Abstract
Pegylated α-interferon plus ribavirin is the current therapy for chronic hepatitis C virus (HCV) infection. Serum HCV-RNA concentration before treatment has been identified as an independent predictive factor of response. We have compared the percentage of HCV-infected hepatocytes with the concentration of serum HCV-RNA in baseline samples as predictors of response. We included 97 patients with chronic HCV infection (genotype 1), treated with pegylated-interferon-α2b plus ribavirin. Of these 97, 38 (39%) were sustained responders and 59 (61%) were not. Statistical differences between responders and nonresponders were found regarding the percentage of infected hepatocytes (6.83 ± 4.50% versus 13.44 ± 10.05%; P = 0.00003) but not in serum HCV-RNA concentration [1.71 ± 2.70 (×106 IU/L) versus 1.32 ± 1.86 (×106 IU/L); P = 0.40694]. Other factors associated with response were age, γ-glutamyl transpeptidase level, and absence of previous therapy. Logistic regression demonstrated that percentage of infected hepatocytes (odds ratio, 1.160; 95% confidence interval, 1.065–1.264) and previous therapy (odds ratio, 0.294; 95% confidence interval, 0.109–0.795) were significant predictive factors for response. Therefore, the percentage of infected hepatocytes in liver biopsy before treatment is a better predictive factor of sustained response to 48 weeks of therapy with pegylated α-interferon plus ribavirin than serum HCV-RNA concentration in baseline serum sample.
Hepatitis C virus (HCV) infection is a major health problem as it is estimated that 170 million people around the world are chronically infected by this virus.1 Pegylated α-interferon (PEG-IFN) plus ribavirin (a purine nucleoside analog) is the current therapy for the treatment of patients with chronic HCV infection. However, the sustained response rate (loss of serum HCV-RNA and normalization of alanine amino transferase (ALT) levels for more than 6 months after the end of the therapy) to these treatments is around 40% for HCV genotype 1-infected patients.2,3,4,5
Because these therapies have important side effects and high cost, it is important to identify which patients have the best chance to respond before the therapy. In this regard, several virological and clinical factors have been identified as associated with the likelihood of response. Among these factors, absence of fibrosis in the liver biopsy, viral genotype, and serum HCV-RNA concentration have been shown to be independent factors associated with a sustained response to the therapy.6,7,8 Regarding serum viremia levels, the threshold for a favorable response to 24 weeks of therapy has been established at 800,000 IU/ml.9 However, measurement of serum HCV-RNA concentration may not be accurate enough because it depends on several factors such as storage conditions of serum samples, efficiency of HCV-RNA extraction procedures or presence of inhibitors of thermo-stable polymerase chain reaction (PCR) enzymes in serum samples.10,11
In situ hybridization is a technique that allows the localization of a target nucleic acid within individual cells in a tissue section12 with a sensitivity of 10–20 copies of a given RNA per cell.13 Because it has been estimated that the number of HCV genomes per infected cell ranges from 7 to 64 molecules,14 in situ hybridization is a technique sensitive enough to detect HCV-infected hepatocytes in liver biopsies. In fact, using this technique, we have detected HCV-infected hepatocytes in liver biopsies from all chronic hepatitis C patients with detectable HCV-RNA in serum analyzed so far,15,16 and we have found that the serum HCV-RNA concentration is related with the percentage of HCV-infected hepatocytes determined by in situ hybridization.17 However, there are no studies comparing serum HCV viremia and the percentage of HCV-infected hepatocytes as predictors of response to 48 weeks of therapy. Thus, in the present study, we have compared the percentage of HCV-infected hepatocytes, determined by in situ hybridization in the pretreatment liver biopsy, with the HCV-RNA concentration in serum samples obtained the same day as the liver biopsy, as predictors of response to 48 weeks of therapy with pegylated α-IFN plus ribavirin.
Materials and Methods
In the present study, individual data from 97 patients (62 males and 37 females) with chronic HCV infection (abnormal ALT values and anti-HCV and serum HCV-RNA positive for at least 6 months) were analyzed. All patients were infected by the HCV genotype 1 as determined with a reverse hybridization assay (INNO LIPA HCV-II; Innogenetics, Gent, Belgium) and were hepatitis B surface antigen negative, and none of them had antibodies against human immunodeficiency virus 1 and 2. Patients were treated with pegylated-IFN-α2b (Peg-Intron; Schering Corporation, Kenilworth, NJ) at doses of 1.5 μg/kg body weight once weekly plus ribavirin (Rebetol; Schering Corporation) at doses of 1000 to 1200 mg/day for 12 months.
Forty-nine of the 97 patients (50.5%) were nonresponders to previous antiviral therapies (44 were treated with 3 MU/thrice weekly (tiw) IFN-α for 6 to 12 months and 5 with 3 MU/tiw IFN-α plus 1000 to 1200 mg ribavirin for 6 to 12 months), but none of them was under antiviral or immunosuppressive therapy for at least 12 months before entry in the present study. The study was performed following the guidelines of the 1975 Declaration of Helsinki, and a written informed consent was obtained from each patient.
Virological response was determined with the Amplicor HCV Monitor 2.0 test (Roche Diagnostics System, Basel, Switzerland) as described below and confirmed by an in-house reverse transcriptase (RT)-PCR with primers derived from the 5′ noncoding region of the viral genome, with a sensitivity of 10 IU/ml.18 Patients were defined as sustained responders when they presented normal ALT values and did not have detectable serum HCV-RNA for at least 6 months after the end of the therapy. The remaining patients were considered as nonresponders.
A baseline liver biopsy was obtained from each patient in the 1-month period before the therapy. Liver biopsies were immersed in 4% paraformaldehyde-phosphate-buffered saline in less than 30 seconds after they were obtained and fixed overnight in this buffer. The next day, tissue samples were dehydrated through successive baths of ethanol and embedded in paraffin, and the paraffin blocks were stored at 4°C until the histological diagnosis and in situ hybridization were performed. Hepatic necroinflammation and fibrosis were assessed according to the METAVIR score system.19,20 After histological diagnosis, the remaining tissue was used for in situ hybridization.
