Abstract
Pleural effusions may result from various inflammatory, hemodynamic, or neoplastic conditions. A common diagnostic problem lies in distinguishing malignant from benign pleural effusions using routine cytological evaluation. We studied pleural fluid samples obtained from 14 patients with histologically confirmed malignancy and from 6 patients with benign pleural effusions using 12 microsatellite markers from 8 different chromosomal regions. Supernatants and cellular sediments of all 20 pleural fluid samples were analyzed. Routine cytological examination was 100% specific for malignancy but was only 57% sensitive. Microsatellite analyses of pleural fluid supernatants showed genetic alterations in tumor patients only. However, 50% of pleural effusions that were considered negative for malignancy by routine cytological analysis showed either loss of heterozygosity or microsatellite instability. The sensitivity of pleural fluid examination rose to 79% when routine cytological assessment was supplemented by molecular studies. Our data suggest that microsatellite analysis increases the sensitivity of cytological pleural fluid examination in assessing potential malignancy and that combining cytological and molecular methods may improve yield and certainty in diagnostically challenging cases.
Pleural effusion is a common manifestation of various underlying inflammatory, hemodynamic, or neoplastic diseases. In healthy individuals, no more than 15 ml of serous, paucicellular, clear fluid lubricates the pleural surface. Accumulation of pleural fluid may occur through several mechanisms: 1) increased hydrostatic pressure in congestive heart failure; 2) increased vascular permeability in inflammation; 3) decreased oncotic pressure in renal disease; 4) decreased intrapleural pressure in atelectasis; and 5) increased fluid production coupled with decreased lymphatic drainage in tumors.1 About 20% of pleural effusions are due to malignancy, and 50% of these are due to primary lung cancer. Pleural carcinomatosis is frequently seen in lung and breast cancer, and a common clinical problem is the distinction between benign and malignant pleural effusions.2,3 Advanced tumors may involve the pleura by direct extension or through lymphatic and hematogenous spread. Serous or serosanguineous pleural effusion can be seen in both malignant and infectious/inflammatory conditions, underscoring the diagnostic value of fluid cytological examination.
Diagnostic accuracy of pleural effusion cytology depends on the volume of liquid examined, the type of preparation and staining, the experience of the examiner, and the number of sufficient specimens investigated. However, the cytological interpretation of pleural fluids can be challenging, and its diagnostic accuracy is limited.4,5 The sensitivity of conventional cytology ranges from 43%6 to 71%7 with an average of 58%.8
To supplement the morphological examination of doubtful cytological effusions, immunocytochemistry and a variety of molecular methods have frequently been applied.9,10 The use of flow cytometry or image analysis11,12 in the diagnosis of malignant effusions were evaluated in several studies. Generally, the sensitivity and specificity of that was not superior to current immunocytochemistry.13
The use of fluorescence in situ hybridization allows the detection of aneuploidy. Approximately one-fourth of the cytologically negative effusions was fluorescence in situ hybridization (FISH) positive and vice versa. From the initially FISH-negative effusions, 18.9% could subsequently be classified positive with dual-color FISH by visualization of intranuclear chromosomal complexity in rare aneuploid cells. Thus, “overall FISH analysis,” including dual-color evaluation, identified tumor cells in significantly more effusions than conventional cytology, implying greater sensitivity.14
Using reverse transcription polymerase chain reaction, other authors have described higher sensitivity than in conventional routine cytology (72 versus 52%). Different expression levels of glycoprotein MUC 1 and MUC 5 were used to discriminate between benign and malignant effusions.15
Studies of peritoneal washing and pleural effusion investigating telomerase activity reported a high sensitivity and specificity for tumor detection (91 versus 94%). However, most studies for telomerase activity did not show results that surpass routine cytology, and one study reported frequent false-positive results in benign effusions, thereby questioning the validity of this test for effusion diagnostics.16 Especially the detection of specific mutations in tumor-associated genes like p53 is very sensitive in detection cells with this specific genetic alteration.9,17,18 However this method is restricted to cases with certain mutations in the tumor.
Promotor hypermethylation is now accepted as a further very frequent occurring mechanism of tumor suppression genes. Ahrendt et al19 could show that methylation assays were more sensitive than microsatellite analysis. However, methylation-specific polymerase chain reaction needs an initial step of amplified bisulfite-modified DNA with flanking polymerase chain reaction.20 In our hands, this method was very problematic when investigating biological samples with minute amounts of DNA or only degraded DNA. Because only small amounts of DNA were detectable in cell-free supernatants, we chose as first method microsatellite analysis, which was very robust in this study.
Loss of heterozygosity (LOH) is a further widely accepted molecular hallmark of malignancy, progressively accumulating during carcinogenesis at various chromosomal regions,21 but less agreement exists on the importance and frequency of microsatellite instability (MSI) in cancers.22 An increased frequency of microsatellite alterations has been observed in various cancers. These elevated microsatellite alterations at selected tetranucleotide repeat markers were related to high frequency of p53 mutations and do not seem to display a defect mismatched repair.22,23 These frequent microsatellite alterations were found predominantly in advanced tumor stages24 and patients with lymph node metastasis.25
Molecular detection of malignancy in body fluids through microsatellite analysis is promising (reviewed in 26), but reliable detection of cancer-related LOH or MSI typically requires 60 to 80% tumor content.27,28 Pleural effusion samples usually contain only a few epithelial cells and show significant numbers of reactive mesothelium and inflammatory cells. Density gradient separation may improve this ratio in favor of epithelial cells, resulting in slightly higher detection rates of LOH in tumor patients (63 versus 58%).29 Increased levels of free DNA and cancer-related genomic alterations have been described in cell-free plasma30 and serum31,32 of cancer patients.33
To clarify whether molecular analysis of cell-free pleural supernatants could improve diagnostic utility and identification of malignant effusions, we compared the frequency of microsatellite instability of the pleural sediment and supernatants with results of the conventional cytological examination and molecular alterations in the matched tumor tissue samples by using 12 microsatellite markers at 8 different chromosomal regions.
