Abstract
Hereditary nonpolyposis colon cancer (HNPCC, Online Mendelian Inheritance in Man (OMIM) 114500) is an autosomal dominant disorder that is genetically heterogeneous because of underlying mutations in mismatch repair genes, primarily MLH1, MSH2, and MSH6. One challenge to correctly diagnosing HNPCC is that the large size of the causative genes makes identification of mutations both labor intensive and expensive. We evaluated the usefulness of denaturing high performance liquid chromatography (DHPLC) for scanning mismatch repair genes (MLH1, MSH2, and MSH6) for point mutations, small deletions, and insertions. Our assay consisted of 51 sets of primers designed to amplify all exons of these genes. All polymerase chain reaction reactions were amplified simultaneously using the same reaction conditions in a 96-well format. The amplified products were analyzed by DHPLC across a range of optimum temperatures for partial fragment denaturation based on the melting profile of each specific fragment. DNA specimens from 23 previously studied HNPCC patients were analyzed by DHPLC, and all mutations were correctly identified and confirmed by sequence analysis. Here, we present our validation studies of the DHPLC platform for HNPCC mutation analysis and compare its merits with other scanning technologies. This approach provides greater sensitivity and more directed molecular analysis for clinical testing in HNPCC.
Colorectal cancer (CRC) is the second leading cause of cancer-related death in the United States, accounting for more than 57,000 deaths per year (in 2002; Cancer Facts and Figures. American Cancer Society. Available at www.cancer.org). Although the majority of colorectal cancer is not inherited, inherited CRC accounts for up to 10% of total cases and primarily consists of familial adenomatous polyposis (OMIM 175100), or hereditary nonpolyposis colon cancer (HNPCC; OMIM 114500). HNPCC is an autosomal dominant syndrome characterized by increased lifetime risk of early-onset colorectal cancer as well as other cancers of the endometrium, stomach, small intestine, hepatobiliary system, kidney, ureter, and ovary.1,2 Although exact data about its prevalence are unknown, it is estimated that HNPCC or Lynch Syndrome accounts for about 5 to 13% of all CRC, making it the most common hereditary colon cancer syndrome.3,4,5 The penetrance of HNPCC mutations has been estimated at approximately 80%, the lifetime risk for a mutation carrier to develop colorectal cancer.6 Identification of HNPCC mutations is important for clinical surveillance in carriers and genetic testing for at-risk relatives.
Germline mutations in mismatch repair (MMR) genes, most commonly human mutL homolog 1 (MLH1) on chromosome 3p22.3, human mutS homolog 2 (MSH2) on chromosome 2p21, and human mutS homolog 6 (MSH6) on chromosome 2p16.3, are causative of HNPCC7,8,9,10,11. To date, hundreds of mutations, distributed in all exons of the MMR genes have been identified in a large number of families (International Society for Gastrointestinal Hereditary Tumors [InSIGHT]; www.insight-group.org/). Additional mismatch repair genes including human mutS homolog 3 (MSH3) on chromosome 5q14.1 and human postmeiotic segregation 1 and 2 (PMS1 and PMS2) on chromosome 2q32.2 and 7q22.1, respectively, are also associated with HNPCC but account for a smaller proportion of cases.12,13,14 The large size of these genes (19 exons for MLH1, 16 exons for MSH2, and 10 exons for MSH6) encompassing 9241 nucleotides of coding sequence makes sequencing for germline mutations both labor intensive and expensive. Before performing germline mutation analysis, denaturing high-performance liquid chromatography (DHPLC) is an efficient method of screening for MMR gene mutations.15
Oefner and others16,17,18,19 developed DHPLC as a rapid, semiautomated tool for detection of sequence variants. The DHPLC platform meets the needs of the clinical molecular laboratories because of its sensitivity, semiautomated operation and cost effectiveness. The principle behind DHPLC is that negatively charged DNA is linked to a neutral column by a positively charged triethylammonium acetate (TEAA). Variants are detected by differential binding of the homo- and heteroduplexes to the column. Detection of sequence variations using DHPLC in numerous disease-related genes has been reported including CFTR, F9, MECP2, RET, and PTEN.19,20,21 In genes such as BRCA1/2, TSC1/2, NF-1, and APC, the large size and presence of few hot spots makes DHPLC a preferred method for rapid mutation screening and directed molecular analysis.22,23,24,25,26,27,28,29 In HNPCC, one report describing the use of DHPLC for mutation analysis in MLH1 and MSH2 yielded a sensitivity of 97% compared with sequence analysis.15 Later studies by Kurzawski et al30 confirmed the analytical sensitivity of this method (>98%) in a series of 46 patients from HNPCC families by similar DHPLC approach.
Several different approaches have previously been described to identify mutations in HNPCC.31,32,33,34 Scan-ning methods including single-strand conformation an-alysis (SSCP), conformation-sensitive gel electropho-resis (CSGE), denaturing gradient gel electrophoresis (DGGE), as well as sequencing the entire coding region of the MLH1 and MSH2 genes are well established techniques for clinical diagnostic purposes. However, SSCP, CSGE, and DGGE lack sensitivity, and SSCP has inherent size limitations of 250 bp for fragment analysis with a sensitivity of 70 to 80%. DGGE offers increased sensitivity yet is also very time consuming and requires GC-clamp primers, and mutation in GC-rich regions may not be detected. In view of the many deficiencies of these scanning approaches in terms of sensitivity and specificity, we analyzed the efficacy of a DHPLC approach to satisfy the following criteria: high test sensitivity and specificity, reliable and reproducible results, assay robustness, cost effective, and high-throughput capability. In this communication, we report identification of sequence variations in the MLH1, MSH2, and MSH6 genes using a DHPLC platform under optimized conditions, and we discuss the implementation and validation of this approach in a clinical laboratory setting.
Materials and Methods
Patient Samples
Anonymous DNAs from 23 HNPCC patients were obtained from clinical and research laboratories (a+ LabPLUS, Auckland, New Zealand; Mayo Clinic, Rochester, MN; Ohio State University, Columbus, OH). These patients have been previously analyzed using CSGE and SSCP, followed by targeted sequence analysis, or by full gene sequence analysis.
DNA Extraction
DNA was extracted from peripheral blood lymphocytes of noncolorectal cancer individuals for 40 negative con-trols using the Puregene DNA Isolation kit (Gentra Systems, Minneapolis, MN).
Primers for Polymerase Chain Reaction (PCR) Amplification of Gene Exons
The transcript and genomic sequence data were accessed from multiple databases (principally through http://genome.ucsc.edu) that carried the reference sequences for the MLH1, MSH2, and MSH6 genes (GenBank Accession nos. NM000249, NM000251, and NM000179 for MLH1, MSH2, and MSH6, respectively). Using this information, primers were designed to contain at least 50 bp of intron. The sequence of the primer pairs, their corresponding amplicon sizes, and optimized conditions for DHPLC analysis are shown in Table 1. All primers were checked against the National Center for Biotechnology Information SNP database (http://www.ncbi.nlm.nih.gov/SNP/) and the HGVbase (http://hgvbase.cgb.ki.se/) to avoid overlapping with single nucleotide polymorphisms (SNP).
Table 1.