Serum HCV-RNA Quantitation
HCV-RNA concentration in the baseline serum sample taken the same day as the liver biopsy was measured with the Amplicor HCV Monitor 2.0 test (Roche Diagnostics System). Serum samples were aliquoted and stored at −80°C until used. When the viral RNA concentration of a given sample was above the upper dynamic range of quantitation of the assay (500,000 IU/ml), the serum sample was retested diluted (1/10 and 1/100) to obtain an accurate quantitation.
In Situ Hybridization
Genomic HCV-RNA was detected with a complementary RNA probe labeled with digoxigenin 11-UTP obtained by in vitro transcription of the pC5′NCR, which contains the complete 5′NC region of the HCV genome. Hybridization conditions for the in situ detection of the HCV-RNA were as described previously.15,16,17 Specificity of the in situ hybridization was assessed by: 1) digestions of the sections with RNase A (0.2 mg/ml) or DNase I (20 U/ml) for 2 hours at 37°C before hybridization; 2) hybridization with an unrelated RNA probe (a 360 base fragment of the chloramphenicol acetyl transferase gene); and 3) omission of the probe in the hybridization mixture. To further demonstrate the specificity of the HCV-RNA detection by in situ hybridization, liver biopsies from 10 patients with chronic hepatitis B virus infection, 10 patients with alcoholic hepatitis, and 5 patients with chronic autoimmune hepatitis (all of them without HCV markers) were hybridized with the same probe and under the same conditions used for the detection of the HCV-RNA. The percentage of infected cells was determined by visual inspection of at least 20 microscopic fields, counting at least 2000 cells from each liver section. To test the reproducibility of our in situ hybridization technique, the percentage of infected hepatocytes was determined in serial sections of two liver biopsies (from which enough material was available) hybridized in different runs (four sections from each biopsy in three different runs carried out on three different days).
Statistical Analysis
All statistical tests described below were carried out with SPSS package release 9.0 (SPSS, Chicago, IL). All tests performed were two-sided, and statistical significance was established at P < 0.05.
Reproducibility Analysis
In each biopsy, the mean percentage of the infected hepatocytes observed in each run and its 95% confidence interval (CI) were estimated and compared by a one-way analysis of variance, once the equality between the variances of the variables was checked with the Levene’s test.
Univariate Analysis
Several parameters were compared between responder and nonresponder patients. Continuous variables included in the analysis were as follows: serum HCV-RNA concentration (IU/ml); percentage of infected hepatocytes; age; body mass index; ALT level (IU/L); aspartate amino transferase level (IU/ml); γ-glutamyl transpeptidase (GGTP) level (IU/L); ferritin level (ng/ml); iron level (μg/dl); necroinflammatory activity; and fibrosis score. The categorical variables analyzed were as follows: gender (0, male; 1, female); and previous antiviral therapy (0, yes; 1, no). After exploring the continuous variables for normality using the Kolmogorov-Smirnov test, the mean was compared with the Student’s t-test in those with normal distribution. In these variables, equality between the variances of the variables was checked with the Levene’s test. In the continuous variables without normal distribution, the median between responder and nonresponder patients was compared using the nonparametric Mann-Whitney U test. The variables with normal distribution were expressed as the mean ± SD, and those without normal distribution were expressed as the median (range). Categorical variables were compared between responders and nonresponders using the χ2 or Fisher’s exact tests. The same univariate analysis was also performed in the subgroup of previously treated and untreated patients, comparing responders and nonresponders.
Logistic Regression Analysis
Binary logistic regression analysis was performed to explore the influence of the above described variables on the response. Dependent variable was defined as “Response” (0, responder patient; 1, nonresponder patient). First, a global model with all of the variables included in the univariate analysis was considered. Nonsignificant variables were excluded from the model one by one. Overall significance was assessed by the log of likelihood ratio with the χ2 test, and goodness-of-fit was studied by the Hosmer-Lemeshow test. Statistical significance of the coefficients in the regression equation was contrasted with the Wald test. Odds ratios (OR) and their respective 95% CI were also estimated.
To study the ability of the definitive model in the discrimination between responder and nonresponder patients, receiver operating characteristic (ROC) curve was constructed using the predicted probability values estimated with this model as the test variable and “Response = 1” as the value of the state variable. A ROC curve is a graphic representation of the trade-off between the false-negative and false-positive rates for every possible cutoff. The accuracy of the model depends on how well it separates the group of patients being tested into responders and nonresponders. Accuracy is measured by the area under the ROC curve (AUC), being an area of 1, a perfect model. The cutoff probability value to discriminate between responder and nonresponder patients was estimated by examining the coordinates of the ROC curve. This cutoff probability value was established at the maximum specificity and sensitivity. Finally, specificity, sensitivity, false-positive and false-negative rates, positive and negative predictive values, and overall accuracy or diagnostic efficiency of the model were also estimated, according to the coordinates of the ROC curve.
Results
Of the 97 patients analyzed in this study, 38 (39%) were sustained responders, whereas the remaining 59 (61%) patients were nonresponders.
Specificity of the in Situ Hybridization Technique
Positive hybridization signals were observed in the liver biopsies from the 97 patients with chronic hepatitis C analyzed in this study, whereas no signals were detected in the liver samples from the 10 patients with chronic hepatitis B, the 10 patients with alcoholic hepatitis, or the 5 patients with autoimmune chronic hepatitis. Furthermore, when the liver biopsies from the patients with chronic hepatitis C were digested with RNase before the hybridization, no signals were observed, whereas no changes in the hybridization pattern were seen when the liver biopsies were predigested with DNase. Finally, no hybridization signals were observed when the liver biopsies were hybridized with an unrelated probe or when the specific probe was omitted in the hybridization mixture. All of these results demonstrated the specificity and the accuracy of the detection of HCV-RNA in liver biopsies by the in situ hybridization technique.
