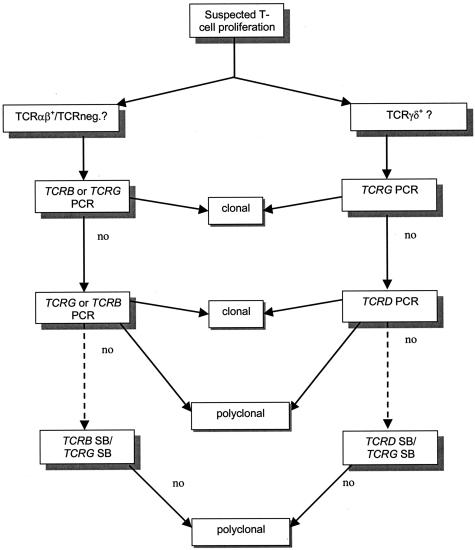
Figure 3.
Strategy for BIOMED-2 multiplex PCR TCR clonality analysis in suspect T-cell proliferations. In TCRαβ+ or TCR-negative T-cell proliferations, there is no clear evidence-based preference for either the TCRB or TCRG locus as the initial target for clonality diagnostics. Thus, either TCRB or TCRG can be used as the first line target, followed by the other locus as the second target. For TCRγδ+ T-cell proliferations, the TCRG locus is the clear first line target for clonality assessment, followed by TCRD as the second target. In the absence of clonal TCR gene rearrangements in PCR, SB analysis might still be considered for all suspect T-cell proliferations, provided that enough high-quality DNA is available. Although the PCR strategy as described here is suitable for fresh or frozen tissue samples, it could possibly be applied to DNA isolated from paraffin-embedded tissue samples, provided that DNA quality is such that PCR products of ∼300 bp can be amplified.
Acknowledgments
We are grateful to Prof. Dr. R. Benner (Department of Immunology, Erasmus MC) for continuous support, Monique Oud and Jos de Vos for technical assistance, and Tar van Os for help in preparing the figures.
References
- Van Dongen JJ, Wolvers-Tettero IL. Analysis of immunoglobulin and T cell receptor genes. Part I: basic and technical aspects. Clin Chim Acta. 1991;198:1–91. doi: 10.1016/0009-8981(91)90246-9. [DOI] [PubMed] [Google Scholar]
- Derksen PW, Langerak AW, Kerkhof E, Wolvers-Tettero IL, Boor PP, Mulder AH, Vrints LW, Coebergh JW, van Krieken JH, Schuuring E, Kluin PM, van Dongen JJ. Comparison of different polymerase chain reaction-based approaches for clonality assessment of immunoglobulin heavy-chain gene rearrangements in B-cell neoplasia. Mod Pathol. 1999;12:794–805. [PubMed] [Google Scholar]
- Arber DA, Braziel RM, Bagg A, Bijwaard KE. Evaluation of T cell receptor testing in lymphoid neoplasms: results of a multicenter study of 29 extracted DNA and paraffin-embedded samples. J Mol Diagn. 2001;3:133–140. doi: 10.1016/S1525-1578(10)60664-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadrach B, Warshawsky I. A comparison of multiplex and monoplex T-cell receptor gamma PCR. Diagn Mol Pathol. 2004;13:127–134. doi: 10.1097/01.pdm.0000126419.92931.a3. [DOI] [PubMed] [Google Scholar]
- Droese J, Langerak AW, Groenen PJ, Bruggemann M, Neumann P, Wolvers-Tettero IL, Van Altena MC, Kneba M, Van Dongen JJ. Validation of BIOMED-2 multiplex PCR tubes for detection of TCRB gene rearrangements in T-cell malignancies. Leukemia. 2004;18:1531–1538. doi: 10.1038/sj.leu.2403428. [DOI] [PubMed] [Google Scholar]
- Beishuizen A, Verhoeven MA, Mol EJ, Breit TM, Wolvers-Tettero IL, van Dongen JJ. Detection of immunoglobulin heavy-chain gene rearrangements by Southern blot analysis: recommendations for optimal results. Leukemia. 1993;7:2045–2053. [PubMed] [Google Scholar]
- Langerak AW, Wolvers-Tettero IL, van Dongen JJ. Detection of T cell receptor beta (TCRB) gene rearrangement patterns in T cell malignancies by Southern blot analysis. Leukemia. 1999;13:965–974. doi: 10.1038/sj.leu.2401427. [DOI] [PubMed] [Google Scholar]
- van Dongen JJ, Langerak AW, Bruggemann M, Evans PA, Hummel M, Lavender FL, Delabesse E, Davi F, Schuuring E, Garcia-Sanz R, van Krieken JH, Droese J, Gonzalez D, Bastard C, White HE, Spaargaren M, Gonzalez M, Parreira A, Smith JL, Morgan GJ, Kneba M, Macintyre EA. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia. 2003;17:2257–2317. doi: 10.1038/sj.leu.2403202. [DOI] [PubMed] [Google Scholar]
- Sandberg Y, Heule F, Lam K, Lugtenburg PJ, Wolvers-Tettero IL, van Dongen JJ, Langerak AW. Molecular immunoglobulin/T-cell receptor clonality analysis in cutaneous lymphoproliferations: experience with the BIOMED-2 standardized polymerase chain reaction protocol. Haematologica. 2003;88:659–670. [PubMed] [Google Scholar]