Primer Sequences and DHPLC Conditions
| Name | Primer sequence (5′ to 3′) | PCR size | Tm* PCR (°C) | DHPLC gradient %B | DHPLC† temperatures (°C) |
|---|---|---|---|---|---|
| MLH1 primer list | |||||
| MLH1-1F | aggtgattggctgaaggcac | 231 | 62 | 47–61 | 62, 63 |
| MLH1-1R | gcccgttaagtcgtagccct | ||||
| MLH1-2F | atgtacattagagtagttgcagactgataaatt | 221 | 57 | 46–60 | 56, 57 |
| MLH1-2R | agtttccagaacagagaaaggtcc | ||||
| MLH1-3F | caagaaaatgggaattcaaagagat | 241 | 55 | 47–61 | 55, 56 |
| MLH1-3R | ctaacaaatgacagacaatgtcatcac | ||||
| MLH1-4F | cctttggtgaggtgacagtgg | 221 | 56 | 46–60 | 57, 58, 59 |
| MLH1-4R | caggattactctgagacctaggcaa | ||||
| MLH1-5F | ttttccccttgggattagtatctatc | 227 | 53 | 47–61 | 55, 57 |
| MLH1-5R | ccctgaaaacttagaagcaattttattt | ||||
| MLH1-6F | ggacatcttgggttttattttcaag | 235 | 56 | 47–61 | 57, 58 |
| MLH1-6R | tgttcaatgtatgagcactagaacaca | ||||
| MLH1-78F | gggctctgacatctagtgtgtgtt | 417 | 56 | 52–66 | 56, 57, 58 |
| MLH1-78R | aaaataatgtgatggaatgataaacca | ||||
| MLH1-9F | tctgattcttttgtaatgtttgagttttg | 241 | 55 | 47–61 | 56, 57 |
| MLH1-9R | cataaaattccctgtgggtgtttc | ||||
| MLH1-10F | ctgaggtgatttcatgactttgtgt | 251 | 59 | 48–62 | 58, 59, 60 |
| MLH1-10R | gaggagagcctgatagaacatctgt | ||||
| MLH1-11F | gtgggctttttctccccct | 281 | 58 | 49–63 | 58, 60, 62 |
| MLH1-11R | ctctcacgtctggccgg | ||||
| MLH1-12 new F | ttttttaatacagactttgctaccaggac | 436 | 55 | 54–68 | 59, 60, 61 |
| MLH1-12 new R | gttttattacagaataaaggaggtaggctg | ||||
| MLH1-13F | ccaaaatgcaacccacaaaatt | 282 | 58 | 49–63 | 58, 59, 60 |
| MLH1-13R | aaccttggcagttgaggcc | ||||
| MLH1-14F | ggtgtctctagttctggtgcctg | 271 | 58 | 48–62 | 59, 60 |
| MLH1-14R | tgcctgtgctccctgga | ||||
| MLH1-15F | cccattttgtcccaactggtt | 203 | 57 | 45–59 | 56, 57, 58 |
| MLH1-15R | gagagctactattttcagaaacgatcag | ||||
| MLH1-16F | tgggaattcaggcttcatttg | 292 | 58 | 49–63 | 58, 59, 60 |
| MLH1-16R | gcacccggctggaaatt | ||||
| MLH1-17F | gcactggagaaatgggatttg | 221 | 59 | 46–60 | 58, 59, 60 |
| MLH1-17R | cctccagcacacatgcatg | ||||
| MLH1-18F | agtctgtgatctccgtttagaatgag | 242 | 57 | 47–61 | 56, 58, 59 |
| MLH1-18R | ttgtatgaggtcctgtcctagtcct | ||||
| MLH1-19F | catcagccaggacaccagtg | 288 | 58 | 49–63 | 58, 59, 60 |
| MLH1-19R | cggaatacagagaaagaagaacaca | ||||
| MSH2 primer list | |||||
| MSH2-1-diag F | ttcgacatggcggtgc | 285 | 67 | 48–62 | 66, 67 |
| MSH2-1 new R | gtccctccccagcacg | ||||
| MSH2-2F | gaagtccagctaatacagtgcttga | 301 | 53 | 49–63 | 52, 54, 55 |
| MSH2-2R | aaacacaattaaattcttcacatttttatttt | ||||
| MSH2-3F | agagtttggatttttcctttttgc | 432 | 57 | 52–66 | 56, 57, 58 |
| MSH2-3R | tcatgtcaattaaagagcctttcc | ||||
| MSH2-4F | ttcatttttgcttttcttattcctttt | 316 | 50 | 50–64 | 52, 55, 57 |
| MSH2-4R | atatgacagaaatatccttctaaaaagtcactat | ||||
| MSH2-5F | actggatccagtggtatagaaatcttc | 285 | 53 | 49–63 | 52, 54, 56 |
| MSH2-5R | gcttcttcagtatatgtcaatgaaaaca | ||||
| MSH2-6F | gcgtagtaaggttttcactaatgagc | 251 | 56 | 48–62 | 56, 57, 58 |
| MSH2-6R | catgtgggtaactgcaggttaca | ||||
| MSH2-7F | tgagacttacgtgcttagttgataaattt | 341 | 53 | 50–64 | 52, 55, 56 |
| MSH2-7R | gcacattgccaagtatatattgtatgag | ||||
| MSH2-8F | tgatgcttgtttatctcagtcaaaatt | 275 | 53 | 48–62 | 53, 54, 55 |
| MSH2-8R | aatctacaaactttcttaaagtggcctt | ||||
| MSH2-9 new F | gtctttacccattatttataggattttgtca | 217 | 56 | 46–60 | 56, 57, 58 |
| MSH2-9 new R | gtatagacaaaagaattattccaacctcc | ||||
| MSH2-10F | attgaaaaatggtagtaggtatttatggaa | 274 | 54 | 48–62 | 54, 55, 56 |
| MSH2-10R | cacatcatgttagagcatttaggga | ||||
| MSH2-11F | atatgtttcacgtagtacacattgcttcta | 249 | 54 | 47–61 | 54, 55 |
| MSH2-11R | tcaaatatcatgatttttcttctgttacc | ||||
| MSH2-13R | tcacaggacagagacatacatttctatct | ||||
| MSH2-14F | tgtggcatatccttcccaatg | 452 | 55 | 52–66 | 55, 56, 57 |
| MSH2-14R | aataatttatactaacttagaataaggcaattactgat | ||||
| MSH2-15F | tacataaattgctgtctcttctcatgc | 311 | 57 | 50–64 | 57, 58 |
| MSH2-15R | aaaaaccttcatcttagtgtcctgttt | ||||
| MSH2-16 new F | taattactaatgggacattcacatgtgt | 230 | 55 | 47–61 | 55, 56 |
| MSH2-16 new R | taccttcattccattactgggattt | ||||
| MSH6 primer list | |||||
| MSH6Exon 1F | tgttgattggccactggg | 463 | 66 | 53–58 | 66, 67, 68 |
| MSH6Exon 1R | caaccccctgtgcgagcctc | ||||
| MSH6Exon 2F | taactgcctttaaggaaacttgacca | 330 | 60 | 50–55 | 59, 60, 61 |
| MSH6Exon 2R | tcatatagaaaaaagtctgcctgtctg | ||||
| MSH6Exon 3F | ctggtcttgaactgctgggat | 289 | 58 | 49–54 | 58, 59, 60 |
| MSH6Exon 3R | cccctttcttcccccatc | ||||
| MSH6Exon 4-1F | tgcacgggtaccattataaagtca | 450 | 58 | 52–57 | 58, 59, 60 |
| MSH6Exon 4-1R | gtattcttggtttctgatgaaatgctag | ||||
| MSH6Exon 4-2F | gaaggaaacgccctcagc | 420 | 58 | 52–57 | 58, 59, 60 |
| MSH6Exon 4-2R | cagttgcctttcatgaataccag | ||||
| MSH6Exon 4-3F | ccacatggatgctcttattgga | 420 | 58 | 52–57 | 58, 59, 60 |
| MSH6Exon 4-3R | tcatctgaaaactgacctatgaaaaact | ||||