Reproducibility of the in Situ Hybridization Technique
To demonstrate the reproducibility of the in situ hybridization for the detection of HCV-RNA, serial sections of two liver biopsies from two different patients were tested in the same run and in different runs. As shown in Table 1, in the two liver biopsies analyzed, no statistical differences in the mean percentage of infected hepatocytes in the sections analyzed in three different runs were found, which indicates that the technique is highly reproducible.
Table 1.
Reproducibility of the in Situ Hybridization Technique
| Infected hepatocytes (%)
|
||
|---|---|---|
| Biopsy A | Biopsy B | |
| Run 1 | ||
| Section 1 | 4.9 | 6.0 |
| Section 2 | 4.4 | 5.5 |
| Section 3 | 5.4 | 5.0 |
| Section 4 | 4.0 | 6.2 |
| Mean (95% CI) | 4.7 (3.7–5.6) | 5.7 (4.8–6.5) |
| Run 2 | ||
| Section 1 | 4.2 | 5.4 |
| Section 2 | 5.0 | 5.2 |
| Section 3 | 3.9 | 5.9 |
| Section 4 | 5.2 | 6.5 |
| Mean (95% CI) | 4.6 (3.6–5.6) | 5.8 (4.8–6.7) |
| Run 3 | ||
| Section 1 | 4.9 | 6.1 |
| Section 2 | 5.3 | 4.9 |
| Section 3 | 4.1 | 6.5 |
| Section 4 | 4.8 | 7.1 |
| Mean (95% CI) | 4.8 (4.0–5.6) | 6.2 (4.7–7.6) |
| P value | 0.88909 | 0.60837 |
Univariate Analysis
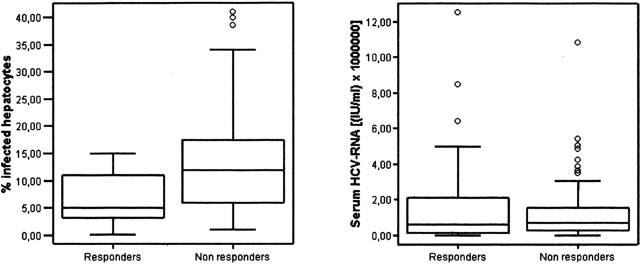
Regarding HCV-RNA concentration in the basal serum sample, there were no statistical differences between sustained responder (1.71 × 106± 2.70 × 106 IU/ml) and nonresponder patients (1.32 × 106± 1.86 × 106 IU/ml) (Figure 1). In contrast, sustained responders had a significantly lower percentage of HCV-infected hepatocytes in the pretreatment liver biopsy than the nonresponder patients (6.83 ± 4.50% versus 13.44 ± 10.05%; P = 0.00003) (Figure 1). Other baseline characteristics significantly associated with a sustained response were age (responder patients were younger), GGTP levels (responder patients had lower levels of this enzyme), and absence of a previous antiviral treatment. The remaining variables analyzed were not significantly associated with a sustained response to the therapy (Table 2).
Figure 1.
Box-plot representations of the percentage of infected hepatocytes and the serum HCV-RNA levels in responder and nonresponder patients. Each box is drawn from the lower quartile (Q1) to the upper quartile (Q3) and the horizontal lines across the boxes indicate the median. The whiskers are drawn from the Q3 to the maximum and from the Q1 to the minimum. Outliers are represented by empty dots.
Table 2.
Results of Univariate Analysis Performed in All of the Patients
| Variable | Responders (n = 38) | Nonresponders (n = 59) | P Value | |
|---|---|---|---|---|
| Age (years)* | 43.92 ± 11.01 | 48.92 ± 8.94 | 0.01611 | |
| Gender | ||||
| Male | 28/38 (73.7%) | 34/59 (57.6%) | } | 0.10795 |
| Female | 10/38 (26.3%) | 25/59 (42.4%) | ||
| Body mass index* | 24.11 ± 2.73 | 25.57 ± 3.45 | 0.07827 | |
| Infected hepatocytes (%)* | 6.83 ± 4.50 | 13.44 ± 10.05 | 0.00003 | |
| Serum HCV-RNA [(IU/ml)× 106]* | 1.71 ± 2.70 | 1.32 ± 1.86 | 0.40694 | |
| ALT (IU/L)* | 114.66 ± 107.37 | 105.31 ± 58.61 | 0.48027 | |
| AST (IU/L)* | 70.29 ± 70.98 | 72.97 ± 37.76 | 0.80948 | |
| GGTP (IU/L)† | 30 (11–239) | 43 (9–326) | 0.01856 | |
| Ferritin (ng/ml)† | 216 (41–1250) | 132 (6–1122) | 0.34071 | |
| Iron (μg/dl)* | 128.27 ± 57.45 | 133.18 ± 56.98 | 0.72552 | |
| Necroinflammatory activity† | 4 (1–6) | 4 (1–7) | 0.60170 | |
| Fibrosis score† | 2 (0–4) | 2 (0–4) | 0.53835 | |
| Previous treatment | ||||
| Yes | 14/38 (36.8%) | 35/59 (59.3%) | } | 0.03065 |
| No | 24/38 (63.2%) | 24/59 (40.7%) |
Statistically significant P values are highlighted in boldface.
AST, aspartate aminotransferase.
Expressed as the mean ± SD.
Expressed as the median (range).
When previously untreated patients were analyzed alone, there were statistical differences between responder and nonresponder patients in the percentage of infected hepatocytes in the basal liver biopsy (6.28 ± 4.29% versus 17.74 ± 11.39%; P = 0.00007) and in the GGTP levels [30 IU/L (range, 11–239 IU/L) versus 43 IU/L (range, 9–326 IU/L); P = 0.01856], whereas no differences were found in the remaining variables, including the baseline serum HCV-RNA concentration (Table 3). The same analysis was performed in previously treated patients, and no significant differences in the variables (including the percentage of hepatocytes) were found (Table 4).
Table 3.