Materials and Methods
Patients
This study was approved by the Institutional Review Board. Twenty individuals (4 female and 16 male) who were treated for pleural effusion from 2003 to 2004 at the University Hospital Regensburg were included. The median age was 67 years of age (range, 45–84). Fourteen patients had a histologically confirmed pulmonary malignancy (median age, 66 years; range, 59–84 years), including non-small-cell lung cancer (n = 6), small-cell lung cancer (n = 1), and metastatic nonpulmonary carcinoma (n = 7). All patients had a long-term smoking history; in one tumor patient, an asbestos exposure was documented. The control group with benign pleural effusions (pleuritis (n = 3) and congestive heart failure (n = 3)) included patients with a median age of 64 years (range, 45–79 years). Five patients had a long-term smoking history (mean, 35 pack years). One patient was nonsmoker. Only eight malignancy-associated pleural effusions were cytologically positive for tumor cells, whereas all six patients with benign pleural effusions were diagnosed correctly as negative for malignant cells in routinely prepared cytology specimens by two independent experienced cytopathologists (Table 1). In all 14 patients, the malignancy was histologically confirmed by biopsies (seven primary intrathoracic cancers: six non-small cell lung cancers (NSCLC) and one small cell lung cancer (SCLC); seven primary extrathoracic cancers: metastasis of one gastric carcinoma, two prostate cancer, one bladder cancer, one breast cancer, one larynx carcinoma, and one thyroid carcinoma). In all patients with malignant diseases, recurrent pleural effusions were diagnosed. In 57% (8 of 14 patients), a positive cytology sample confirmed the suspicion of malignant pleural effusion. In six patients, there was no positive cytology in repeated pleural samples. During the clinical course in all six patients with underlying malignant disease (two patients with primary intrathoracic cancer and four patients with primary extrathoracic cancer), distant metastases were diagnosed by CT or X-ray. The clinical diagnosis of recurrent pleural effusions without therapeutic response in patients with multiple distant metastases strongly suggests the malignancy of the pleural effusions of these patients.34 However, there was no final proof of this. In the six patients of the control group, there was never a consideration of malignancy during the clinical course and radiological controls. In one patient, an additionally performed biopsy confirmed only a pneumonic infiltration.
Table 1.
Clinical and Histopathological Data and Results of Microsatellite Analyses of Pleural Supernatants and Corresponding Tumor Samples
| Clinical data | ||||||||||||||||||||
| Patient ID | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 |
| Sex* | m | m | m | m | m | m | m | f | f | m | f | m | m | m | m | f | m | m | m | m |
| Age at diagnosis (years) | 64 | 59 | 65 | 46 | 67 | 66 | 70 | 71 | 75 | 65 | 68 | 69 | 84 | 56 | 79 | 45 | 69 | 64 | 79 | 48 |
| Cases with histological confirmed malignancy† | 1‡ | 1§ | 1¶ | 1§ | 1§ | 1§ | 1|| | 1** | 1†† | 1§ | 1‡‡ | 1¶ | 1§§ | 1§ | 0 | 0 | 0 | 0 | 0 | 0 |
| Cytologic classification¶¶ | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Microsatellite analysis of cell-free pleural supernatant | ||||||||||||||||||||
| D2S1266 | □ | na | □ | □ | ○ | □ | ○ | □ | □ | □ | □ | □ | ○ | □ | ○ | □ | ○ | □ | ○ | ○ |
| D2S424 | □ | • | □ | □ | □ | □ | ○ | na | na | na | na | na | na | na | na | □ | na | na | na | □ |
| D3S1300 | □ | na | na | ○ | na | □ | na | ○ | □ | ○ | □ | □ | na | □ | □ | □ | □ | □ | ○ | ○ |
| D3S2304 | □ | na | • | ○ | ○ | ○ | □ | ○ | • | • | □ | ○ | □ | □ | □ | ○ | ○ | □ | □ | □ |
| D3S4597 | □ | • | □ | □ | □ | □ | • | □ | □ | □ | □ | □ | □ | □ | □ | ○ | □ | □ | ○ | □ |
| D3S4623 | □ | na | □ | □ | □ | • | na | □ | □ | □ | □ | □ | □ | □ | □ | ○ | □ | □ | □ | □ |
| D3S1539 | □ | na | ○ | □ | □ | □ | □ | na | na | na | na | na | na | na | na | □ | na | na | na | □ |
| D12S363 | □ | □ | • | ○ | □ | □ | ○ | • | ○ | □ | ▴ | ○ | □ | □ | □ | □ | □ | ○ | na | ○ |
| D12S827 | □ | • | na | • | □ | □ | □ | na | ○ | ○ | na | ○ | ○ | □ | □ | □ | □ | □ | na | □ |
| D12S335 | □ | na | na | □ | □ | ○ | na | na | ○ | ○ | □ | ○ | na | □ | □ | □ | □ | □ | na | □ |
| D5S346 | □ | • | • | □ | □ | □ | • | ○ | □ | □ | ▴ | □ | □ | □ | □ | □ | □ | □ | □ | □ |
| TP53.Alu | □ | na | na | □ | • | • | na | □ | ○ | □ | ○ | ○ | □ | □ | □ | ○ | ○ | □ | na | □ |
| Total number of markers showing LOH/MSI | 0 | 4 | 3 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Microsatellite analysis of corresponding tumor samples | ||||||||||||||||||||
| D2S1266 | ▴ | □ | ○ | □ | ▴ | □ | na | ○ | • | ○ | • | ○ | □ | na | ||||||
| D2S424 | ○ | □ | ○ | □ | □ | □ | na | na | na | ○ | na | na | □ | na | ||||||
| D3S1300 | na | ○ | ○ | ▴ | na | □ | na | □ | • | • | na | ○ | na | na | ||||||
| D3S2304 | • | □ | na | ○ | □ | ○ | na | ○ | • | □ | ○ | □ | na | na | ||||||
| D3S4597 | ▴ | • | na | □ | na | □ | na | □ | □ | ○ | □ | ○ | na | na | ||||||
| D3S4623 | na | • | ○ | ▴ | ▴ | • | na | □ | • | □ | □ | □ | na | na | ||||||
| D3S1539 | □ | na | na | • | ○ | □ | na | na | na | na | na | □ | na | na | ||||||
| D12S363 | ▴ | □ | na | ○ | □ | □ | na | na | ○ | ○ | ▴ | ▴ | □ | na | ||||||
| D12S827 | na | ○ | ▴ | ○ | na | □ | na | □ | ○ | ○ | na | na | na | na | ||||||
| D12S335 | na | ▴ | ○ | □ | ○ | ○ | na | □ | ○ | ○ | na | na | na | na | ||||||
| D5S346 | ▴ | □ | • | □ | ○ | □ | na | • | □ | □ | na | □ | □ | na | ||||||
| TP53.Alu | na | • | • | □ | ○ | • | na | □ | ○ | ○ | na | □ | □ | na | ||||||
| Total number of markers showing LOH/MSI | 5 | 4 | 3 | 3 | 2 | 2 | na | 2 | 4 | 1 | 2 | 1 | 0 | na | ||||||
•, LOH; ○, noninformative; ▴, MSI; □, no microsatellite alteration; na, data not available.
m, male; f, female.
0, nonmalignant; 1, histological confirmed malignancy.
‡¶||**††§§ Lung metastasis of gastric, prostate, bladder, breast, thyroid, and larynx carcinoma, respectively.
Primary NSCLC.
Primary SCLC.
0, negative cytological result; 1, positive cytological result.
Cytological Samples and DNA Isolation
Pleural fluid was centrifuged (1200 × g for 10 minutes) at 4°C. The two paired fractions, cellular pellet and corresponding supernatant, were used for DNA isolation. Matched control DNA either was isolated from peripheral blood (EDTA blood) or from benign archival tissue (normal tissue) of each patient as described previously,35 using the MagNa Pure LC DNA Isolation kit (Roche Diagnostics GmbH, Mannheim, Germany) according to the manufacturer’s instructions.