| MSH6Exon 4-4F | tttgttgatacttcactgggaaagtt | 420 | 57 | 52–57 | 57, 58, 59 |
| MSH6Exon 4-4R | ctcctgatcaataaggcattttttg | ||||
| MSH6Exon 4-5F | ctctaggtggttgtgtcttctacctc | 420 | 57 | 52–57 | 57, 58, 59 |
| MSH6Exon 4-5R | tgagtagcctctcaagatctggaa | ||||
| MSH6Exon 4-6F | cgaagttgtagagcttctaaagaagct | 480 | 57 | 53–58 | 56, 57, 58 |
| MSH6Exon 4-6R | gtcctacagccaattctgttgc | ||||
| MSH6Exon 4-7F | agcctcctggaatacctagagaaac | 420 | 58 | 52–57 | 58, 59, 60 |
| MSH6Exon 4-7R | acttatttttagggataatatacagctggc | ||||
| MSH6Exon 5F | cacttaggctgataaaaccccc | 386 | 57 | 51–56 | 57, 58, 59 |
| MSH6Exon 5R | gtatgttattcctaatgtcacaaatgacttt | ||||
| MSH6Exon 6F | aagacaaaagtttatgaaactgttactacca | 250 | 56 | 48–53 | 56, 57, 59 |
| MSH6Exon 6R | agaagcaaatatcttttatcacatctaaatg | ||||
| MSH6Exon 7F | taacctagaagatgaatttatgtaatatgattt | 224 | 53 | 46–51 | 53, 54, 55 |
| MSH6Exon 7R | ttcagataatcttctataaaaatagttatttgt | ||||
| MSH6Exon 8F | tgagttacttccttatgcatattttact | 275 | 57 | 48–53 | 56, 57, 58 |
| MSH6Exon 8R | aatattagcgatacatgtgctagca | ||||
| MSH6Exon 9F | tgctagcacatgtatcgctaatatt | 320 | 56 | 50–55 | 56, 57, 58 |
| MSH6Exon 9R | gcatcatcccttcccctttta | ||||
| MSH6Exon 10F | gaagggatgatgcactatgaaaaa | 296 | 52 | 49–54 | 52, 56, 57 |
| MSH6Exon 10R | gtagaaggtagataagaattaaaagggtttaattt |
Tm for the fragment.
Optimum temperatures for DHPLC analysis were empirically determined using the predicted fragment melting profile generated by WAVEMAKER software. (table continued)
PCR Analysis
The complete coding regions of the MLH1, MSH2, and MSH6 genes, including all splice junctions, were amplified in a total of 51 fragments using primers designed in our laboratory. All exonic fragments of each gene, including intron junctions, were amplified individually using primers designed with Primer Express 1.0 (PE Applied Biosystems, Foster City, CA) with a Tm of approximately 60°C (19 fragments for MLH1 and 16 fragments each for MSH2) and 58°C (16 fragments for MSH6). PCR analysis was performed using standard reaction conditions of 50 ng DNA, 200 μmol/L primers, 200 μmol/L dNTPs, 1× PCR buffer with 1.5 μmol/L MgCl2, and 1.25 units/reaction Faststart TaqDNA polymerase (Roche) in a 96-well plate format. Amplification was performed in the Eppendorf Mastercycler using the following conditions: 95°C for 30 seconds, annealing at X°C (MLH1-58, MSH2-58.8, MSH6-57.8) for 30 seconds, and extension at 72°C for 1 minute, for a total of 35 cycles, and with a final extension at 72°C for 7 minutes, and then held at 4°C. All PCR products were examined by gel electrophoresis before analysis on DHPLC.
DHPLC Analysis
A Transgenomic WAVE DNA Fragment Analysis System (Transgenomic, Inc., Omaha, NE) and associated WAVEMAKER software were used. An aliquot (5 μl) of the PCR product was directly injected into a DNASep column. Each fragment was analyzed using two to three partially denaturing temperatures (Table 1) to maximize detection of unknown mutations located in various positions throughout the fragment. The optimum conditions were determined empirically, based on fragment melting profile at each selected temperature. The column mobile phase for sample elution consisted of a mixture of the following: buffer A, 0.1 mol/L TEAA; and buffer B, 0.1 mol/L TEAA with 25% acetonitrile. Samples were eluted at a linear gradient of buffer B over a 4.5-minute period at a constant flow rate of 0.9 ml/minute. The starting gradient varied among fragments, depending on the DNA sequence and fragment size, and was determined by the WAVEMAKER software. The chromatograms of each fragment were compared with those of the wild type, and fragments containing heteroduplexes with a shorter retention time compared with wild-type fragments were sequenced to confirm putative sequence variations.
Sequence Analysis
PCR products (20 μl) were purified using a QIAquick PCR Purification kit (Qiagen Inc., Germany). The subsequent purified products were subjected to cycle sequencing in both forward and reverse directions using a BigDye Terminator Cycle Sequencing version 3.1 kit (PE Applied Biosystems). Sequencing reaction contained purified PCR product (1.5 ng/μl), Terminator Ready Reaction mix (4 μl), and primer (3.3 pmol) in a 10-μl reaction. Cycle sequencing was performed according to the manufacturer’s instructions. The products from each reaction were electrophoresed in an Applied Biosystems PRISM 3100 DNA Sequencer. The sequences for each fragment were aligned to the wild-type sequences obtained from GenBank and analyzed for sequence variation using Sequencher software (Gene Codes Corp., Ann Arbor, MI). GenBank nos. NM000249, NM000251, and NM000179 were used as the MLH1, MSH2, and MSH6 wild-type cDNA reference sequences, respectively, and American College of Medical Genetics (ACMG) guidelines were followed for interpretation of sequence variation (www.acmg.net).