Results of Univariate Analysis Performed in Previously Untreated Patients
| Variable | Responders (n = 24) | Nonresponders (n = 24) | P value | |
|---|---|---|---|---|
| Age (years)* | 43.67 ± 12.22 | 46.42 ± 9.72 | 0.39270 | |
| Gender | ||||
| Male | 16/24 (66.7%) | 11/24 (45.8%) | } | 0.14573 |
| Female | 8/24 (33.7%) | 13/24 (54.2%) | ||
| Body mass index* | 24.10 ± 2.82 | 25.78 ± 3.90 | 0.10149 | |
| Infected hepatocytes(%)* | 6.28 ± 4.29 | 17.74 ± 11.39 | 0.00007 | |
| Serum HCV-RNA [(IU/ml)× 106]* | 1.46 ± 1.82 | 1.86 ± 1.74 | 0.24816 | |
| ALT(IU/L)* | 110.17 ± 84.19 | 117.29 ± 69.87 | 0.50932 | |
| AST(IU/L)* | 64.71 ± 41.72 | 84.92 ± 47.25 | 0.12312 | |
| GGTP(IU/L)† | 32 (11–239) | 54 (13–263) | 0.01332 | |
| Ferritin(ng/ml)† | 236 (41–1250) | 128 (6–884) | 0.57109 | |
| Iron(μg/dl)† | 134 (38–214) | 116 (42–250) | 1.00000 | |
| Necroinflammatory activity* | 4.08 ± 1.35 | 3.96 ± 1.27 | 0.74226 | |
| Fibrosis score* | 1.71 ± 1.33 | 1.67 ± 1.17 | 0.90884 |
Statistically significant P values are highlighted in boldface.
AST, aspartate aminotransferase.
Expressed as the mean ± SD.
Expressed as the median (range).
Table 4.
Results of Univariate Analysis Performed in Previously Treated Patients
| Variable | Responders (n = 14) | Nonresponders (n = 35) | P value | |
|---|---|---|---|---|
| Age (years)* | 44.36 ± 8.98 | 50.63 ± 8.07 | 0.02134 | |
| Gender | ||||
| Male | 12/14 (85.7%) | 23/35 (65.7%) | } | 0.29361 |
| Female | 2/14 (14.3%) | 12/35 (34.3%) | ||
| Body mass index* | 24.13 ± 2.30 | 25.19 ± 2.59 | 0.63773 | |
| Infected hepatocytes(%)* | 7.79 ± 4.84 | 10.49 ± 7.91 | 0.40603 | |
| Serum HCV-RNA [(IU/ml)× 106]* | 2.12 ± 3.83 | 0.96 ± 1.86 | 0.73154 | |
| ALT(IU/L)* | 122.36 ± 142.36 | 97.06 ± 48.85 | 0.45837 | |
| AST(IU/L)* | 79.86 ± 105.38 | 64.78 ± 27.43 | 0.14698 | |
| GGTP(IU/L)† | 28.5 (15–175) | 38 (9–326) | 0.28791 | |
| Ferritin(ng/ml)† | 197 (83–451) | 160 (33–1122) | 0.36006 | |
| Iron(μg/dl)* | 117.43 ± 67.24 | 136.03 ± 62.52 | 0.50088 | |
| Necroinflammatory activity† | 3 (1–5) | 4 (1–7) | 0.08681 | |
| Fibrosis score† | 1 (0–3) | 2 (0–4) | 0.32197 |
Statistically significant P values are highlighted in boldface.
AST, aspartate aminotransferase.
Expressed as the mean ± SD.
Expressed as the median (range).
When a viremia level of 800,000 IU/ml and 7% of infected hepatocytes (the mean of infected hepatocytes in responder patients) were chosen as the threshold for sustained response, it was found that 54 patients had a basal serum HCV-RNA concentration lower than 800,000 IU/ml, and 21 (38.9%) of them were sustained responders (Table 5). In contrast, 40 patients had 7% or fewer infected hepatocytes, and 21 (52.5%) of them were sustained responders. On the other hand, of the 43 patients with viremia levels higher than 800,000 IU/ml, 17 (39.5%) were sustained responders; whereas 57 patients had more than 7% infected hepatocytes in the basal liver biopsy, and 17 (29.8%) of them were responders (Table 5).
Table 5.
Percentage of Responder and Nonresponder Patients, according to the HCV-RNA Concentration or the Percentage of Infected Hepatocytes
| Responders | Nonresponders | P value | ||
|---|---|---|---|---|
| HCV-RNA concentration | ||||
| <800,000 IU/ml (n = 54) | 21/54 (38.9%) | 33/54 (61.1%) | } | 0.94837 |
| ≥800,000 IU/ml (n = 43) | 17/43 (39.5%) | 26/43 (60.5%) | ||
| Infected hepatocytes (%) | ||||
| ≤7% (n = 40) | 21/40 (52.5%) | 19/40 (47.5%) | } | 0.02431 |
| >7% (n = 57) | 17/57 (29.8%) | 40/57 (70.2%) |
Multivariate Analysis
Logistic Regression Analysis
A global binary logistic analysis model was constructed to study the effect of all of the variables analyzed in the univariate analysis on the “Response” as dependent variable (0, responder patients; 1, nonresponder patients). Table 6 shows the best fitted model obtained after the exclusion of the nonsignificant variables (one by one). Log of likelihood ratio contrasted by the χ2 test demonstrated that the model was highly significant [χ2 = 31.212; degrees of freedom (df) = 4; P = 0.0000028]. The Hosmer-Lemeshow test, which evaluates the differences between the probabilities predicted by the model and those observed, showed that the goodness-of-fit of the model was acceptable (χ2 = 10.605; df = 8; P = 0.225).
