Abstract
Modification by SUMO-1 is proposed to play a role in protein targeting and/or stability. The SUMO-1-conjugating enzyme Ubc9 interacts with androgen receptor (AR), a ligand-activated transcription factor belonging to the steroid receptor superfamily. We show here that AR is covalently modified by SUMO-1 (sumoylated) in an androgen-enhanced fashion and identify the principal acceptor site in the N-terminal domain of AR. Substitutions of sumoylated Lys residues enhanced transcriptional activity of AR without influencing its transrepressing activity. Interestingly, the same Lys residues form the cores of the recently described transcriptional synergy control motifs in AR [Iñiguez-Lluhi, J. A. & Pearce, D. (2000) Mol. Cell. Biol. 20, 6040–6050]. These motifs, which match perfectly with the sumoylation consensus sequence, are also present in the N-terminal domains of glucocorticoid, mineralocorticoid, and progesterone receptor. Taken together, our data suggest that reversible sumoylation is a mechanism for regulation of steroid receptor function.
Steroid receptors such as androgen receptor (AR) are ligand-regulated transcription factors belonging to the nuclear receptor superfamily. They convey the effects of steroid hormones on the regulation of cell growth, differentiation, and homeostasis (1). To regulate transcription, the receptors bind to specific hormone response elements of target genes and/or exhibit crosstalk with other transcription factors through protein–protein interactions. A plethora of coregulatory proteins recognized by different functional domains of the receptors [the N-terminal transactivation region, the central DNA-binding domain (DBD), and the C-terminal ligand-binding domain] mediate transactivation and transrepression functions of nuclear receptors (1–3). The mechanisms by which steroid receptors compartmentalize in the cell nuclei and find their specific binding motifs, hormone response elements, from a vast number of base pairs of chromosomal DNA have remained elusive. Even though the transcriptional activity of the receptors is mainly controlled by ligand binding, covalent modifications such as phosphorylation also play an important role in regulation (4, 5). Posttranslational modifications often elicit fast alterations in protein–protein interactions in multiprotein complexes and subcellular structures. Some nuclear receptors are also known to be ubiquitinylated, which targets them to degradation (5–7). Ubc9, the conjugating enzyme for SUMO-1, has recently been shown to interact with at least two steroid receptors, AR and glucocorticoid receptor (GR) (8, 9). However, coexpressed Ubc9 enhanced AR-dependent transcription in a fashion that appeared to be independent of its ability to catalyze SUMO-1 conjugation (9).
The SUMO-1 modification (sumoylation) pathway resembles that of ubiquitin conjugation, but the enzymes involved in the two processes are distinct (10). SUMO-1 (also known as sentrin, GMP1, PIC1, and Ubl1, or in yeast as Smt3) is activated for conjugation by E1 enzymes and subsequently transferred to the E2-conjugating enzyme Ubc9 (11–15). Sumoylation is reversible (16–18). The genes encoding all key proteins of the modification process are essential in yeast, and the conjugation machinery is well conserved (13, 16, 19). The sumoylation appears to play multiple roles, including (i) protein targeting, (ii) protein stabilization, and (iii) transcriptional activation. Conjugation of SUMO-1 to RanGAP1 targets the otherwise cytosolic protein to the nuclear pore complex (20–22), and modification of promyelocytic leukemia protein by SUMO-1 has been reported to direct it to subnuclear domains termed promyelocytic leukemia protein nuclear bodies (23, 24). This latter modification may also be needed for promyelocytic leukemia protein-mediated recruitment of other proteins to these structures (25). Sumoylation can prevent degradation of the target protein, as exemplified by IκBα (26). The potential importance of sumoylation in transcriptional regulation is highlighted by recent reports that modification of p53 by SUMO-1 can enhance its transcriptional activity (27, 28). The present study shows that a nuclear receptor, AR, is covalently modified by SUMO-1. We have identified the sumoylation sites in the N-terminal domain of AR and show that the SUMO-1 modification in certain contexts indeed inhibits the activity of AR. Intriguingly, the sumoylation sites are identical to the negative motifs in AR and GR, which have recently been shown to restrict the transcriptional synergy of these receptors on promoters containing compound response elements (29).
Materials and Methods
Materials.