Results
Summary of DHPLC Detection of Known Mutations and Polymorphisms
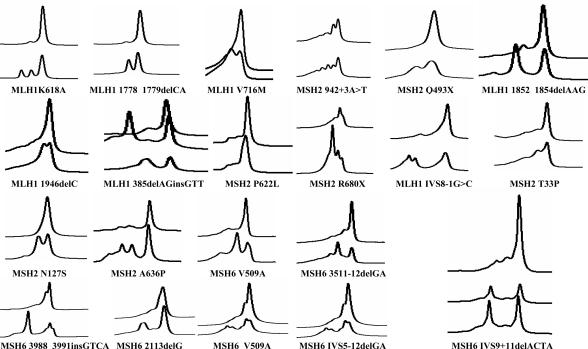
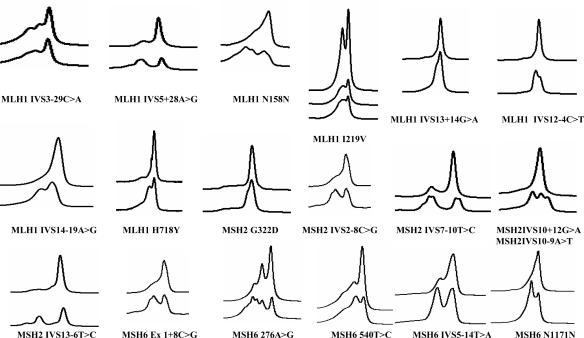
A total of 23 known positive controls from HNPCC patients containing sequence variations in the MLH1, MSH2, and MSH6 genes were analyzed, and the results are shown in Table 2 and Figure 1. Of these, 5 represented unique mutations within the sample set, whereas 18 were recurrent mutations that had been identified in different laboratories reported in the literature. This sample set represented seven missense, six deletion, two nonsense, two splicing, one insertion, and one indel type mutation. All were detected by DHPLC analysis using temperature algorithm for analysis (Table 1). Thus, the sensitivity of DHPLC analysis for these known mutations was 100%. Sequence analysis of this sample set as well as the 40 wild-type controls identified several additional variants (missense mutations), representing polymorphisms, which are commonly found in these genes (Figure 2). The temperatures for detection of mutations are shown in the Table 2. Most of the mutations were detected at all three temperatures, but some were detected at only the low and the medium temperatures. This is dependent on the sequence composition and position of the mutation within the sequence. AT-rich sequences are easier to melt and are hence detected at lower temperatures compared with GC-rich sequences, which melt at high temperatures. Exon 1 of MSH2 and MSH6 was found to be particularly GC rich and difficult to design primers.
Table 2.
Summary of Mutations Detected by DHPLC
| Sample | Gene | Exon | Mutation name (based on)
|
Mutation | Method* | DHPLC†
|
|||
|---|---|---|---|---|---|---|---|---|---|
| Codon | Nucleotide | TL | TMD | TH | |||||
| 153 | MLH1 | 5 | delAGinsGTT | 385 | 385delAGinsGTT | SSCP | + | + | + |
| 242 | MLH1 | 9 | G>C | IVS8-1 | IVS8-1G>C | SSCP | + | + | − |
| 151 | MLH1 | 16 | delAAG | 1852-1854 | 1852-1854delAAG | SSCP | + | + | + |
| 144 | MLH1 | 16 | AA>GC | 1852-1853 | K618A | SSCP | + | + | + |
| 145 | MLH1 | 16 | ddCA | 1778-1779 | 1778-1779delCA | SSCP | + | + | + |
| 152 | MLH1 | 17 | delC | 1946 | 1946GdelC | SSCP | + | + | − |
| 146 | MLH1 | 19 | G>A | 2146 | V716M | SSCP | + | + | − |
| 233 | MSH2 | 1 | A>C | 97 | T33P | SSCP | + | + | − |
| 234 | MSH2 | 3 | A>G | 380 | N127S | SSCP | + | + | − |
| 147 and 610 | MSH2 | 5 | A>T | 3′ intron | 942+3A>T | SSCP | + | + | − |
| 148 | MSH2 | 9 | C>T | 493 | Q493X | SSCP | + | + | − |
| 609 | MSH2 | 10 | delG | 2113 | 2113delG | CSGE | + | + | − |
| 154 | MSH2 | 12 | C>T | 1865 | P622L | SSCP | + | + | + |
| 235 | MSH2 | 12 | G>C | 1906 | A636P | SSCP | + | + | − |
| 243 | MSH2 | 13 | C>T | 2038 | R680X | SSCP | + | + | + |
| 264 | MSH6 | 4-3 | T>C | 1526 | V509A | SSCP | + | + | + |
| 265 | MSH6 | 6 | delGA | 3511 | 3511-3512delGA | SSCP | + | + | + |
| 266 | MSH6 | 9 | insGTCA | 3988 | 3988-3991insGTCA | SSCP | + | + | + |
| 267 | MSH6 | 9 | delACTA | – | IVS9+11delACTA | SSCP | + | + | + |
| 268 | MSH6 | 9 | delACTA | – | IVS9+11delACTA | SSCP | + | + | + |
Method previously used to identify mutation.
Three temperatures of DHPLC analysis are designated low, medium, and high (TL, TMD, and TH, respectively). +, positive; −, negative.
Figure 1.
DHPLC profiles for MLH1, MSH2, and MSH6 mutations. The DHPLC profiles of HNPCC patients with mutations in mismatch repair genes are shown for all of the mutations in MLH1, MSH2, and MSH6 used for assay validation compared with the wild-type profiles, respectively. The wild-type profile is shown on the top and mutant profile at the bottom of individual profiles.
Figure 2.
DHPLC profiles for MLH1, MSH2, and MSH6 polymorphisms. The DHPLC profiles of HNPCC patients with polymorphisms in mismatch repair genes are shown for all of the mutations in MLH1, MSH2, and MSH6 used for assay validation compared with the wild-type profiles, respectively.
Reproducibility of Elution Profiles
The elution profiles shown in Figure 2 were found to be reproducible under different conditions such as change of new buffer, change of column, and amount of PCR product injected. The reproducibility of the elution profiles is represented in Figure 1 in MSH2 942 + 3A>T and MSH6 for the mutation IVS9 + 11delACTA in which more than one positive control was available. This region is flanked on either side by SNPs and also a repeat of the ACTA sequence. Careful design of primers is important to be able to detect changes in sequence in this region.
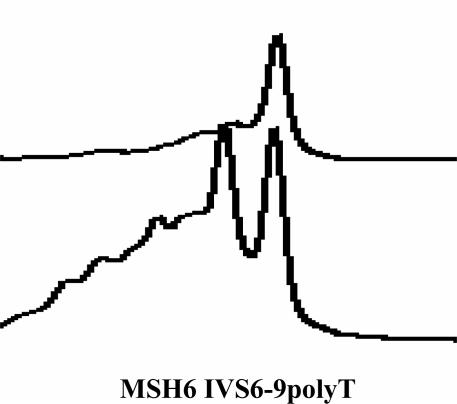
Repeat Sequence Regions
Figure 3 represents the elution profile for the poly(T) variable tract in MSH6 intron 6. The elution profile is complex because of the variable region, and a single peak is not observed for this exon. It is important to examine the elution profile for this exon carefully to avoid a false-negative result. The forward sequencing primer for this exon is designed 5′ of the repeat region to avoid the detection of heterozygosity in this region. The reverse sequencing primer detects the variability of this region.
Figure 3.
DHPLC profiles for MSH6 intron 6 variable poly(T) region. Note the nonspecific heteroduplexes immediately before the main peak, indicating a highly polymorphic region. The wild-type profile is on the top and the poly(T) profile is shown below it.
Discussion
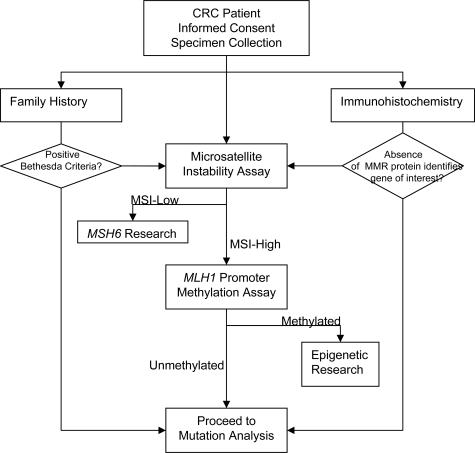
We have developed a hierarchical mutation detection strategy that has been designed for ease of use and sensitivity but that is also cost effective, thus achieving a balance between comprehensiveness and labor/running costs. The contribution to current practice lies in streamlining a testing regimen such that each tier of the process is manageable, while providing acceptable timeliness for reporting. This strategy compares favorably with the mutation detection strategies that have been used by many testing laboratories in examining the mismatch repair genes, with a reported detection frequency of about 60 to 70%35 (Figure 4).