As shown in Table 6, only the percentage of infected hepatocytes and previous antiviral therapy were statistically significant (P = 0.001 and P = 0.016, respectively), whereas age and GGTP levels were not (P = 0.111 and P = 0.101, respectively). Thus, the percentage of infected hepatocytes and the previous antiviral therapy could be considered as significant prognostic factors for response, adjusting for age and GGTP levels. In this sense, the OR estimated for the percentage of infected hepatocytes was 1.160 (95% CI, 1.065–1.264) (Table 6), indicating that the probability of being nonresponder is higher in the patients with a high percentage of infected hepatocytes. On the other hand, the OR for the previous antiviral treatment was 0.294 (95% CI, 0.109–0.795) (Table 4), which indicates that patients with previous antiviral therapy have a higher probability of being a nonresponder than those without a previous treatment.
TABLE 6.
Results of Logistic Regression Analysis
| Variable | Coefficient (B) | P value | Odds ratio | 95% CI of odds ratio |
|---|---|---|---|---|
| Percentage of infected hepatocytes | 0.149 | 0.001 | 1.160 | 1.065–1.264 |
| Age | 0.043 | 0.111 | 1.044 | 0.990–1.100 |
| Previous treatment* | −1.225 | 0.016 | 0.294 | 0.109–0.795 |
| GGTP level | 0.008 | 0.101 | 1.008 | 0.999–1.017 |
| Constant | −2.791 | 0.048 |
Dependent variable analyzed was ′Response′ and was defined as 0, responders and 1, nonresponders.
Categorical variable (0, yes; 1, no).
CI, confidence interval; GGTP, gamma glutamyl transpeptidase.
According to the estimated parameters of the model, the probability of no response for a given patient could be predicted by substituting the value of the factors into the following equation:
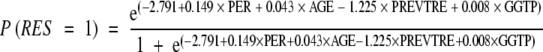 |
where P (RES = 1) is the probability of no response; PER is the percentage of infected hepatocytes; PREVTRE indicates whether the patient had received previous antiviral therapy or not (0, yes; 1:, no); AGE is the age of the patient expressed in years; and GGTP is the GGTP level.
ROC Curve
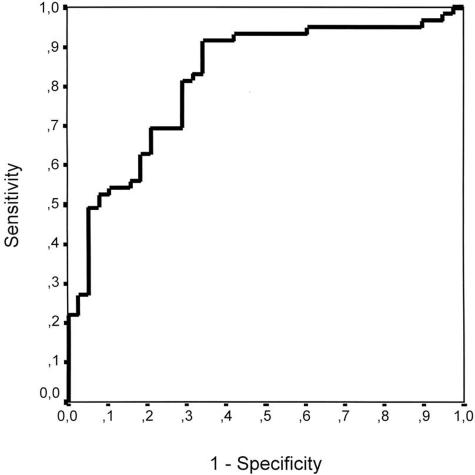
The probability values estimated with the equation described above were used to construct a ROC curve taking no response as the state variable (Figure 2). AUC was 0.819 (95% CI, 0.733–0.906). The contrast of null hypothesis of the true AUC = 0.5 rendered a P value = 0.0000001, leading to rejection of the null hypothesis. The ROC curve demonstrated that the regression model was able to discriminate between responder and nonresponder patients with a relatively high accuracy.
Figure 2.
ROC curve constructed with the probability values predicted with the logistic regression model, using no response as the state variable. AUC = 0.819; 95% confidence interval of AUC, 0.733–0.906.
Coordinates of the ROC curve were examined to look for the threshold probability value giving the maximum specificity and sensitivity. The threshold was found at a probability value of 0.46, so patients with a predicted probability value of 0.46 or more were considered as nonresponders by the model. Table 7 shows the number of patients correctly and incorrectly classified by the model, taking a predicted probability value of 0.46 as a cutoff. Taking the data depicted on Table 7 into account, the model had 65.8% specificity and 91.5% sensitivity for detecting nonresponder patients. False-positive and false-negative rates were 34.2 and 8.5%, respectively. Predictive positive value was 80.6%, and predictive negative value 83.3%. Finally, the overall accuracy or diagnostic efficiency of the model was 81.4% in the identification of nonresponder patients.
TABLE 7.
Number of Patients Correctly and Incorrectly Classified by the Logistic Regression Model
| Observed (n = 97) | Predicted
|
Percentage of patients classified correctly | |
|---|---|---|---|
| Responders (n = 30) | Nonresponders (n = 67) | ||
| Responders (n = 38) | 25 | 13 | 65.8 |
| Nonresponders (n = 59) | 5 | 54 | 91.5 |
| Overall percentage of correctly classified patients | 81.4 | ||
Patients with a predicted probability value ≥0.46 were considered as nonresponders.
Discussion
Between 40% and 60% of the patients treated with PEG-IFN plus ribavirin achieve a sustained response with normalization of ALT levels and disappearance of HCV-RNA from serum.2,3,4,5 The viremia threshold for a favorable response at 24 weeks of therapy has been established at 800,000 IU/ml.12 However, when therapy is prolonged to 48 weeks, this viremia level does not predict the response, because prolongation of therapy increases the response rates in patients with high viral load.21 Thus, although the threshold of 800,000 IU/ml may be useful to tailor the therapy duration, it is not a predictive factor of sustained response when patients have to be treated for 48 weeks.
In this report, we have evaluated the basal characteristics of 97 patients with chronic hepatitis C infected with the HCV genotype 1 treated for 48 weeks with standard doses of pegylated IFN plus ribavirin, 38 (39%) of whom were sustained responders. The univariate analysis of these characteristics showed that the variables associated with a sustained response were younger age, low GGTP levels, absence of a previous antiviral treatment, and a low percentage of infected hepatocytes in the basal liver biopsy. When the subgroup of previously untreated patients were analyzed separately, the only statistical differences found between responder and nonresponder patients were the percentage of infected hepatocytes and the GGTP levels. In previously treated patients, the percentage of infected hepatocytes was also lower in responder than in nonresponder patients, although the difference did not reach statistical significance probably due to the small number of patients included in this study.