The rabbit polyclonal anti-AR antibody K183 has been described (30). Mouse monoclonal anti-Flag antibody M2, rabbit polyclonal anti-green fluorescent protein (anti-GFP), and mouse monoclonal anti-GMP-1 were from Sigma, Santa Cruz Biotechnology, and Zymed, respectively. Horseradish peroxidase-conjugated anti-rabbit IgG and anti-mouse IgG were from Zymed. Lissamine-rhodamine-conjugated goat anti-mouse IgG was from Jackson ImmunoResearch.
Plasmid Constructions.
pcDNA-Flag vector was constructed by inserting a double-stranded oligonucleotide encoding the Flag epitope into the KpnI/BamHI site of the mammalian expression vector pcDNA3.1(+) (Invitrogen). To create pcDNA-Flag-hAR, the region encoding the N-terminal region of hAR was cloned in-frame with BglII/EcoRI into the BamHI/EcoRI site of pcDNA-Flag. Subsequently, the EcoRI/XbaI fragment encoding the C-terminal part of hAR was inserted to generate full-length receptor expression vector. Point mutations K386R and K520R were generated in hAR by overlapping PCR, and the mutated fragments were introduced into pcDNA-Flag-hAR by using Eco91I/Eco47III and KpnI/HindIII sites in the hAR cDNA, respectively, to create pcDNA-Flag-K386R, pcDNA-Flag-K520R, and pcDNA-Flag-K386R/K520R expression constructs. pcDNA-Flag-hARΔDBD, lacking the amino acids 576– 657, was created by ligating the BglII/BamHI fragment from pEGFP-hARΔDBD to the BamHI site of pcDNA-Flag. Glutathione S-transferase (GST)-Ubc9 was constructed by inserting a PCR-amplified fragment encoding Ubc9 amino acids 2–158 into the BamHI/EcoRI site of pGEX-5X-1 (Amersham Pharmacia). pFlag-SUMO-1 was generated by inserting a PCR-amplified cDNA fragment encoding SUMO-1 amino acids 2–101 to the HindIII/SalI site of pFlag-CMV-2. For the pEGFP-SUMO-1GA mutant, the PCR primer carried a point mutation that converts G97A. For expression of GST-SUMO-1 and enhanced GFP (EGFP)-SUMO-1, the SUMO-1 cDNA fragment was inserted into the BamHI/SalI site of pGEX-5X-1 or the BglII/SalI site of pEGFP-C2 (CLONTECH). All PCR-amplified fragments were verified by sequencing. pARE2-TATA-LUC and pFLAG-Ubc9 have been described (9, 31). pCMV-RelA and pκB6tk-LUC were from Patrick Baeuerle (Alberts-Ludwig Universität, Freiburg, Germany) (32). pGEM3Z-hGR was provided by Eckardt Treuter (Karolinska Institute, Huddinge, Sweden). pSG5-TRα1, pCMV5-hERα, and pCMV5-ERβ vectors were gifts from Paul M. Yen (National Institutes of Health, Bethesda, MD), Benita Katzenellenbogen (University of Illinois, Urbana), and Jan-Åke Gustafsson (Karolinska Institute, Huddinge, Sweden), respectively. For pSG5-hERα and pSG5-hERβ, the corresponding cDNAs from the pCMV5 constructs were inserted into pSG5.
Cell Culture and Transfections.
COS-1 and HeLa cells were obtained from the American Type Culture Collection and cultured in DMEM supplied with 10% FBS/25 units/ml penicillin and streptomycin. For HeLa cells, nonessential amino acids were also added. For immunoprecipitation experiments, 3.5 × 105 COS-1 cells were seeded on 6-cm dishes 24 h before transfection. Two hours before transfection, the cells received fresh medium containing 10% charcoal-stripped FBS. By using the FuGene 6 reagent (Roche Molecular Biochemicals), 0.5 μg of Flag-AR expression plasmids and 1.5 μg of GFP-SUMO-1 or EGFP-C2 were transfected. Eight hours after transfection, the cells were supplied with 100 nM testosterone. For transactivation and transrepression assays, 5 × 104 COS-1 cells were seeded on 12-well plates 24 h before transfection, and 4 h before transfection, the cells received fresh medium containing 10% charcoal-stripped FBS. Reporter plasmid (150 ng), 50 ng of pCMVβ (CLONTECH), and indicated amounts of AR expression constructs were transfected. Eighteen hours after transfection, the cells received fresh medium containing 2% charcoal-stripped FBS and 100 nM testosterone or vehicle. After a 30-h culture, the cells were harvested, and luciferase (LUC) and β-galactosidase activities were assayed as described (33, 34). pCMV-ARE2-LUC that contains the CMV-ARE2-TATA sequence (30) in KpnI/HindIII site of pGL3-Basic (Promega) was used for the promoter interference assay.