Figure 4.
Proposed strategy for genetic testing in colorectal cancer patients. A comprehensive strategy incorporating 1) family history and clinical data; 2) microsatellite instability; and 3) immunohistochemistry as screening tools mutation for targeted analysis. Methylation testing further focuses the process on samples most likely to carry a germline mutation (ie, microsatellite unstable, unmethylated, and IHC-positive tumors).
Here, we report the successful identification of mutations in the MLH1, MSH2, and MSH6 genes in a characterized cohort of 23 HNPCC patients using a DHPLC platform. The patients reported here were previously tested in other clinical laboratories and found to have mutations that were detected by the SSCP, CSGE, DGGE, Protein Truncation Test (PTT), and sequencing assays.
DHPLC is a rapid method for screening for mutations, which are confirmed subsequently by sequencing the relevant exon. This finding is in general agreement with the view that DHPLC is more sensitive than SSCP, DGGE, CSGE, and Heteroduplex analysis (HA) analyses for identifying sequence variants, with sensitivities ranging from 95 to 100%. Although our work shows that DHPLC is an effective platform for diagnostic laboratories, especially for screening large genes, it is important to identify and carefully address the caveats in the test design using this technology. The American College of Medical Genetics has recently put out the standard guidelines for use of DHPLC in clinical laboratories for critical review (www.acmg.net). DHPLC-based assays for mismatch repair genes have been recently described by Holinski-Feder et al,15 Kurzawski et al,30 and Young et al36. The assays described do not have identical PCR conditions, and profiles for mutations are not easily distinguishable from the wild type. We believe that the published methods are not suitable for a diagnostic laboratory setting, in which it is important to have a streamlined assay with high sensitivity and accuracy. Generally, most of the profiles on DHPLC are clearly different, although we have observed that an insertion may not result in an obvious deviant profile (data not presented), which is highly dependent on the position of the variant within the fragment.
Several groups have found that DHPLC is effective and has advantages over other methods such as DGGE, SSCP, and PTT assays for mutation screening in different genes.37,38,39,40,41,42,43,44,45,46 The disadvantages for SSCP and PTT assays include that the results are dependent on the quality of the blood sample, transcript stability, and extraction method, whereas DGGE involves use of labeled primers; and all of these techniques are labor intensive.22,34,39,42 Furthermore, a high percentage of the mutations reported in MLH1, MSH2, and MSH6 are missense mutations Human Genome Mutation Database; InSIGHT [http://www.insight-group.org/]), which cannot be detected by PTT. The amplification and mutation screening protocol described here involves a reduction in both handling errors and operator fatigue, compared with other assays, because all amplifications are carried out in a 96-well microtiter tray format using a multichannel pipette. In addition, MLH1 and MSH6 gene exons for more than one patient can be amplified in the same 96-well tray. It is essential that the clinical molecular genetic laboratories using this technology consider the following points. Because the quality of the profile on the DHPLC is dependent on the quality of the PCR product, it is important to employ high-stringency PCR conditions to avoid nonspecificity in the PCR reactions. Although WAVEMAKER software is the primary tool used to design the PCR fragments, it is necessary to carefully check the temperature predictions made by the software. It may be necessary to employ higher temperatures to melt the desired regions in the fragments, especially if the fragment is GC rich.
Another important issue regarding the use of DHPLC analysis for HNPCC mutational analysis is interpretation of missense mutations and polymorphisms. Our clinical laboratory protocol requires that any heteroduplex detected by DHPLC be confirmed by sequence analysis. Protocols involving sequencing alone have been described as expensive, cumbersome, and labor intensive, and they yield unnecessary data. Our results have shown that even in the presence of a polymorphism in the wild-type control, sequence variations in the patient sample can be identified with complex heteroduplex patterns (data not shown). It is important to select wild type, which is homozygous at positions of known variation in the genes. We have observed such complex profiles in the MSH6 gene, which is highly polymorphic, and thus it is important to use a wild type that is homozygous for the known variants. A run of the same nucleotide, especially T or A, will also affect the elution profile. For example MSH2 intron 1 and MSH6 intron 6 have a tract of variable poly(T) at the 5′ end, which severely affects the elution profiles especially if a variant is present very close the poly(T) tail. Among the 40 wild types analyzed for the MSH6 intron 6 variant, we found only two wild types homozygous for this variant indicating that this variant is common (Figure 3). The MSH6 intron 6 variant has not been previously reported as a polymorphism in InSIGHT on HNPCC Database (http://www.insight-group.org/; Table 3).
Table 3.
Summary of Polymorphisms Detected by DHPLC
| Gene | Exon | Nucleotide | Change | Codon | Name | DNA no. | In database* |
|---|---|---|---|---|---|---|---|
| MLH1 | 4 | 5′ Intron | C>A | IVS3−29 C>A | 7 | Yes | |
| 5 | 5′ Intron | A>G | IVS5+28 A>G | 7 | No | ||
| 6 | 474 | C>T | 158 | N158N | 18 | No | |
| 7 and 8 | 655 | A>G | 219 | I219V | 6 | Yes | |
| 13 | 3′ Intron | G>A | IVS13+14 G>A | 2, 4 | Yes | ||
| 13 | 5′ Intron | C>T | IVS12−54 C>T | 7 | No | ||
| 15 | 5′ Intron | A>G | IVS14−19 A>G | 21 | No | ||
| 19 | 2152 | C>T | 718 | H718Y | 20, 21, 18 | Yes | |
| MSH2 | 2 | 5′ Intron | C>G | IVS2−8C>G | 634 | No | |
| 6 | 965 | G>A | 322 | G322D | 19 | Yes | |
| 7 | 5′ Intron | T>C | IVS7−10 T>C | 21 | Yes | ||
| 10 | 3′ Intron | G>A | IVS10+12 G>A | 21 | Yes | ||
| 10 | 5′ Intron | A>T | IVS10−9 A>T | 21 | Yes | ||
| 13 | 5′ Intron | T>C | IVS13−6 T>C | 6, 19, 21 | Yes | ||
| MSH6 | 1 | 116 | G>A | 39 | G39E | 0222 | Yes |
| 1 | 186 | C>A | 62 | R62R | 0222 | Yes | |
| 2 | 276 | A>G | 92 | P92P | 634 | Yes | |
| 3 | 540 | T>C | 180 | N180N | 634 | Yes | |
| 5 | 3′ Intron | T>A | IVS5+14 T>A | 007 | Yes | ||
| 6 | 3513 | T>C | 1171 | N1171N | 005 | No | |
| 7 | 5′ Intron | PolyT variable | IVS6−9polyT | 0222 | No |
Database: InSIGHT (www.insight-group.org).
Other researchers have suggested that unique heteroduplex patterns representing polymorphisms are reproducible and can be reliably interpreted, resulting in a reduction of sequencing effort.24,26 Our laboratory is developing a database containing DHPLC profiles for known polymorphisms and variants within the MLH1, MSH2, and MSH6 genes. We identified five missense variants, four in MLH1 and one in MSH2, which have been reported as polymorphisms in the InSIGHT database. This database serves as a cross-reference for polymorphisms/mutations detected. Our results indicated a 100% mutation detection rate for the various types of mutations tested using our DHPLC scanning strategy.