In the present study, there were no differences in the HCV-RNA concentration in the baseline serum sample between sustained responders and nonresponder patients, indicating that serum viral load is not a predictive factor of response to 48 weeks of therapy with PEG-IFN plus ribavirin. It may be argued that the inclusion of patients who were previously treated could influence the results obtained regarding serum HCV-RNA concentration. However, this is not the case, because in previously untreated patients, there were no differences in the basal serum HCV-RNA concentration between responder and nonresponder patients. In addition, only 21 of the 54 (38.8%) patients with a HCV-RNA concentration lower than 800,000 IU/ml were sustained responders. In contrast, using 7% of infected hepatocytes (the mean percentage of infected cells in sustained responders) as the threshold for a sustained response, it was found that 21 of 40 patients (52.5%) with 7% or fewer infected hepatocytes in the pretreatment liver biopsy were sustained responders. Furthermore, 43 patients had a level of viremia more than 800,000 IU/ml, and 17 (39.5%) of them were sustained responders. On the contrary, 57 cases had more than 7% of infected hepatocytes, and 17 (29.8%) of them were sustained responders. Considering all of these data as a whole, it is suggested that serum HCV-RNA concentration in the pretreatment serum sample may not be accurate enough to predict which patients will or will not respond to the pegylated interferon plus ribavirin therapy. The reason why the percentage of infected hepatocytes is predictive of response while the viremia levels are not predictive even though there is a relationship between the percentage of HCV-infected hepatocytes and the serum HCV-RNA concentration is not clear. It may be speculated that this discrepancy is due to underestimation of viremia in serum samples with a high HCV-RNA concentration due to a plateau effect. However, this explanation is unlikely because serum samples with viremia levels above the dynamic range of quantitation of the test used in this study (500,000 IU/ml) were retested diluted 1/10 and 1/100 to obtain an accurate HCV-RNA quantitation. Another explanation is that the relationship between the percentage of infected hepatocytes and serum HCV-RNA concentration may not be linear because viremia may depend not only on the release of viral particles from infected cells (and thus on the percentage of HCV-containing hepatocytes) but also on the rate of virion clearance from circulation and on the contribution of HCV replication in extrahepatic sites. However, this hypothesis should be demonstrated in future research. On the other hand, our findings agree with those reported by Gervais et al22 who had also found that the intrahepatic HCV-RNA content in the basal liver biopsy, measured by a competitive RT-PCR assay, was statistically lower in responder than in nonresponder patients to the IFN therapy, while no differences were found in the basal serum HCV-RNA concentration, although the threshold of intrahepatic HCV-RNA concentration for a favorable response was not established. However, it should be stated that quantitation of HCV-RNA in liver samples by RT-PCR may not be accurate not only because of the above-mentioned problems inherent to the technique, including the efficiency of HCV-RNA extraction but also because of the presence of blood contaminating the liver biopsy. Furthermore, if the liver sample is not frozen immediately after it is obtained and if it is not stored properly in liquid nitrogen, the intrahepatic HCV-RNA content may be underestimated because of the degradation of the viral RNA, as has been reported.23 Degradation of viral RNA may be also a problem in the in situ hybridization technique. However, this problem is avoided if the liver sample is placed in paraformaldehyde-phosphate-buffered saline in less than 3 minutes,24 because this fixative not only preserves the tissue quality but also retains RNA within the tissue and allows good recognition of the target RNA by the probes.25,26 Furthermore, this fixation inactivates RNases, and thus tissue slides may be stored at 4°C until use, without RNA degradation. Under these conditions, the results obtained in the reproducibility assays show that our in situ hybridization technique is highly reproducible without the potential problems of the RT-PCR.
On the other hand, it may be argued that assessing the percentage of infected hepatocytes in a liver biopsy may be inaccurate due to a nonuniform distribution of infected cells in the liver. However, this fact does not seem to be the case because it has been demonstrated that HCV-RNA levels are similar in both the left and the right hepatic lobes using quantitative RT-PCR.27,28,29 Furthermore, in situ hybridization studies performed by different groups have shown that HCV-infected hepatocytes are randomly distributed along the liver biopsies.30,31,32 Thus, as a whole, all of these data suggest that determination of the percentage of infected hepatocytes at one site is representative of this percentage at other sites.
In the multivariate analysis, the percentage of infected hepatocytes (OR, 1.160; 95% CI, 1.065–1.264) and the previous antiviral therapy (OR, 0.294; 95% CI, 0.1109–0.795) were the only significant variables associated with the response. Therefore, the percentage of infected hepatocytes and the previous antiviral treatment can be considered as prognostic factors for a sustained response adjusting for age and GGTP.
The probability of being a nonresponder for a given patient can be calculated using the equation derived from the multivariable analysis (see Results). The ROC curve constructed using the probability values estimated with the above-mentioned equation demonstrated that the regression model was able to discriminate between responders and nonresponders with accuracy (AUC = 0.819; 95% CI, 0.733–0.906; P value of the contrast null hypothesis = 0.0000001). The threshold P value that provides the highest specificity and sensitivity was 0.46. This probability has a positive predictive value of 80.6% and a negative predictive value of 83.3%, being the overall accuracy of the model 81.4%. That means that 81.4% of the patients will be accurately identified as sustained responders or nonresponders when the proposed equation is used.
Finally, it may be argued that a disadvantage of measuring the percentage of infected hepatocytes is that patients must undergo an invasive procedure such as a liver biopsy, whereas measurement of serum HCV-RNA concentration can be done noninvasively and repeatedly to monitor the response to the antiviral therapy. However, in this regard, two aspects must be considered. First, determination of the percentage of HCV-containing hepatocytes is useful in identifying patients who have the best chance to respond to the antiviral treatment but not to control this response during therapy. Second, although liver biopsy is an invasive procedure, expert consensus has recommended the performance of liver biopsy before initiation of antiviral therapy31,32,33,34,35, and in clinical practice, most patients undergo a liver biopsy before treatment. In conclusion, in the present study, we have demonstrated that in patients infected with HCV genotype 1, the determination of the percentage of infected hepatocytes in the pretreatment liver biopsy is a predictive factor of sustained response to 48 weeks of therapy with pegylated interferon plus ribavirin.