SUMO-1 Conjugation in Vitro.
GST-Ubc9 and GST-SUMO-1 were purified as described (35), eluted in TBS [137 mM NaCl/20 mM Tris⋅HCl (pH 7.6)]/10 mM reduced glutathione, and dialyzed overnight against TBS. SUMO-1-activating enzyme fraction was purified from HeLa cells essentially as described (15), except that Q Sepharose Fast Flow (Amersham Pharmacia) was used instead of Mono Q. For the conjugation assay, receptor constructs were translated in vitro by using the TNT rabbit reticulocyte system (Promega) in the presence of [35S]Met. One microliter of the in vitro translation product was incubated with 5 μl of GST-SUMO-1, 4 μl of GST-Ubc9, and 2 μl of HeLa cell fraction at 30°C for 1 h in the presence of 1 mM DTT/4 mM MgCl2/2 mM ATP. The reaction was terminated by adding 15 μl of 2 × SDS sample buffer. The samples were heated at 95°C for 5 min, resolved by electrophoresis on 7.5% polyacrylamide gels under denaturing conditions (SDS/PAGE), and visualized by fluorography.
Immunoprecipitation and Immunoblotting.
COS-1 cells were collected in PBS containing 20 mM N-ethylmaleimide, and cell extracts were prepared in modified RIPA buffer [50 mM Tris⋅HCl (pH 7.8)/150 mM NaCl/5 mM EDTA/15 mM MgCl2/1% NP-40/0.75% sodium deoxycholate/1 mM DTT], 1:100 diluted protease inhibitor mixture (Sigma), and 20 mM N-ethylmaleimide. Immunoprecipitation with mouse monoclonal anti-Flag antibody was performed as described (31). Bound proteins were released in 2 × SDS sample buffer, resolved on 7.5% SDS/PAGE, transferred onto Hybond enhanced chemiluminescence nitrocellulose membrane, and visualized by using the enhanced chemiluminescence detection reagents (Amersham Pharmacia) according to the manufacturer's instructions.
Results
AR Is Modified by SUMO-1.
Even though our previous results suggested that Ubc9 is capable of activating AR in a fashion that is independent of its sumoylation activity (9), it was still pertinent to examine the possibility that AR is covalently modified by SUMO-1. Flag-tagged human AR and SUMO-1 fused to GFP were transiently expressed in COS-1 cells, and cell extracts were prepared in the presence of N-ethylmaleimide, which is known to inhibit SUMO-1 deconjugation in vitro (16, 17). Immunoblotting of the cell extracts with a polyclonal anti-AR antibody showed three immunoreactive bands, a major ≈115-kDa band corresponding to the unmodified AR and two less intense more slowly migrating bands (≥220 kDa) (Fig. 1A). To find out whether the slowly migrating bands represent AR modified by GFP-tagged SUMO-1, immunoprecipitations were performed with Flag antibody. Both anti-GFP and anti-SUMO-1 antibody detected the two slowly migrating bands of higher molecular mass in the immunoprecipitate, whereas the 115-kDa unmodified AR band was not detected (Fig. 1 B and C). When the GFP-tagged SUMO-1G97A mutant, which cannot be conjugated to target proteins, was used instead of wild-type (wt) SUMO-1 (36), no sumoylated AR forms were found (Fig. 1, lanes 4 and 5). These results indicate that the slowly migrating bands represent SUMO-1-modified forms of AR and suggest that there are at least two sumoylation sites in the receptor.
Figure 1.