All fragments showing an altered profile on DHPLC in comparison with the wild-type profile are confirmed by sequence analysis to identify the precise alteration. Although the majority of HNPCC patients will have a mutation detected using this approach, up to 12 to 15% may have a false-negative result due to possible gross alterations,47,48 and these patients will require additional testing approaches. The GMP conversion technology is indicated for patients who have a strong family history.49 This technology facilitates the separation of the two alleles so that they can be screened individually for mutations; however, it is not clinically available at present. Newer technologies such as the multiplex ligation-dependent probe amplification methodology allow detection of large gene deletions, which may comprise approximately 10% or even greater percentage of the mutations in MMR genes.48,50,51
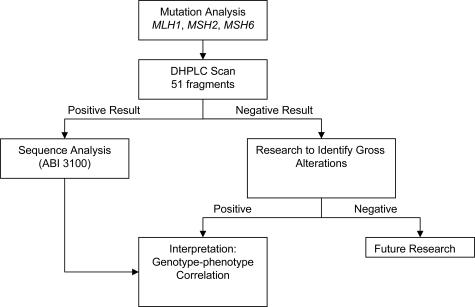
We propose the following tiered strategy for genetic testing for HNPCC, which is shown in Figure 5. This strategy is based on careful evaluation of the literature and development of an optimal testing strategy for a diagnostic test. The substratification of the testing strategy suits the Amsterdam criteria so that they may provide increased patient selectivity for mutation screening. Methylation of the MLH1 promoter has been shown to occur in a majority of sporadic CRC cases,52,53 and we propose that this should be undertaken along with microsatellite instability (MSI) and immunohistochemistry (IHC) before proceeding to mutation analysis. MSI and IHC are important diagnostic tools that are easy to perform and may provide an indicator for the possible causative gene and reduce the cost of mutation analysis. This approach is cost effective because the samples the number of assays that need to performed can be reduced if a positive result is obtained at any step. If a mutation has been identified in the targeted gene after MSI/IHC has been performed previously, then screening can be stopped, and the sample can be reported. If no mutation is obtained in targeted gene, then screening for mutations in the gene should be continued for other types of mutations. In our experience, it is difficult for the referring centers to access tumor tissue to perform MSI/IHC analysis, and only blood samples were obtained from patients. In these cases, mutation scanning for all three genes MLH1, MSH2, and MSH6 was recommended. Furthermore, the cost of DHPLC is about $1/injection × 3 (3 temperatures/samples) = $153. Subsequently adding sequencing of variants, the cost is $153 + $120 (∼15 DHPLC fragments selected for bidirectional sequencing/$4/sequence) = $273. This contrast with the cost of full bidirectional sequencing of all 51 fragments = $408. This cost includes only cost of reagents and running the machines. The labor cost is not included because it can vary from institution to institution. In conclusion, we have reported our approach to clinical genetic testing in HNPCC patients and at-risk family members. Our data provide validation of this approach for both known and unknown patient specimens, including high test sensitivity and specificity, compared with other currently used methods in clinical diagnostic laboratories.
Figure 5.
Algorithm for DHPLC screening for identification of mutations in mismatch repair genes in colorectal cancer patients.
Acknowledgments
We thank the technical staff of the molecular genetics laboratories at Mayo Clinic and Molecular Genetics Laboratory, Auckland Hospital (Auckland, New Zealand) for their efforts in characterizing the mutations that were presented in this manuscript. We also thank the HNPCC families that participated in this project through genetic testing.
Footnotes
Supported by National Cancer Institute grant U24-CA78142-03 to Texas Cancer Genetics Network.
References
- Burt RW. Familial risk and colorectal cancer. Gastroenterol Clin North Am. 1996;25:793–803. doi: 10.1016/s0889-8553(05)70275-2. [DOI] [PubMed] [Google Scholar]
- Burt RW. Familial association. Adv Exp Med Biol. 1999;470:99–104. doi: 10.1007/978-1-4615-4149-3_11. [DOI] [PubMed] [Google Scholar]
- Coughlin SS, Miller DS. Public health perspectives on testing for colorectal cancer susceptibility genes. Am J Prev Med. 1999;16:99–104. doi: 10.1016/s0749-3797(98)00137-8. [DOI] [PubMed] [Google Scholar]
- de Leon MP, Pedroni M, Benatti P, Percesepe A, Di Gregorio C, Foroni M, Rossi G, Genuardi M, Neri G, Leonardi F, Viel A, Capozzi E, Boiocchi M, Roncucci L. Hereditary colorectal cancer in the general population: from cancer registration to molecular diagnosis. Gut. 1999;45:32–38. doi: 10.1136/gut.45.1.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cederquist K, Golovleva I, Emanuelsson M, Stenling R, Gronberg H. A population based cohort study of patients with multiple colon and endometrial cancer: correlation of microsatellite instability (MSI) status, age at diagnosis and cancer risk. Int J Cancer. 2001;91:486–491. doi: 10.1002/1097-0215(200002)9999:9999<::aid-ijc1093>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Bocker T, Ruschoff J, Fishel R. Molecular diagnostics of cancer predisposition: hereditary non-polyposis colorectal carcinoma and mismatch repair defects. Biochim Biophys Acta. 1999;1423:1–10. doi: 10.1016/s0304-419x(99)00008-6. [DOI] [PubMed] [Google Scholar]
- Fishel R, Lescoe MK, Rao MR, Copeland NG, Jenkins NA, Garber J, Kane M, Kolodner R. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1993;75:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- Aaltonen LA, Peltomaki P. Genes involved in hereditary nonpolyposis colorectal carcinoma. Anticancer Res. 1994;14:1657–1660. [PubMed] [Google Scholar]
- Bronner CE, Baker SM, Morrison PT, Warren G, Smith LG, Lescoe MK, Kane M, Earabino C, Lipford J, Lindblom A, Tannergård P, Bollag RJ, Godwin AR, Ward DC, Nordenskjld M, Fishel R, Kolodner R, Liskay RM. Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature. 1994;368:258–261. doi: 10.1038/368258a0. [DOI] [PubMed] [Google Scholar]
- Papadopoulos N, Nicolaides NC, Wei Y-F, Ruben SM, Carter KC, Rosen CA, Haseltine WA, Fleischmann RD, Fraser CM, Adams MD, Venter JC, Hamilton SR, Petersen GM, Watson P, Lynch HT, Peltomaki P, Mecklin J-P, de la Chapelle A, Kinzler KW, Vogelstein B. Mutation of a mutL homolog in hereditary colon cancer. Science. 1994;263:1625–1629. doi: 10.1126/science.8128251. [DOI] [PubMed] [Google Scholar]