Histological Samples and DNA Isolation
Although the malignancy was histologically confirmed in all 14 patients by biopsies in 2 patients, sufficient paraffin embedded tumor tissue was not available to perform further molecular examinations. All available 12 matched tumor tissues were examined by the same 12 microsatellite markers. DNA was isolated from matched formalin-fixed paraffin-embedded tissue sections as described previously.36 Deparaffinized tissue sections (5 μm) were stained with methylene blue for 15 seconds and subsequently microdissected either manually or by laser microdissection (P.A.L.M., Laser Microdissection System, Microlaser Technologies AG, Bernried, Germany) by a pathologist (M.W.). DNA was extracted using the MagNa Pure LC DNA Isolation kit II and the MagNa Pure LC with corresponding software 2.0 (Roche Diagnostics) following the manufacturer’s instructions. As control, DNA was isolated from peripheral blood (EDTA blood) or from benign archival tissue (normal tissue) of each patient.
Microsatellite Markers, Polymerase Chain Reaction (PCR) Protocol, and Microsatellite Analysis
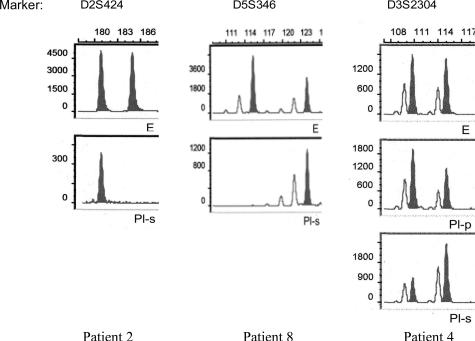
LOH and MSI were evaluated using 12 fluorescence-labeled primer pairs chosen from the Genome Database (http://www.gdb.org) and manufactured by metaBIOn GmbH (Martinsried, Germany). The microsatellite repeat polymorphisms were located at eight chromosomal regions: 2q35-36 (D2S424 and D2S1266), 3p14 (D3S1300), 3p21 (D3S2304, D3S4597, and D3S4623), 3p25 (D3S1539), 12p12 (D12S363 and D12S827), 12q14 (D12S335), 5q21-q22 (D3S346 at the APC gene), and 17p13 (TP53.Alu at the p53 gene) (Table 2). Paired DNA samples were amplified by PCR in a final volume of 25 μl containing 1.5 mmol/L MgCl2, 10 mmol/L dNTPs, 15 μmol/L primer, and 0.2 U of Taq polymerase (Roche Diagnostics), using 2 μl of isolated DNA as template. The reaction mixture was denatured 3 minutes at 94°C, followed by 40 cycles at 94°C, at specific annealing temperature (Table 2), and at 72°C for 1 minute each. A final extension step (72°C for 8 minutes) was included. One microliter of labeled amplified DNA was mixed with formamide and GeneScan Rox-400 size standard (Applied Biosystems, Warrington, UK). The samples were denatured for 2 minutes at 95°C and subsequently analyzed by capillary electrophoresis using an ABI Prism 3100 Genetic Analyzer and GeneScan Analysis software 3.7 (Applied Biosystems, Foster City, CA). LOH was determined by comparing the calculated allele ratios of normal tissue (EDTA blood, normal tissue), supernatant, and cell pellet in each pleural effusion sample (Figure 1). LOH was determined by calculating the ratio of quotient 1 ((peak height of normal allele 2)/(peak height of normal allele 1)) and quotient 2 ((peak height of abnormal allele 2)/(peak height of abnormal allele 1)). LOH was defined by a ratio of less than 0.5 or greater than 2.0.
Table 2.
Microsatellite Markers
| Chromosomal location | Marker | Primer sequence (5′ to 3′) | PCR-Tm (°C) | bp |
|---|---|---|---|---|
| 2q35 | D2S424 | s: Hex-TGGTGGAGATGTTTAAAGGC | 56 | ∼177 |
| a: GGGCAGGAAAAGGATTGTAT | ||||
| D2S1266 | s: Fam-GACGTAAATGTAAGACACCAA | 58 | ∼254 | |
| a: TCAGGATATTTAGGGTATCCA | ||||
| 3p14.2 | D3S1300 | s: Ned-AGCTCACATTCTAGTCAGCCT | 52 | ∼217–241 |
| a: GCCAATTCCCCAGATG | ||||
| 3p21.3 | D3S4597 | s: Fam-TGTTCCCTTCCCTATAAACAGAT | 53 | ∼167 |
| a: GAAAGCAAGGAAGGCACATG | ||||
| D3S4623 | s: Hex-GGCAGTGTTTGAGCTTACATGGG | 69 | ∼172 | |
| a: CAGGCTCTGGAAACCAAGC | ||||
| D3S2304 | s: Hex-GCGACACAGCGAGACTCTA | 56 | ∼113 | |
| a: AATATGGGGGTGGGAGGCA | ||||
| 3p25 | D3S1539 | s: Hex-CTTTCCATTACTCTCTCCATA | 53 | ∼285 |
| a: CCAGTGCTGTTTTAGCTTC | ||||
| 12p12.3 | D12S827 | s: Hex-GCTAAGGTGGGAGGATCAC | 56 | ∼255 |
| a: TCTGGACTGATGCTCGCTT | ||||
| D12S363 | s: Fam-GAGGGGTGGCATCTCT | 56 | ∼208 | |
| a: CTCAAATGAAATCAGCATAAA | ||||
| 12q14.1 | D12S335 | s: Fam-TCATCCAGGCTTCACC | 57 | ∼255 |
| a: TGGCAAGGACAGACACA | ||||
| 5q21-q22 | D5S346 | s: Hex-ACTCACTCTAGTGATAAATCGGG | 55 | ∼96–122 |
| (APC) | a: AGCAGATAAGACAGTATTACTAGTT | |||
| 17p13.1 | TP53.Alu | s: Hex-TCGAGGAGGTTGCAGTAAGCGGA | 59 | ∼150 |
| a. AACAGCTCCTTTAATGGCAG |
Figure 1.
Examples demonstrating LOH in pleural supernatants; E, DNA from EDTA blood samples; Pl-s, pleural supernatant; Pl-p, pleural cellular pellet.
Alternative LOH criteria (ratios of <0.6 or <0.7, and >1.67 or >1.43, respectively) were tested as well.37,38 MSI was defined as the occurrence of additional peaks compared with control DNA. Loci with MSI were not scored for LOH. All samples showing either LOH or MSI were confirmed in an independent PCR reaction to exclude false-positive results due to preferential amplification of one allele during the PCR reaction.
Statistical Analyses
Statistical analyses were performed using SPSS version 10.0 (SPSS, Chicago, IL), using a P values of <0.05 as the significance level. For simple testing procedures, P values were analyzed in relation to clinical and pathological data using the two-sided Fisher’s exact test. The McNemar test for dependent variables was used to compare results of cytology and LOH analysis and to compare the molecular analysis of pleural effusion samples with tumor tissue samples.
Results
Cytological Examination of Benign and Malignant Pleural Effusions
A positive cytology sample was seen in 8 of 14 cases with histologically confirmed pulmonary malignancy. All six patients with benign disease were cytologically negative for malignant cells. The specificity of pleural fluid cytology was 100% with a sensitivity of only 57%.