- Jaffe ES HN, Stein H, Vardiman JW. Lyon: IARC Press; World Health Organization Classification of Tumours. Pathology and Genetics of Tumours of Haematopoietic and Lymphoid Tissues. 2001 [Google Scholar]
- Moreau EJ, Langerak AW, van Gastel-Mol EJ, Wolvers-Tettero IL, Zhan M, Zhou Q, Koop BF, van Dongen JJ. Easy detection of all T cell receptor gamma (TCRG) gene rearrangements by Southern blot analysis: recommendations for optimal results. Leukemia. 1999;13:1620–1626. doi: 10.1038/sj.leu.2401540. [DOI] [PubMed] [Google Scholar]
- Breit TM, Wolvers-Tettero IL, Beishuizen A, Verhoeven MA, van Wering ER, van Dongen JJ. Southern blot patterns, frequencies, and junctional diversity of T-cell receptor-delta gene rearrangements in acute lymphoblastic leukemia. Blood. 1993;82:3063–3074. [PubMed] [Google Scholar]
- Raab-Traub N, Flynn K. The structure of the termini of the Epstein-Barr virus as a marker of clonal cellular proliferation. Cell. 1986;47:883–889. doi: 10.1016/0092-8674(86)90803-2. [DOI] [PubMed] [Google Scholar]
- Drexler H. The Leukemia-Lymphoma Cell Line Facts Book. San Diego, CA: Academic Press,; 2001 [Google Scholar]
- Langerak AW, Wolvers-Tettero LM, van Dongen JJM. Immunoglobulin and T-cell receptor gene analysis in the diagnosis of lymphoid malignancies. Rev Clin Exp Hematol. 1997;3:3–27. [Google Scholar]
- Linke B, Bolz I, Fayyazi A, von Hofen M, Pott C, Bertram J, Hiddemann W, Kneba M. Automated high resolution PCR fragment analysis for identification of clonally rearranged immunoglobulin heavy chain genes. Leukemia. 1997;11:1055–1062. doi: 10.1038/sj.leu.2400736. [DOI] [PubMed] [Google Scholar]
- Kneba M, Bolz I, Linke B, Hiddemann W. Analysis of rearranged T-cell receptor beta-chain genes by polymerase chain reaction (PCR) DNA sequencing and automated high resolution PCR fragment analysis. Blood. 1995;86:3930–3937. [PubMed] [Google Scholar]
- Kitano K, Ichikawa N, Shimodaira S, Ito T, Ishida F, Kiyosawa K. Eosinophilia associated with clonal T-cell proliferation. Leuk Lymphoma. 1997;27:335–342. doi: 10.3109/10428199709059688. [DOI] [PubMed] [Google Scholar]
- Simon HU, Plotz SG, Dummer R, Blaser K. Abnormal clones of T cells producing interleukin-5 in idiopathic eosinophilia. N Engl J Med. 1999;341:1112–1120. doi: 10.1056/NEJM199910073411503. [DOI] [PubMed] [Google Scholar]
- Brugnoni D, Airo P, Rossi G, Bettinardi A, Simon HU, Garza L, Tosoni C, Cattaneo R, Blaser K, Tucci A. A case of hypereosinophilic syndrome is associated with the expansion of a CD3−CD4+ T-cell population able to secrete large amounts of interleukin-5. Blood. 1996;87:1416–1422. [PubMed] [Google Scholar]
- Cogan E, Schandene L, Crusiaux A, Cochaux P, Velu T, Goldman M. Brief report: clonal proliferation of type 2 helper T cells in a man with the hypereosinophilic syndrome. N Engl J Med. 1994;330:535–538. doi: 10.1056/NEJM199402243300804. [DOI] [PubMed] [Google Scholar]
- Roufosse F, Schandene L, Sibille C, Willard-Gallo K, Kennes B, Efira A, Goldman M, Cogan E. Clonal Th2 lymphocytes in patients with the idiopathic hypereosinophilic syndrome. Br J Haematol. 2000;109:540–548. doi: 10.1046/j.1365-2141.2000.02097.x. [DOI] [PubMed] [Google Scholar]
- O’Shea JJ, Jaffe ES, Lane HC, MacDermott RP, Fauci AS. Peripheral T cell lymphoma presenting as hypereosinophilia with vasculitis: clinical, pathologic, and immunologic features. Am J Med. 1987;82:539–545. doi: 10.1016/0002-9343(87)90458-x. [DOI] [PubMed] [Google Scholar]
- Nador RG, Chadburn A, Gundappa G, Cesarman E, Said JW, Knowles DM. Human immunodeficiency virus (HIV)-associated polymorphic lymphoproliferative disorders. Am J Surg Pathol. 2003;27:293–302. doi: 10.1097/00000478-200303000-00002. [DOI] [PubMed] [Google Scholar]