Footnotes
Supported by a grant from the Fundación de Investigaciones Biomédicas.
References
- World Health Organization Hepatitis C. Wkly Epidemiol Rec. 1997;72:65–69. [Google Scholar]
- Manns MP, McHutchison JG, Gordon SC, Rustgi VK, Shiffman M, Reindollar R, Goodman ZD, Koury K, Ling M, Albrecht JK. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001;358:958–965. doi: 10.1016/s0140-6736(01)06102-5. [DOI] [PubMed] [Google Scholar]
- Bruno S, Camma C, Di Marco V, Rumi M, Vinci M, Camozzi M, Rebucci C, Di Bona D, Colombo M, Craxi A, Mondelli MU, Pinzello G. Peginterferon alfa-2b plus ribavirin for naive patients with genotype 1 chronic hepatitis C: a randomized controlled trial. J Hepatol. 2004;41:474–481. doi: 10.1016/j.jhep.2004.05.012. [DOI] [PubMed] [Google Scholar]
- Shiffman ML, Di Bisceglie AM, Lindsay KL, Morishima C, Wright EC, Everson GT, Lok AS, Morgan TR, Bonkovsky HL, Lee WM, Dienstag JL, Ghany MG, Goodman ZD, Everhart JE. Hepatitis C Antiviral Long-Term Treatment Against Cirrhosis Trial Group: Peginterferon alfa-2a and ribavirin in patients with chronic hepatitis C who have failed prior treatment. Gastroenterology. 2004;126:1015–1923. doi: 10.1053/j.gastro.2004.01.014. [DOI] [PubMed] [Google Scholar]
- Hadziyannis SJ, Sette H, Jr, Morgan TR, Balan V, Diago M, Marcellin P, Ramadori G, Bodenheimer H, Jr, Bernstein D, Rizzetto M, Zeuzem S, Pockros PJ, Lin A, Ackrill AM, PEGASYS International Study Group Peginterferon-alpha2a and ribavirin combination therapy in chronic hepatitis C: a randomized study of treatment duration and ribavirin dose. Ann Intern Med. 2004;140:346–355. doi: 10.7326/0003-4819-140-5-200403020-00010. [DOI] [PubMed] [Google Scholar]
- Yuki N, Hayashi N, Kasahara A, Hagiwara H, Takehara T, Oshita M, Katayama K, Fusamoto H, Kamada T. Pretreatment viral load and response to prolonged interferon-alpha course for chronic hepatitis C. J Hepatol. 1995;22:457–463. doi: 10.1016/0168-8278(95)80109-x. [DOI] [PubMed] [Google Scholar]
- Garson JA, Brillanti S, Whitby K, Foli M, Deaville R, Masci C, Miglioli M, Barbara L. Analysis of clinical and virological factors associated with response to alpha interferon therapy in chronic hepatitis C. J Med Virol. 1995;45:348–353. doi: 10.1002/jmv.1890450320. [DOI] [PubMed] [Google Scholar]
- Lindsay KL, Davis GL, Schiff ER, Bodenheimer HC, Balart LA, Dienstag JL, Perrillo RP, Tamburro CH, Goff JS, Everson GT, Silva M, Katkov WN, Goodman Z, Lau JY, Maertens G, Gogate J, Sanghvi B, Albrecht J. Response to higher doses of interferon alfa-2b in patients with chronic hepatitis C: a randomized multicenter trial. Hepatitis Interventional Therapy Group. Hepatology. 1996;24:1034–1040. doi: 10.1002/hep.510240509. [DOI] [PubMed] [Google Scholar]
- Pawlotsky JM, Bouvier-Alias M, Hezode C, Darthuy F, Remire J, Dhumeaux D. Standardization of hepatitis C virus RNA quantification. Hepatology. 2000;32:654–659. doi: 10.1053/jhep.2000.16603. [DOI] [PubMed] [Google Scholar]
- Davis GL, Lau JY, Urdea MS, Neuwald PD, Wilber JC, Lindsay K, Perrillo RP, Albrecht J. Quantitative detection of hepatitis C virus RNA with a solid-phase signal amplification method: definition of optimal conditions for specimen collection and clinical application in interferon-treated patients. Hepatology. 1994;19:1337–1341. [PubMed] [Google Scholar]
- Lau JY, Simmonds P, Urdea MS. Implications of variations of “conserved” regions of hepatitis C virus genome. Lancet. 1995;346:425–426. doi: 10.1016/s0140-6736(95)92786-7. [DOI] [PubMed] [Google Scholar]
- McNicol AM, Farquharson MA. In situ hybridization and its diagnostic applications in pathology. J Pathol. 1997;182:250–261. doi: 10.1002/(SICI)1096-9896(199707)182:3<250::AID-PATH837>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Höfler H, Childers H, Montminy MR, Lecham RM, Goodmann RH, Wolfe HJ. In situ hybridization methods for the detection of somatostain mRNA in tissue sections using antisense RNA probes. Histochem J. 1986;18:597–604. doi: 10.1007/BF01675295. [DOI] [PubMed] [Google Scholar]
- Chang M, Williams O, Mittler J, Quintanilla A, Carithers RL, Jr, Perkins J, Corey L, Gretch DR. Dynamics of hepatitis C virus replication in human liver. Am J Pathol. 2003;163:433–444. doi: 10.1016/S0002-9440(10)63673-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Inigo E, Bartolome J, de Lucas S, Manzarbeitia F, Pardo M, Arocena C, Gosalvez J, Oliva H, Carreno V. Histological damage in chronic hepatitis C is not related to the extent of infection in the liver. Am J Pathol. 1999;154:1877–1881. doi: 10.1016/S0002-9440(10)65445-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lucas S, Bartolome J, Rodriguez-Inigo E, Casqueiro M, Millan A, Ruiz-Moreno M, Oliva H, Carreno V. Distribution of hepatitis C virus infection in liver biopsies from children and adults with chronic hepatitis C. J Med Virol. 2001;64:1–5. doi: 10.1002/jmv.1009. [DOI] [PubMed] [Google Scholar]