Microsatellite and LOH Analyses in the Cellular Sediment of Pleural Effusions
Only one tumor-associated case showed LOH with the microsatellite markers D2S1266, D3S4623, and D12S363 in the cellular pellet fraction of the pleural effusion, translating into a specificity of 100% and a sensitivity of 7% (1 of 14). The corresponding cytological examination was positive for malignant cells.
MSI and LOH Analyses in Supernatants of Pleural Effusions
The frequency of LOH at the investigated markers ranged from 20% (4 of 20; D5S346) to 6% (1 of 18; D3S4623). Four markers (D2S1266, D3S1300, D3S1539, and D12S335) did not show any molecular alterations (Table 1). Patients with pulmonary malignancy showed significantly more molecular alterations in the cell-free supernatant compared with patients with benign underlying disease (P = 0.014). LOH and MSI were detectable in supernatants in 71% (10 of 14) of malignant cases. Sixty-four percent (9 of 14) showed LOH only, whereas one patient showed MSI with two markers (D5S346 and D12S363). Cases with positive tumor cytology showed LOH in 87% (7 of 8), and four cases showed allelic losses at two or more chromosomal loci. Fifty percent (3 of 6) of tumor patients with negative pleural fluid cytology revealed LOH and MSI. One of those cases was found to be cytologically positive for tumor cells during a follow-up visit. In three patients with histologically confirmed malignancy, both cytological examination and molecular analyses revealed normal results. No microsatellite alterations were seen in a single case with positive tumor cytology. All nonmalignancy-associated cases revealed negative results in molecular examinations. Molecular examination revealed a specificity of 100% and a sensitivity of 71% using all 12 markers. Consolidating the test panel to only five frequently affected markers (D3S2304, D3S4597, D12S363, D5S346, and TP53.Alu) did not decrease sensitivity. We also tested previously published alternative LOH cut-off criteria (ratios of <0.6 and >1.67),37,38 which improved the sensitivity of the molecular analysis to 79%, while retaining a 100% specificity. Application of less stringent criteria for LOH (ratio of <0.7 and >1.43) resulted in increased sensitivity (86%), but the specificity decreased to 83%. Combination of routine cytological and molecular analyses increased the sensitivity to 79% and did not change the 100% specificity.
Microsatellite and LOH Analyses in Matched Histological Samples
Although the malignancy was histologically confirmed in all 14 patients by biopsies in 2 patients, paraffin-embedded tumor tissue was not sufficiently available to perform further molecular examinations. All 12 matched tumor tissues were examined by applying the same 12 microsatellite markers (Table 1). The percentage of marker-detectable molecular alterations (LOH and/or MSI) in matched paraffin-embedded tumor tissue samples ranged from 41% (5 of 12; D11S2364) to 8.3% (1 of 12; D12S335). The marker D2S424 did not show any molecular alterations. In 11 of 12 tumors (91.6%), allelic imbalances were detectable, including 100% of the primary intrathoracic tumors (6 of 6) and 83% (5 of 6) of the primary extrathoracic tumors. An accumulation of two or more LOH/MSI in various chromosomal regions was found in 75% (9 of 12) of the tumor samples.
Comparison between Microsatellite and LOH Analyses in Matched Histological Samples and Pleural Effusion Samples
Comparing the results of microsatellite analysis with the molecular analysis of the pleural supernatants statistically using the McNemar test for dependent variables, we found no significant differences between the frequency of molecular alterations in the matched probes (P = 1.0). In five primary NSCLCs, at least some of the mutations detectable in the tumor tissue and the matched supernatants of pleural effusions overlapped. Comparing the tumor tissue of primary extrathoracic cancers with the matched cell-free pleural supernatants revealed different molecular alterations.
Discussion
Pleural effusions are the result of various underlying conditions such as congestion, inflammation, and cancer. Cytological examination is of considerable diagnostic value to distinguish between benign and malignant pleural effusions, but interpretation can be challenging if inflammation or reactive mesothelial cells obscure the sample. The sensitivity of conventional cytology is described with an average of 58%.8 In our study, conventional cytological examination of pleural fluid detected tumor cells with a sensitivity of 57%.
Allelic losses at chromosomes 339 (3p21.3, 3p14.2, and 3p25), 5q21-q22,40 and at 17p13.141,42 have been described as further early genetic changes in various pulmonary malignancies, such as lung cancer,36,43 metastatic breast cancer,44 or other cancers.45,46,47 Confirming this, we found molecular alterations in 91.6% of the matched tumor tissue samples, and in 75%, an accumulation of two or more LOH/MSI in each case. Statistically comparing the microsatellite analysis of tumor samples with matched pleural supernatants by using the McNemar test for dependent variables did not show significant differences between the frequency of molecular alterations in the matched probes (P = 1,0). However, there were not the same specific mutations in pleural supernatants like in the matched tumor tissue detectable in every case. Differences in molecular alterations between cells in solid tumors and body fluids were also reported before.48,49,50,51 Besides the same specific mutations in cytological smears as those found in resected paraffin-embedded tissue, Yuichi et al52 observed additional molecular alterations in tumor tissue in comparison with pleural fluid. On the other hand Nara de Matos Grania et al53 found more frequent LOHs in pleural effusions than in the matched tumor tissues in breast cancer patients. In contrast, in our study, patients with various malignant extra- and intrathoracic cancers were included. Even though we found similar specific alterations in all primary NSCLCs and matched supernatants of pleural effusions, we also found different single mutations especially in primary extrathoracic cancers with manifestation of malignant pleural effusion. These inconsistent results in pleural effusions and matched tumors could be caused by tumor heterogeneity during progression and distant spread of tumor cells.54,55
Using our microsatellite marker panel, we were able to detect molecular alterations in 91.6% of the matched tumor tissue samples and in 71% of the malignant pleural supernatants. Four markers (D2S1266, D3S1300, D3S1539, and D12S335) did not show any molecular alterations in pleural supernatants. In one matched tumor tissue sample, we found no corresponding LOH/MSI. One of this markers (D3S1539) was noninformative in the majority of cases (six cases with tumor and two benign), and two others (D3S1300 and D2S335) were only informative in 70 to 75% of the cases. Therefore the molecular analysis could be optimized by using further microsatellite markers at chromosomal loci 3p25, 3p14.2, and 12q14.1. The most frequently affected markers in our study include D3S2304, D3S4597, D12S363, D5S346, and TP53.Alu. Considering the fact that malignant cells in effusions accumulate metastatic attributes, the use of further microsatellite markers that are involved in cell adhesion and infiltration could be added.53
Besides allelic losses, other mechanisms than aberrant promoter hypermethylation are accepted for silencing of tumor suppressor genes. However, methylation-specific polymerase chain reaction needs bisulfite-modified DNA as template. Because we have only small amounts of DNA in cell-free supernatants and microsatellite analysis has no intermediary further modifying step like bisulfite-modification, we used microsatellite analysis with various microsatellite markers for molecular detection of cancer-related alterations. By increasing the sensitivity of a PCR approach to detect methylated DNA sequences, Palmisano et al56,57 demonstrated that aberrant methylation of the p16 and/or O6-methyl-guanine-DNA methyltransferase promoters can be detected in small DNA samples. The use of aberrant gene methylation as a molecular marker system seems to offer a potentially powerful additional target for following studies.