- Nihal M, Mikkola D, Wood GS. Detection of clonally restricted immunoglobulin heavy chain gene rearrangements in normal and lesional skin: analysis of the B cell component of the skin-associated lymphoid tissue and implications for the molecular diagnosis of cutaneous B cell lymphomas. J Mol Diagn. 2000;2:5–10. doi: 10.1016/S1525-1578(10)60609-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elenitoba-Johnson KS, Bohling SD, Mitchell RS, Brown MS, Robetorye RS. PCR analysis of the immunoglobulin heavy chain gene in polyclonal processes can yield pseudoclonal bands as an artifact of low B cell number. J Mol Diagn. 2000;2:92–96. doi: 10.1016/S1525-1578(10)60622-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muche JM, Sterry W, Gellrich S, Rzany B, Audring H, Lukowsky A. Peripheral blood T-cell clonality in mycosis fungoides and nonlymphoma controls. Diagn Mol Pathol. 2003;12:142–150. doi: 10.1097/00019606-200309000-00005. [DOI] [PubMed] [Google Scholar]
- Mitterbauer-Hohendanner G, Mannhalter C, Winkler K, Mitterbauer M, Skrabs C, Chott A, Simonitsch-Klupp I, Gleiss A, Lechner K, Jaeger U. Prognostic significance of molecular staging by PCR-amplification of immunoglobulin gene rearrangements in diffuse large B-cell lymphoma (DLBCL). Leukemia. 2004;18:1102–1107. doi: 10.1038/sj.leu.2403376. [DOI] [PubMed] [Google Scholar]
- Scarisbrick JJ, Whittaker S, Evans AV, Fraser-Andrews EA, Child FJ, Dean A, Russell-Jones R. Prognostic significance of tumor burden in the blood of patients with erythrodermic primary cutaneous T-cell lymphoma. Blood. 2001;97:624–630. doi: 10.1182/blood.v97.3.624. [DOI] [PubMed] [Google Scholar]
- Theodorou I, Bigorgne C, Delfau MH, Lahet C, Cochet G, Vidaud M, Raphael M, Gaulard P, Farcet JP. VJ rearrangements of the TCR gamma locus in peripheral T-cell lymphomas: analysis by polymerase chain reaction and denaturing gradient gel electrophoresis. J Pathol. 1996;178:303–310. doi: 10.1002/(SICI)1096-9896(199603)178:3<303::AID-PATH475>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Posnett DN, Sinha R, Kabak S, Russo C. Clonal populations of T cells in normal elderly humans: the T cell equivalent to “benign monoclonal gammapathy.”. J Exp Med. 1994;179:609–618. doi: 10.1084/jem.179.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delfau-Larue MH, Laroche L, Wechsler J, Lepage E, Lahet C, Asso-Bonnet M, Bagot M, Farcet JP. Diagnostic value of dominant T-cell clones in peripheral blood in 363 patients presenting consecutively with a clinical suspicion of cutaneous lymphoma. Blood. 2000;96:2987–2992. [PubMed] [Google Scholar]
- Hodges E, Krishna MT, Pickard C, Smith JL. Diagnostic role of tests for T cell receptor (TCR) genes. J Clin Pathol. 2003;56:1–11. doi: 10.1136/jcp.56.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assaf C, Hummel M, Steinhoff M, Geilen CC, Orawa H, Stein H, Orfanos CE. Early TCR-{beta} and TCR-{gamma} PCR detection of T-cell clonality indicates minimal tumor disease in lymph nodes of cutaneous T-cell lymphoma: diagnostic and prognostic implications. Blood. 2005;105:503–510. doi: 10.1182/blood-2004-06-2220. [DOI] [PubMed] [Google Scholar]
- Fraser-Andrews EA, Woolford AJ, Russell-Jones R, Seed PT, Whittaker SJ. Detection of a peripheral blood T cell clone is an independent prognostic marker in mycosis fungoides. J Invest Dermatol. 2000;114:117–121. doi: 10.1046/j.1523-1747.2000.00830.x. [DOI] [PubMed] [Google Scholar]
- Vega F, Luthra R, Medeiros LJ, Dunmire V, Lee SJ, Duvic M, Jones D. Clonal heterogeneity in mycosis fungoides and its relationship to clinical course. Blood. 2002;100:3369–3373. doi: 10.1182/blood.V100.9.3369. [DOI] [PubMed] [Google Scholar]
- Loughran TP, Jr, Starkebaum G, Aprile JA. Rearrangement and expression of T-cell receptor genes in large granular lymphocyte leukemia. Blood. 1988;71:822–824. [PubMed] [Google Scholar]
- Vonderheid EC, Bernengo MG, Burg G, Duvic M, Heald P, Laroche L, Olsen E, Pittelkow M, Russell-Jones R, Takigawa M, Willemze R. Update on erythrodermic cutaneous T-cell lymphoma: report of the International Society for Cutaneous Lymphomas. J Am Acad Dermatol. 2002;46:95–106. doi: 10.1067/mjd.2002.118538. [DOI] [PubMed] [Google Scholar]