- Wu G, Wu W, Hegde M, Fawkner M, Chong B, Love D, Su LK, Lynch P, Snow K, Richards CS. Detection of sequence variations in the adenomatous polyposis coli (APC) gene using denaturing high-performance liquid chromatography. Genet Test. 2001;5:281–290. doi: 10.1089/109065701753617408. [DOI] [PubMed] [Google Scholar]
- Papadopoulos N, Lindblom A. Molecular basis of HNPCC: mutations of MMR genes. Hum Mutat. 1997;10:89–99. doi: 10.1002/(SICI)1098-1004(1997)10:2<89::AID-HUMU1>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Peltomaki P. DNA mismatch repair gene mutations in human cancer. Environ Health Perspect. 1997;105(Suppl 4):775–780. doi: 10.1289/ehp.105-1470030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltomaki P, de la Chapelle A. Mutations predisposing to hereditary nonpolyposis colorectal cancer. Adv Cancer Res. 1997;71:93–119. doi: 10.1016/s0065-230x(08)60097-4. [DOI] [PubMed] [Google Scholar]
- Holinski-Feder E, Muller-Koch Y, Friedl W, Moeslein G, Keller G, Plaschke J, Ballhausen W, Gross M, Baldwin-Jedele K, Jungck M, Mangold E, Vogelsang H, Schackert HK, Lohsea P, Murken J, Meitinger T. DHPLC mutation analysis of the hereditary nonpolyposis colon cancer (HNPCC) genes hMLH1 and hMSH2. J Biochem Biophys Methods. 2001;47:21–32. doi: 10.1016/s0165-022x(00)00148-2. [DOI] [PubMed] [Google Scholar]
- Oefner PJ, Bonn GK, Huber CG, Nathakarnkitkool S. Comparative study of capillary zone electrophoresis and high-performance liquid chromatography in the analysis of oligonucleotides and DNA. J Chromatogr. 1992;625:331–340. doi: 10.1016/0021-9673(92)85217-h. [DOI] [PubMed] [Google Scholar]
- Liu WO, Oefner PJ, Qian C, Odom RS, Francke U. Denaturing HPLC-identified novel FBN1 mutations, polymorphisms, and sequence variants in Marfan syndrome and related connective tissue disorders. Genet Test. 1997;1:237–242. doi: 10.1089/gte.1997.1.237. [DOI] [PubMed] [Google Scholar]
- Underhill PA, Jin L, Lin AA, Mehdi SQ, Jenkins T, Vollrath D, Davis RW, Cavalli-Sforza LL, Oefner PJ. Detection of numerous Y chromosome biallelic polymorphisms by denaturing high-performance liquid chromatography. Genome Res. 1997;7:996–1005. doi: 10.1101/gr.7.10.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donovan MC, Oefner PJ, Roberts SC, Austin J, Hoogendoorn B, Guy C, Speight G, Upadhyaya M, Sommer SS, McGuffin P. Blind analysis of denaturing high-performance liquid chromatography as a tool for mutation detection. Genomics. 1998;52:44–49. doi: 10.1006/geno.1998.5411. [DOI] [PubMed] [Google Scholar]
- Liu W, Smith DI, Rechtzigel KJ, Thibodeau SN, James CD. Denaturing high performance liquid chromatography (DHPLC) used in the detection of germline and somatic mutations. Nucleic Acids Res. 1998;26:1396–1400. doi: 10.1093/nar/26.6.1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AC, Austin J, Hansen N, Hoogendoorn B, Oefner PJ, Cheadle JP, O’Donovan MC. Optimal temperature selection for mutation detection by denaturing HPLC and comparison to single-stranded conformation polymorphism and heteroduplex analysis. Clin Chem. 1999;45:1133–1140. [PubMed] [Google Scholar]
- Arnold N, Gross E, Schwarz-Boeger U, Pfisterer J, Jonat W, Kiechle M. A highly sensitive, fast, and economical technique for mutation analysis in hereditary breast and ovarian cancers. Hum Mutat. 1999;14:333–339. doi: 10.1002/(SICI)1098-1004(199910)14:4<333::AID-HUMU9>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Choy YS, Dabora SL, Hall F, Ramesh V, Niida Y, Franz D, Kasprzyk-Obara J, Reeve MP, Kwiatkowski DJ. Superiority of denaturing high performance liquid chromatography over single-stranded conformation and conformation-sensitive gel electrophoresis for mutation detection in TSC2. Ann Hum Genet. 1999;63:383–391. doi: 10.1046/j.1469-1809.1999.6350383.x. [DOI] [PubMed] [Google Scholar]
- Gross E, Arnold N, Goette J, Schwarz-Boeger U, Kiechle M. A comparison of BRCA1 mutation analysis by direct sequencing, SSCP and DHPLC. Hum Genet. 1999;105:72–78. doi: 10.1007/s004399900092. [DOI] [PubMed] [Google Scholar]
- Wagner T, Stoppa-Lyonnet D, Fleischmann E, Muhr D, Pages S, Sandberg T, Caux V, Moeslinger R, Langbauer G, Borg A, Oefner P. Denaturing high-performance liquid chromatography detects reliably BRCA1 and BRCA2 mutations. Genomics. 1999;62:369–376. doi: 10.1006/geno.1999.6026. [DOI] [PubMed] [Google Scholar]
- Gross E, Arnold N, Pfeifer K, Bandick K, Kiechle M. Identification of specific BRCA1 and BRCA2 variants by DHPLC. Hum Mutat. 2000;16:345–353. doi: 10.1002/1098-1004(200010)16:4<345::AID-HUMU7>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Jones AC, Sampson JR, Hoogendoorn B, Cohen D, Cheadle JP. Application and evaluation of denaturing HPLC for molecular genetic analysis in tuberous sclerosis. Hum Genet. 2000;106:663–668. doi: 10.1007/s004390000316. [DOI] [PubMed] [Google Scholar]
- Benit P, Bonnefont JP, Kara Mostefa A, Francannet C, Munnich A, Ray PF. Denaturing high-performance liquid chromatography (DHPLC)-based prenatal diagnosis for tuberous sclerosis. Prenat Diagn. 2001;21:279–283. doi: 10.1002/pd.55. [DOI] [PubMed] [Google Scholar]
- Roberts PS, Jozwiak S, Kwiatkowski DJ, Dabora SL. Denaturing high-performance liquid chromatography (DHPLC) is a highly sensitive, semi-automated method for identifying mutations in the TSC1 gene. J Biochem Biophys Methods. 2001;47:33–37. doi: 10.1016/s0165-022x(00)00149-4. [DOI] [PubMed] [Google Scholar]
- Kurzawski G, Safranow K, Suchy J, Chlubek D, Scott RJ, Lubinski J. Mutation analysis of MLH1 and MSH2 genes performed by denaturing high-performance liquid chromatography. J Biochem Biophys Methods. 2002;51:89–100. doi: 10.1016/s0165-022x(02)00003-9. [DOI] [PubMed] [Google Scholar]
- Panariello L, Scarano MI, de Rosa M, Capasso L, Renda A, Riegler G, Rossi GB, Salvatore F, Izzo P. hMLH1 mutations in hereditary nonpolyposis colorectal cancer kindreds: mutations in brief no. 182: online. Hum Mutat. 1998;12:216–217. [PubMed] [Google Scholar]
- Fidalgo P, Almeida MR, West S, Gaspar C, Maia L, Wijnen J, Albuquerque C, Curtis A, Cravo M, Fodde R, Leitao CN, Burn J. Detection of mutations in mismatch repair genes in Portuguese families with hereditary non-polyposis colorectal cancer (HNPCC) by a multi-method approach. Eur J Hum Genet. 2000;8:49–53. doi: 10.1038/sj.ejhg.5200393. [DOI] [PubMed] [Google Scholar]