In our study, the molecular analysis of the cellular sediment revealed a sensitivity of only 7%, far below the sensitivity level of conventional cytology. This is likely to reflect the need of large numbers of intrapleural tumor cells for successful microsatellite analysis, whereas only few malignant cells are required for a positive cytological diagnosis. Lee et al29 increased the sensitivity of molecular examination of cell pellets up to 63% using extensive cell segregation by density gradient separation. The sensitivity of pleural effusion cytology also depends on the number of sufficient specimens investigated. Whereas 53% tumor-cell positive effusions were detected examining one specimen, the sensitivity increased to 73% when three specimens were examined. Other studies reported that immunocytochemical detection of Ber-EP4 and CD44 in doubtful cases could improve the sensitivity of cytology up to 78%8 and 83%.58,59 This suggests that an additionally performed molecular diagnostic test should ideally not fractionate the cell pellets, which could diminish the likelihood to detect few tumor cells in cytological examinations.
Immunocytochemistry is helpful in cytologically doubtful cases. We did not compare immunocytochemistry with molecular detection in this study, because after cytology and DNA isolation, no additional material was available for further studies. However, the combination of cytology and immunocytochemistry on the cells in the specimens and microsatellite analyses using the free DNA could potentially complement each other and improve the sensitivity of tumor detection in pleural fluid.
The current study used a new finding that tumor DNA is also detectable in cell-free body fluid.60 Extensive research has been undertaken to diagnose bladder cancer by DNA analysis of exfoliated tumors cells in urine samples. Eisenberger et al61 reported detection of tumor DNA in urine samples in 76% of their patients with renal cell carcinoma but did not separate urinary fluid from cells. It is unclear whether cellular or cell-free tumor-DNA in the urine accounted for their positive test results. Uttinger et al62 described that microsatellite analysis of cell-free urinary supernatants might identify more cancer cases than analysis of urine cell sediments, consistent with results of the current study. Wang et al63,64 could also improve the detection for p53 mutations with high cancer specificity by analyzing the supernatants of pure pancreatic juice. It is possible that in supernatants, the percentage of tumor DNA is larger when compared with “contaminating” normal DNA from benign inflammatory or mesothelial cells, thus making LOH more detectable. The higher concentration of free nucleic acid of tumor cells in cell-free pleural supernatants can be explained by the selective growth advances of tumor cells, possible passive leakage of tumor DNA, or active secretion during tumor progression.65 Coulet et al66 have discussed PCR artifacts attributable to limited amounts of template DNA. However, our study detected PCR products in all cases, and an independent amplification was performed in all positive cases to avoid false-positive results due to preferential amplification of one allele.
The standard criterion for LOH analysis using the ABI Prism Genetic Analyzer was a reproducible ratio of <0.5 or >2.0.67 Other studies examining free nuclear acids by capillary electrophoresis did not always apply this commonly used ratio. We also tested previously published alternative LOH cut-off criteria (ratios of <0.6 and >1.67),37,38 which improved the sensitivity of the molecular analysis to 79%, while retaining a 100% specificity. Decreasing the stringency of LOH criteria to a ratio of <0.7 and >1.43 resulted in further increased sensitivity of 86%. However, the specificity decreased to 83%, which is not desirable in clinical practice. Five microsatellite markers (D3S2304, D3S4597, TP53, D5S346, and D12S363) detected tumor-associated molecular alterations most frequently, consistent with previously described allelic losses at 3p21.3 (RASSF1), 3p25, and 17p13.1 (TP53) in breast carcinomas, non-small-cell lung cancers,68 and squamous cell cancers of the head and neck.39 LOH at 5q21-22 (APC) is frequent in gastrointestinal and other cancers.69,70 MGP, located at chromosome 12p12.3-p13.1, is an important regulating gene during the process of embryonic lung branching morphogenesis and has recently been considered a tumor suppressor gene in colon cancer.71,72,73 The current study found LOH at the chromosomal region of MGP. In summary, combining molecular analysis of supernatants of pleural effusions and cytological examination might be a useful diagnostic tool in the assessment of pleural fluid for malignancy, significantly increasing sensitivity without jeopardizing specificity. Further studies involving larger patient cohorts are needed to confirm our findings.
Footnotes
Supported by the Wilhelm-Sander Foundation (2000.127.1).
This research was performed at the Department of Pathology, University of Regensburg, Regensburg, Germany.
The manuscript has been read and approved by all authors. The requirements for authorship have been met and each author believes that the manuscript represents honest work.
References
- Chapman SJ, Davies RJ. Pleural effusions. Clin Med. 2004;4:207–210. doi: 10.7861/clinmedicine.4-3-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light RW. Diagnostic principles in pleural disease. Eur Respir J. 1997;10:476–481. doi: 10.1183/09031936.97.10020476. [DOI] [PubMed] [Google Scholar]
- Reithineier A, Lydtin H. Pleural effusion. Internist (Berl) 1996;37:959–968. [PubMed] [Google Scholar]
- Garcia-Bonafe M, Moragas A. Differential diagnosis of malignant and reactive cells from serous effusions: image and texture analysis study. Anal Cell Pathol. 1996;12:85–98. [PubMed] [Google Scholar]
- Kjellberg SI, Dresler CM, Goldberg M. Pleural cytologies in lung cancer without pleural effusions. Ann Thorac Surg. 1997;64:941–944. doi: 10.1016/s0003-4975(97)00817-5. [DOI] [PubMed] [Google Scholar]