- Gosalvez J, Rodriguez-Inigo E, Ramiro-Diaz JL, Bartolome J, Tomas JF, Oliva H, Carreno V. Relative quantification and mapping of hepatitis C virus by in situ hybridization and digital image analysis. Hepatology. 1998;27:1428–1434. doi: 10.1002/hep.510270534. [DOI] [PubMed] [Google Scholar]
- Castillo I, Pardo M, Bartolome J, Ortiz-Movilla N, Rodriguez-Iñigo E, de Lucas S, Salas C, Jimenez-Heffernan JA, Perez-Mota A, Graus J, Lopez-Alcorocho JM, Carreño V. Occult hepatitis C virus infection in patients in whom the etiology of persistently abnormal results of liver-function tests is unknown. J Infect Dis. 2004;189:7–14. doi: 10.1086/380202. [DOI] [PubMed] [Google Scholar]
- Bedossa P. Intraobserver and interobserver variations in liver biopsy interpretation in patients with chronic hepatitis C: METAVIR Cooperative Study Group. Hepatology. 1994;20:15–20. [PubMed] [Google Scholar]
- Bedossa P, Poynard T. An algorithm for the grading of activity in chronic hepatitis C: The METAVIR Cooperative Study Group. Hepatology. 1996;24:289–293. doi: 10.1002/hep.510240201. [DOI] [PubMed] [Google Scholar]
- McHutchison JG, Gordon SC, Schiff ER, Shiffman ML, Lee WM, Rustgi VK, Goodman ZD, Ling MH, Cort S, Albrecht JK. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C: Hepatitis Interventional Therapy Group. N Engl J Med. 1998;339:1485–1492. doi: 10.1056/NEJM199811193392101. [DOI] [PubMed] [Google Scholar]
- Gervais A, Martinot M, Boyer N, Auperin A, Le Breton W, Degott C, Valla D, Marcellin P. Quantitation of hepatic hepatitis C virus RNA in patients with chronic hepatitis C: relationship with severity of disease, viral genotype and response to treatment. J Hepatol. 2001;35:399–405. doi: 10.1016/s0168-8278(01)00138-6. [DOI] [PubMed] [Google Scholar]
- Madejon A, Manzano ML, Arocena C, Castillo I, Carreno V. Effects of delayed freezing of liver biopsies on the detection of hepatitis C virus RNA strands. J Hepatol. 2000;32:1019–1025. doi: 10.1016/s0168-8278(00)80107-5. [DOI] [PubMed] [Google Scholar]
- Angerer LM, Angerer RC. Detection of poly A+ RNA in sea urchin eggs and embryos by quantitative in situ hybridization. Nucleic Acids Res. 1981;9:2819–2840. doi: 10.1093/nar/9.12.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer RH, Ward DC. Actin gene expression visualized in chicken muscle tissue culture by using in situ hybridization with a biotinated nucleotide analog. Proc Natl Acad Sci USA. 1982;79:7331–7335. doi: 10.1073/pnas.79.23.7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence JB, Singer RH. Quantitative analysis of in situ hybridization methods for the detection of actin gene expression. Nucleic Acids Res. 1985;13:1777–1799. doi: 10.1093/nar/13.5.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idrovo V, Dailey PJ, Jeffers LJ, Coelho-Little E, Bernstein D, Bartholomew M, Alvarez L, Urdea MS, Collins ML, Schiff ER. Hepatitis C virus RNA quantification in right and left lobes of the liver in patients with chronic hepatitis C. J Viral Hepat. 1996;3:239–246. doi: 10.1111/j.1365-2893.1996.tb00049.x. [DOI] [PubMed] [Google Scholar]
- Terrault NA, Dailey PJ, Ferrell L, Collins ML, Wilber JC, Urdea MS, Bhandari BN, Wright TL. Hepatitis C virus: quantitation and distribution in liver. J Med Virol. 1997;51:217–224. [PubMed] [Google Scholar]
- Fanning L, Loane J, Kenny-Walsh E, Sheehan M, Whelton M, Kirwan W, Collins JK, Shanahan F. Tissue viral load variability in chronic hepatitis C. Am J Gastroenterol. 2001;96:3384–3389. doi: 10.1111/j.1572-0241.2001.05271.x. [DOI] [PubMed] [Google Scholar]
- Agnello V, Abel G, Knight GB, Muchmore E. Detection of widespread hepatocyte infection in chronic hepatitis C. Hepatology. 1998;28:573–584. doi: 10.1002/hep.510280240. [DOI] [PubMed] [Google Scholar]
- Negro F, Pacchioni D, Shimizu Y, Miller RH, Bussolati G, Purcell RH, Bonino F. Detection of intrahepatic replication of hepatitis C virus RNA by in situ hybridization and comparison with histopathology. Proc Natl Acad Sci USA. 1992;89:2247–2251. doi: 10.1073/pnas.89.6.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamas E, Baccarini P, Housset C, Kremsdorf D, Brechot C. Detection of hepatitis C virus (HCV) RNA sequences in liver tissue by in situ hybridization. J Hepatol. 1992;16:219–223. doi: 10.1016/s0168-8278(05)80119-9. [DOI] [PubMed] [Google Scholar]
- National Institute of Health Consensus development conference panel statement: management of hepatitis C. Hepatology. 1997;26(Suppl 1):2S–10S. doi: 10.1002/hep.510260701. [DOI] [PubMed] [Google Scholar]
- The British Society of Gastroenterology Guidelines on the use of liver biopsy in clinical practice. Gut. 1999;45(Suppl 4):1–11. doi: 10.1136/gut.45.2008.iv1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Association for the Study of the Liver Consensus statement EASL: International Conference on Hepatitis C. J Hepatol. 1999;31(Suppl 1):3–8. [PubMed] [Google Scholar]