- Parc YR, Halling KC, Burgart LJ, McDonnell SK, Schaid DJ, Thibodeau SN, Halling AC. Microsatellite instability and hMLH1/hMSH2 expression in young endometrial carcinoma patients: associations with family history and histopathology. Int J Cancer. 2000;86:60–66. doi: 10.1002/(sici)1097-0215(20000401)86:1<60::aid-ijc9>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Godino J, de La Hoya M, Diaz-Rubio E, Benito M, Caldes T. Eight novel germline MLH1 and MSH2 mutations in hereditary non-polyposis colorectal cancer families from Spain. Hum Mutat. 2001;18:549. doi: 10.1002/humu.1240. [DOI] [PubMed] [Google Scholar]
- Jass JR. Diagnosis of hereditary non-polyposis colorectal cancer. Histopathology. 1998;32:491–497. doi: 10.1046/j.1365-2559.1998.00442.x. [DOI] [PubMed] [Google Scholar]
- Young J, Barker M, Fraser L, Walsh MD, Spring K, Biden KG, Hopper JL, Leggett BA, Jass JR. Mutation searching in colorectal cancer studies: experience with a denaturing high-pressure liquid chromatography system for exon-by-exon scanning of tumour suppressor genes. Pathology. 2002;34:529–533. doi: 10.1080/0031302021000035965-1. [DOI] [PubMed] [Google Scholar]
- Bellacosa A, Genuardi M, Anti M, Viel A, Ponz de Leon M. Hereditary nonpolyposis colorectal cancer: review of clinical, molecular genetics, and counseling aspects. Am J Med Genet. 1996;62:353–364. doi: 10.1002/(SICI)1096-8628(19960424)62:4<353::AID-AJMG7>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Genuardi M, Anti M, Capozzi E, Leonardi F, Fornasarig M, Novella E, Bellacosa A, Valenti A, Gasbarrini GB, Roncucci L, Benatti P, Percesepe A, Ponz de Leon M, Coco C, de Paoli A, Valentini M, Boiocchi M, Neri G, Viel A. MLH1 and MSH2 constitutional mutations in colorectal cancer families not meeting the standard criteria for hereditary nonpolyposis colorectal cancer. Int J Cancer. 1998;75:835–839. doi: 10.1002/(sici)1097-0215(19980316)75:6<835::aid-ijc4>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- Liu T, Wahlberg S, Rubio C, Holmberg E, Gronberg H, Lindblom A. DGGE screening of mutations in mismatch repair genes (hMSH2 and hMLH1) in 34 Swedish families with colorectal cancer. Clin Genet. 1998a;53:131–135. doi: 10.1111/j.1399-0004.1998.tb02660.x. [DOI] [PubMed] [Google Scholar]
- Planck M, Koul A, Fernebro E, Borg A, Kristoffersson U, Olsson H, Wenngren E, Mangell P, Nilbert M. hMLH1, hMSH2 and hMSH6 mutations in hereditary non-polyposis colorectal cancer families from southern Sweden. Int J Cancer. 1999;83:197–202. doi: 10.1002/(sici)1097-0215(19991008)83:2<197::aid-ijc9>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Borg A, Isola J, Chen J, Rubio C, Johansson U, Werelius B, Lindblom A. Germline BRCA1 and HMLH1 mutations in a family with male and female breast carcinoma. Int J Cancer. 2000;85:796–800. doi: 10.1002/(sici)1097-0215(20000315)85:6<796::aid-ijc10>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Beck NE, Tomlinson IP, Homfray T, Frayling I, Hodgson SV, Harocopos C, Bodmer WF. Use of SSCP analysis to identify germline mutations in HNPCC families fulfilling the Amsterdam criteria. Hum Genet. 1997;99:219–224. doi: 10.1007/s004390050343. [DOI] [PubMed] [Google Scholar]
- Glasl S, Papatheodorou L, Baretton G, Jung C, Gross M. Novel germline mutation (300–305delAGTTGA) in the human MSH2 gene in hereditary non-polyposis colorectal cancer (HNPCC). Hum Mutat. 2000;16:91–92. doi: 10.1002/1098-1004(200007)16:1<91::AID-HUMU22>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Caluseriu O, Cordisco EL, Viel A, Majore S, Nascimbeni R, Pucciarelli S, Genuardi M. Four novel MSH2 and MLH1 frameshift mutations and occurrence of a breast cancer phenocopy in hereditary nonpolyposis colorectal cancer. Hum Mutat. 2001;17:521. doi: 10.1002/humu.1137. [DOI] [PubMed] [Google Scholar]
- Berends MJ, Wu Y, Sijmons RH, Mensink RG, van der Sluis T, Hordijk-Hos JM, de Vries EG, Hollema H, Karrenbeld A, Buys CH, van der Zee AG, Hofstra RM, Kleibeuker JH. Molecular and clinical characteristics of MSH6 variants: an analysis of 25 index carriers of a germline variant. Am J Hum Genet. 2002;70:26–37. doi: 10.1086/337944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldes T, Godino J, de la Hoya M, Garcia Carbonero I, Perez Segura P, Eng C, Benito M, Diaz-Rubio E. Prevalence of germline mutations of MLH1 and MSH2 in hereditary nonpolyposis colorectal cancer families from Spain. Int J Cancer. 2002;98:774–779. doi: 10.1002/ijc.10240. [DOI] [PubMed] [Google Scholar]
- Wang Y, Friedl W, Lamberti C, Jungck M, Mathiak M, Pagenstecher C, Propping P, Mangold E. Hereditary nonpolyposis colorectal cancer: frequent occurrence of large genomic deletions in MSH2 and MLH1 genes. Int J Cancer. 2003;103:636–641. doi: 10.1002/ijc.10869. [DOI] [PubMed] [Google Scholar]
- Wang Y, Friedl W, Sengteller M, Jungck M, Filges I, Propping P, Mangold E. A modified multiplex PCR assay for detection of large deletions in MSH2 and MLH1. Hum Mutat. 2002;19:279–286. doi: 10.1002/humu.10042. [DOI] [PubMed] [Google Scholar]
- Yan H, Kinzler KW, Vogelstein B. Tech.sight. Genetic testing: present and future. Science. 2000;289:1890–1892. doi: 10.1126/science.289.5486.1890. [DOI] [PubMed] [Google Scholar]
- Charbonnier F, Raux G, Wang Q, Drouot N, Cordier F, Limacher JM, Saurin JC, Puisieux A, Olschwang S, Frebourg T. Detection of exon deletions and duplications of the mismatch repair genes in hereditary nonpolyposis colorectal cancer families using multiplex polymerase chain reaction of short fluorescent fragments. Cancer Res. 2000;60:2760–2763. [PubMed] [Google Scholar]
- Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30:e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menigatti M, Di Gregorio C, Borghi F, Sala E, Scarselli A, Pedroni M, Foroni M, Benatti P, Roncucci L, Ponz de Leon M, Percesepe A. Methylation pattern of different regions of the MLH1 promoter and silencing of gene expression in hereditary and sporadic colorectal cancer. Genes Chromosomes Cancer. 2001;31:357–361. doi: 10.1002/gcc.1154. [DOI] [PubMed] [Google Scholar]
- Yuen ST, Chan TL, Ho JW, Chan AS, Chung LP, Lam PW, Tse CW, Wyllie AH, Leung SY. Germline, somatic and epigenetic events underlying mismatch repair deficiency in colorectal and HNPCC-related cancers. Oncogene. 2002;21:7585–7592. doi: 10.1038/sj.onc.1205968. [DOI] [PubMed] [Google Scholar]