- Martensson G, Pettersson K, Thiringer G. Differentiation between malignant and non-malignant pleural effusion. Eur J Respir Dis. 1985;67:326–334. [PubMed] [Google Scholar]
- Nance KV, Shermer RW, Askin FB. Diagnostic efficacy of pleural biopsy as compared with that of pleural fluid examination. Mod Pathol. 1991;4:320–324. [PubMed] [Google Scholar]
- Motherby H, Nadjari B, Friegel P, Kohaus J, Ramp U, Bocking A. Diagnostic accuracy of effusion cytology. Diagn Cytopathol. 1999;20:350–357. doi: 10.1002/(sici)1097-0339(199906)20:6<350::aid-dc5>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Ahrendt SA, Sidransky D. The potential of molecular screening. Surg Oncol Clin N Am. 1999;8:641–656. [PubMed] [Google Scholar]
- Davidson B. Malignant effusions: from diagnosis to biology. Diagn Cytopathol. 2004;31:246–254. doi: 10.1002/dc.20133. [DOI] [PubMed] [Google Scholar]
- Musilova J, Michalova K. Cytogenetic study of cancer cells in effusions. Cancer Genet Cytogenet. 1986;19:271–279. doi: 10.1016/0165-4608(86)90056-7. [DOI] [PubMed] [Google Scholar]
- Motherby H, Pomjanski N, Kube M, Boros A, Heiden T, Tribukait B, Bocking A. Diagnostic DNA-flow- vs. -image-cytometry in effusion cytology. Anal Cell Pathol. 2002;24:5–15. doi: 10.1155/2002/840210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motherby H, Kube M, Friedrichs N, Nadjari B, Knops K, Donner A, Baschiera B, Dalquen P, Bocking A. Immunocytochemistry and DNA-image cytometry in diagnostic effusion cytology I. Prevalence of markers in tumour cell positive and negative smears. Anal Cell Pathol. 1999;19:7–20. doi: 10.1155/1999/459158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiegl M, Kaufmann H, Zojer N, Schuster R, Wiener H, Mullauer L, Roka S, Huber H, Drach J. Malignant cell detection by fluorescence in situ hybridization (FISH) in effusions from patients with carcinoma. Hum Pathol. 2000;31:448–455. doi: 10.1053/hp.2000.6550. [DOI] [PubMed] [Google Scholar]
- Yu CJ, Shew JY, Liaw YS, Kuo SH, Luh KT, Yang PC. Application of mucin quantitative competitive reverse transcription polymerase chain reaction in assisting the diagnosis of malignant pleural effusion. Am J Respir Crit Care Med. 2001;164:1312–1318. doi: 10.1164/ajrccm.164.7.2102067. [DOI] [PubMed] [Google Scholar]
- Braunschweig R, Guilleret I, Delacretaz F, Bosman FT, Mihaescu A, Benhattar J. Pitfalls in TRAP assay in routine detection of malignancy in effusions. Diagn Cytopathol. 2001;25:225–230. doi: 10.1002/dc.2043. [DOI] [PubMed] [Google Scholar]
- Ahrendt SA, Chow JT, Yang SC, Wu L, Zhang MJ, Jen J, Sidransky D. Alcohol consumption and cigarette smoking increase the frequency of p53 mutations in non-small cell lung cancer. Cancer Res. 2000;60:3155–3159. [PubMed] [Google Scholar]
- Hu Y, McDermott MP, Ahrendt SA. The p53 codon 72 proline allele is associated with p53 gene mutations in non-small cell lung cancer. Clin Cancer Res. 2005;11:2502–2509. doi: 10.1158/1078-0432.CCR-04-1913. [DOI] [PubMed] [Google Scholar]
- Ahrendt SA, Chow JT, Xu LH, Yang SC, Eisenberger CF, Esteller M, Herman JG, Wu L, Decker PA, Jen J, Sidransky D. Molecular detection of tumor cells in bronchoalveolar lavage fluid from patients with early stage lung cancer. J Natl Cancer Inst. 1999;91:332–339. doi: 10.1093/jnci/91.4.332. [DOI] [PubMed] [Google Scholar]
- Dahl C, Guldberg P. DNA methylation analysis techniques. Biogerontology. 2003;4:233–250. doi: 10.1023/a:1025103319328. [DOI] [PubMed] [Google Scholar]
- Janne PA, Li C, Zhao X, Girard L, Chen TH, Minna J, Christiani DC, Johnson BE, Meyerson M. High-resolution single-nucleotide polymorphism array and clustering analysis of loss of heterozygosity in human lung cancer cell lines. Oncogene. 2004;23:2716–2726. doi: 10.1038/sj.onc.1207329. [DOI] [PubMed] [Google Scholar]
- Ahrendt SA, Decker PA, Doffek K, Wang B, Xu L, Demeure MJ, Jen J, Sidransky D. Microsatellite instability at selected tetranucleotide repeats is associated with p53 mutations in non-small cell lung cancer. Cancer Res. 2000;60:2488–2491. [PubMed] [Google Scholar]
- Xu L, Chow J, Bonacum J, Eisenberger C, Ahrendt SA, Spafford M, Wu L, Lee SM, Piantadosi S, Tockman MS, Sidransky D, Jen J. Microsatellite instability at AAAG repeat sequences in respiratory tract cancers. Int J Cancer. 2001;91:200–204. doi: 10.1002/1097-0215(200002)9999:9999<::aid-ijc1031>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- Singer G, Kallinowski T, Hartmann A, Dietmaier W, Wild PJ, Schraml P, Sauter G, Mihatsch MJ, Moch H. Different types of microsatellite instability in ovarian carcinoma. Int J Cancer. 2004;112:643–646. doi: 10.1002/ijc.20455. [DOI] [PubMed] [Google Scholar]
- Woenckhaus M, Stoehr R, Dietmaier W, Wild PJ, Zieglmeier U, Foerster J, Merk J, Blaszyk H, Pfeifer M, Hofstaedter F, Hartmann A. Microsatellite instability at chromosome 8p in non-small cell lung cancer is associated with lymph node metastasis and squamous differentiation. Int J Oncol. 2003;23:1357–1363. [PubMed] [Google Scholar]
- Sidransky D. Emerging molecular markers of cancer. Nat Rev Cancer. 2002;2:210–219. doi: 10.1038/nrc755. [DOI] [PubMed] [Google Scholar]
- Dietmaier W, Hartmann A, Wallinger S, Heinmoller E, Kerner T, Endl E, Jauch KW, Hofstadter F, Ruschoff J. Multiple mutation analyses in single tumor cells with improved whole genome amplification. Am J Pathol. 1999;154:83–95. doi: 10.1016/S0002-9440(10)65254-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoehr R, Wild P, Hartmann A. Lasermicrodissection: an important prerequisite for the molecular-genetic analysis of bladder cancer. Pathol Res Pract. 2003;199:355–362. doi: 10.1078/0344-0338-00431. [DOI] [PubMed] [Google Scholar]
- Lee JH, Hong YS, Ryu JS, Chang JH. p53 and FHIT mutations and microsatellite alterations in malignancy-associated pleural effusion. Lung Cancer. 2004;44:33–42. doi: 10.1016/j.lungcan.2003.10.007. [DOI] [PubMed] [Google Scholar]
- Nawroz H, Koch W, Anker P, Stroun M, Sidransky D. Microsatellite alterations in serum DNA of head and neck cancer patients. Nat Med. 1996;2:1035–1037. doi: 10.1038/nm0996-1035. [DOI] [PubMed] [Google Scholar]
- Sanchez-Cespedes M, Monzo M, Rosell R, Pifarre A, Calvo R, Lopez-Cabrerizo MP, Astudillo J. Detection of chromosome 3p alterations in serum DNA of non-small-cell lung cancer patients. Ann Oncol. 1998;9:113–116. doi: 10.1023/a:1008230331221. [DOI] [PubMed] [Google Scholar]
- Chen X, Bonnefoi H, Diebold-Berger S, Lyautey J, Lederrey C, Faltin-Traub E, Stroun M, Anker P. Detecting tumor-related alterations in plasma or serum DNA of patients diagnosed with breast cancer. Clin Cancer Res. 1999;5:2297–2303. [PubMed] [Google Scholar]
- Anker P, Mulcahy H, Chen XQ, Stroun M. Detection of circulating tumour DNA in the blood (plasma/serum) of cancer patients. Cancer Metastasis Rev. 1999;18:65–73. doi: 10.1023/a:1006260319913. [DOI] [PubMed] [Google Scholar]
- Ang P, Tan EH, Leong SS, Koh L, Eng P, Agasthian T, Cheah FK. Primary intrathoracic malignant effusion: a descriptive study. Chest. 2001;120:50–54. doi: 10.1378/chest.120.1.50. [DOI] [PubMed] [Google Scholar]
- Dietmaier W, Wallinger S, Bocker T, Kullmann F, Fishel R, Ruschoff J. Diagnostic microsatellite instability: definition and correlation with mismatch repair protein expression. Cancer Res. 1997;57:4749–4756. [PubMed] [Google Scholar]
- Wistuba II, Behrens C, Milchgrub S, Bryant D, Hung J, Minna JD, Gazdar AF. Sequential molecular abnormalities are involved in the multistage development of squamous cell lung carcinoma. Oncogene. 1999;18:643–650. doi: 10.1038/sj.onc.1202349. [DOI] [PubMed] [Google Scholar]
- Sozzi G, Conte D, Mariani L, Lo Vullo S, Roz L, Lombardo C, Pierotti MA, Tavecchio L. Analysis of circulating tumor DNA in plasma at diagnosis and during follow-up of lung cancer patients. Cancer Res. 2001;61:4675–4678. [PubMed] [Google Scholar]
- Ahmed M, Giles F, Joe Y, Weber DM, Jilani I, Manshouri T, Giralt S, De Lima M, Keating M, Albitar M. Use of plasma DNA in detection of loss of heterozygosity in patients with multiple myeloma. Eur J Haematol. 2003;71:174–178. doi: 10.1034/j.1600-0609.2003.00125.x. [DOI] [PubMed] [Google Scholar]
- Braga E, Senchenko V, Bazov I, Loginov W, Liu J, Ermilova V, Kazubskaya T, Garkavtseva R, Mazurenko N, Kisseljov F, Lerman MI, Klein G, Kisselev L, Zabarovsky ER. Critical tumor-suppressor gene regions on chromosome 3P in major human epithelial malignancies: allelotyping and quantitative real-time PCR. Int J Cancer. 2002;100:534–541. doi: 10.1002/ijc.10511. [DOI] [PubMed] [Google Scholar]
- Miyaki M, Seki M, Okamoto M, Yamanaka A, Maeda Y, Tanaka K, Kikuchi R, Iwama T, Ikeuchi T, Tonomura A. Genetic changes and histopathological types in colorectal tumors from patients with familial adenomatous polyposis. Cancer Res. 1990;50:7166–7173. [PubMed] [Google Scholar]
- Sanz-Ortega J, Valor C, Saez MC, Ortega L, Sierra E, Poch J, Hernandez S, Sanz-Esponera J. 3p21, 5q21, 9p21 and 17p13 allelic deletions accumulate in the dysplastic spectrum of laryngeal carcinogenesis and precede malignant transformation. Histol Histopathol. 2003;18:1053–1057. doi: 10.14670/HH-18.1053. [DOI] [PubMed] [Google Scholar]
- Tsuji N, Furuse K, Asanuma K, Furuya M, Kondoh K, Kamagata C, Sasaki M, Kobayashi D, Yagihashi A, Takahashi H, Watanabe N. Mutations of the p53 gene and loss of heterozygosity at chromosome 17p13.1 are associated with increased survivin expression in breast cancer. Breast Cancer Res Treat. 2004;87:23–31. doi: 10.1023/B:BREA.0000041575.73262.aa. [DOI] [PubMed] [Google Scholar]
- Kohno H, Hiroshima K, Toyozaki T, Fujisawa T, Ohwada H. p53 mutation and allelic loss of chromosome 3p, 9p of preneoplastic lesions in patients with nonsmall cell lung carcinoma. Cancer. 1999;85:341–347. [PubMed] [Google Scholar]
- Matsumoto S, Kasumi F, Sakamoto G, Onda M, Nakamura Y, Emi M. Detailed deletion mapping of chromosome arm 3p in breast cancers: a 2-cM region on 3p14.3-21.1 and a 5-cM region on 3p24.3-25.1 commonly deleted in tumors. Genes Chromosomes Cancer. 1997;20:268–274. [PubMed] [Google Scholar]
- Wistuba II, Thomas B, Behrens C, Onuki N, Lindberg G, Albores-Saavedra J, Gazdar AF. Molecular abnormalities associated with endocrine tumors of the uterine cervix. Gynecol Oncol. 1999;72:3–9. doi: 10.1006/gyno.1998.5248. [DOI] [PubMed] [Google Scholar]
- Sanz-Ortega J, Sanz-Esponera J, Caldes T, Gomez de, la Concha E, Sobel ME, Merino MJ. LOH at the APC/MCC gene (5Q21) in gastric cancer and preneoplastic lesions: prognostic implications. Pathol Res Pract. 1996;192:1206–1210. doi: 10.1016/S0344-0338(96)80152-X. [DOI] [PubMed] [Google Scholar]
- Huang JS, Chiang CP, Kok SH, Kuo YS, Kuo MY. Loss of heterozygosity of APC and MCC genes in oral squamous cell carcinomas in Taiwan. J Oral Pathol Med. 1997;26:322–326. doi: 10.1111/j.1600-0714.1997.tb00223.x. [DOI] [PubMed] [Google Scholar]
- Davidson B, Risberg B, Reich R, Berner A. Effusion cytology in ovarian cancer: new molecular methods as aids to diagnosis and prognosis. Clin Lab Med. 2003;23:729–754. doi: 10.1016/s0272-2712(03)00058-1. [DOI] [PubMed] [Google Scholar]
- Davidson B, Reich R, Lazarovici P, Ann Florenes V, Nielsen S, Nesland JM. Altered expression and activation of the nerve growth factor receptors TrkA and p75 provide the first evidence of tumor progression to effusion in breast carcinoma. Breast Cancer Res Treat. 2004;83:119–128. doi: 10.1023/B:BREA.0000010704.17479.8a. [DOI] [PubMed] [Google Scholar]
- Grunewald K, Haun M, Fiegl M, Urbanek M, Muller-Holzner E, Massoner A, Riha K, Propst A, Marth C, Gastl G. Mammaglobin expression in gynecologic malignancies and malignant effusions detected by nested reverse transcriptase-polymerase chain reaction. Lab Invest. 2002;82:1147–1153. doi: 10.1097/01.lab.0000027840.16064.8a. [DOI] [PubMed] [Google Scholar]
- Patel IS, Madan P, Getsios S, Bertrand MA, MacCalman CD. Cadherin switching in ovarian cancer progression. Int J Cancer. 2003;106:172–177. doi: 10.1002/ijc.11086. [DOI] [PubMed] [Google Scholar]
- Dai Y, Morishita Y, Mase K, Sato N, Akaogi E, Mitsui T, Noguchi M. Application of the p53 and K-ras gene mutation patterns for cytologic diagnosis of recurrent lung carcinomas. Cancer. 2000;90:258–263. doi: 10.1002/1097-0142(20000825)90:4<258::aid-cncr10>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- de Matos Granja N, Soares R, Rocha S, Paredes J, Longatto Filho A, Alves VA, Wiley E, Schmitt FC, Bedrossian C. Evaluation of breast cancer metastases in pleural effusions by molecular biology techniques. Diagn Cytopathol. 2002;27:210–213. doi: 10.1002/dc.10171. [DOI] [PubMed] [Google Scholar]
- Davidson B, Konstantinovsky S, Nielsen S, Dong HP, Berner A, Vyberg M, Reich R. Altered expression of metastasis-associated and regulatory molecules in effusions from breast cancer patients: a novel model for tumor progression. Clin Cancer Res. 2004;10:7335–7346. doi: 10.1158/1078-0432.CCR-04-0183. [DOI] [PubMed] [Google Scholar]
- Elloul S, Bukholt Elstrand M, Nesland JM, Trope CG, Kvalheim G, Goldberg I, Reich R, Davidson B. Snail, Slug, and Smad-interacting protein 1 as novel parameters of disease aggressiveness in metastatic ovarian and breast carcinoma. Cancer. 2005;103:1631–1643. doi: 10.1002/cncr.20946. [DOI] [PubMed] [Google Scholar]
- Palmisano WA, Divine KK, Saccomanno G, Gilliland FD, Baylin SB, Herman JG, Belinsky SA. Predicting lung cancer by detecting aberrant promoter methylation in sputum. Cancer Res. 2000;60:5954–5958. [PubMed] [Google Scholar]
- Belinsky SA, Palmisano WA, Gilliland FD, Crooks LA, Divine KK, Winters SA, Grimes MJ, Harms HJ, Tellez CS, Smith TM, Moots PP, Lechner JF, Stidley CA, Crowell RE. Aberrant promoter methylation in bronchial epithelium and sputum from current and former smokers. Cancer Res. 2002;62:2370–2377. [PubMed] [Google Scholar]
- Lozano MD, Panizo A, Toledo GR, Sola JJ, Pardo-Mindan J. Immunocytochemistry in the differential diagnosis of serous effusions: a comparative evaluation of eight monoclonal antibodies in Papanicolaou stained smears. Cancer. 2001;93:68–72. [PubMed] [Google Scholar]
- Fetsch PA, Abati A. Immunocytochemistry in effusion cytology: a contemporary review. Cancer. 2001;93:293–308. doi: 10.1002/cncr.9044.abs. [DOI] [PubMed] [Google Scholar]
- Goessl C, Muller M, Straub B, Miller K. DNA alterations in body fluids as molecular tumor markers for urological malignancies. Eur Urol. 2002;41:668–676. doi: 10.1016/s0302-2838(02)00126-4. [DOI] [PubMed] [Google Scholar]
- Eisenberger CF, Schoenberg M, Enger C, Hortopan S, Shah S, Chow NH, Marshall FF, Sidransky D. Diagnosis of renal cancer by molecular urinalysis. J Natl Cancer Inst. 1999;91:2028–2032. doi: 10.1093/jnci/91.23.2028. [DOI] [PubMed] [Google Scholar]
- Utting M, Werner W, Dahse R, Schubert J, Junker K. Microsatellite analysis of free tumor DNA in urine, serum, and plasma of patients: a minimally invasive method for the detection of bladder cancer. Clin Cancer Res. 2002;8:35–40. [PubMed] [Google Scholar]
- Wang Y, Yamaguchi Y, Watanabe H, Ohtsubo K, Wakabayashi T, Sawabu N. Usefulness of p53 gene mutations in the supernatant of bile for diagnosis of biliary tract carcinoma: comparison with K- ras mutation. J Gastroenterol. 2002;37:831–839. doi: 10.1007/s005350200137. [DOI] [PubMed] [Google Scholar]
- Wang Y, Yamaguchi Y, Watanabe H, Ohtsubo K, Motoo Y, Sawabu N. Detection of p53 gene mutations in the supernatant of pancreatic juice and plasma from patients with pancreatic carcinomas. Pancreas. 2004;28:13–19. doi: 10.1097/00006676-200401000-00002. [DOI] [PubMed] [Google Scholar]
- Nawroz-Danish H, Eisenberger CF, Yoo GH, Wu L, Koch W, Black C, Ensley JF, Wei WZ, Sidransky D. Microsatellite analysis of serum DNA in patients with head and neck cancer. Int J Cancer. 2004;111:96–100. doi: 10.1002/ijc.20240. [DOI] [PubMed] [Google Scholar]
- Coulet F, Blons H, Cabelguenne A, Lecomte T, Lacourreye O, Brasnu D, Beaune P, Zucman J, Laurent-Puig P. Detection of plasma tumor DNA in head and neck squamous cell carcinoma by microsatellite typing and p53 mutation analysis. Cancer Res. 2000;60:707–711. [PubMed] [Google Scholar]
- Wang Y, Hung SC, Linn JF, Steiner G, Glazer AN, Sidransky D, Mathies RA. Microsatellite-based cancer detection using capillary array electrophoresis and energy-transfer fluorescent primers. Electrophoresis. 1997;18:1742–1749. doi: 10.1002/elps.1150181007. [DOI] [PubMed] [Google Scholar]
- Wistuba II, Lam S, Behrens C, Virmani AK, Fong KM, LeRiche J, Samet JM, Srivastava S, Minna JD, Gazdar AF. Molecular damage in the bronchial epithelium of current and former smokers. J Natl Cancer Inst. 1997;89:1366–1373. doi: 10.1093/jnci/89.18.1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piard F, Martin L, Chapusot C, Ponnelle T, Faivre J. Genetic pathways in colorectal cancer: interest for the pathologist. Ann Pathol. 2002;22:277–288. [PubMed] [Google Scholar]
- Dolan K, Garde J, Walker SJ, Sutton R, Gosney J, Field JK. LOH at the sites of the DCC, APC, and TP53 tumor suppressor genes occurs in Barrett’s metaplasia and dysplasia adjacent to adenocarcinoma of the esophagus. Hum Pathol. 1999;30:1508–1514. doi: 10.1016/s0046-8177(99)90175-2. [DOI] [PubMed] [Google Scholar]
- Fan C, Sheu D, Fan H, Hsu K, Allen Chang C, Chan E. Down-regulation of matrix Gla protein messenger RNA in human colorectal adenocarcinomas. Cancer Lett. 2001;165:63–69. doi: 10.1016/s0304-3835(01)00416-5. [DOI] [PubMed] [Google Scholar]
- Sheikh MS, Shao ZM, Chen JC, Fontana JA. Differential regulation of matrix Gla protein (MGP) gene expression by retinoic acid and estrogen in human breast carcinoma cells. Mol Cell Endocrinol. 1993;92:153–160. doi: 10.1016/0303-7207(93)90002-2. [DOI] [PubMed] [Google Scholar]
- Levedakou EN, Strohmeyer TG, Effert PJ, Liu ET. Expression of the matrix Gla protein in urogenital malignancies. Int J Cancer. 1992;52:534–537. doi: 10.1002/ijc.2910520406. [DOI] [PubMed] [Google Scholar]