AR is modified by SUMO-1. COS-1 cells grown on 6-cm dishes were transfected with vectors encoding Flag-tagged wt AR (wt) or AR lacking most of the DBD and the hinge region (ΔDBD), and GFP-tagged SUMO-1 (SUMO) or SUMO-1GA (SUMOGA) expression vector. The cells received 100 nM testosterone (T) or vehicle 12 h before harvesting as indicated. Five percent of the cell extracts were immunoblotted with AR antibody (A), and the rest were subjected to immunoprecipitation with anti-Flag antibody. The immunoprecipitates were immunoblotted with anti-GFP antibody (B) or anti-SUMO-1 antibody (C). Arrowheads depict the slowly migrating sumoylated forms of AR.
Ligand treatment of COS-1 cells enhanced the SUMO-1 modification of AR (Fig. 1, lanes 2 vs. 3). This is not explained by differences in the amount of AR protein in the cells (Fig. 1A). Rather, the role of the ligand is likely to be coupled to the transfer of holo-AR to the nucleus, where Ubc9 is primarily located (U.K., unpublished work). Only a relatively small proportion of apo-AR resides in COS-1 cell nuclei, and ligand treatment induces a complete transfer of holo-AR to nuclei (37–39). The AR mutant ΔDBD, lacking most of the DBD and the flanking hinge region (amino acid residues 576–657), which contains the bipartite nuclear localization signal (37, 38), is transferred poorly to the nucleus on ligand exposure and is totally unable to recognize DNA (39). The ΔDBD mutant was sumoylated less efficiently than wt AR, although the pattern of modification remained unchanged (Fig. 1A, lanes 6 and 7). Because the missing AR region is known to contribute to the AR–Ubc9 interaction (9), the less efficient attachment of SUMO-1 to the ΔDBD mutant could also derive from its inefficient interaction with Ubc9.
K386 Is the Major AR Site Modified by SUMO-1.
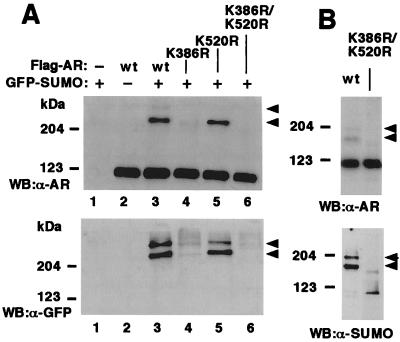
Sumoylation of proteins has been shown to occur at specific Lys residues, which are in most cases embedded in a consensus sequence (I/L/V)KXE, where X represents any amino acid (40, 41). Expression of a series of AR deletion mutants in COS-1 cells indicated that the two major slowly migrating forms of AR depended on the presence of the region encompassing amino acids 337–553 in the N-terminal region (data not shown). This region contains three Lys residues, K347, K386, and K520. Of these three residues, the neighboring amino acid residues of K386 (sequence IKLE) and K520 (sequence VKSE) match with the proposed SUMO-1 attachment consensus sequence. To test whether these residues were indeed sumoylated, point mutations K386R and K520R were introduced into the Flag-tagged human AR. Immunoprecipitations of AR from extracts of COS-1 cells expressing these constructs revealed that the K386R mutant of AR is only weakly sumoylated (Fig. 2A, lane 4). Even though the K520R mutation was not as deleterious to the modification as the K386R mutation, the K520R mutant of AR was nevertheless less efficiently sumoylated than wt AR (Fig. 2A, lane 5). When the point mutations were combined, no SUMO-1 was attached to the double mutant (Fig. 2A, lane 6). Likewise, in the absence of ectopically expressed SUMO-1, immunoblotting revealed slower migrating AR forms in cells expressing the wt AR but not in those expressing the compound mutant (Fig. 2B Upper). Immunoblotting of immunoprecipitated ARs with anti-SUMO-1 antibody confirmed that the slower migrating AR forms indeed represented sumoylated AR (Fig. 2B Lower).
Figure 2.
K386 is the major sumoylation site in AR. COS-1 cells were transfected with pGFP-SUMO-1 or empty expression vector as indicated along with wt Flag-hAR, Flag-K386R, Flag-K520R, or Flag-K386R/K520R. The cells received 100 nM testosterone 12 h before harvesting. Immunoblots and immunoprecipitations were performed as described in Fig. 1. (A Upper) Immunoblotting (WB) of cell extracts with anti-AR antibody (α-AR); (A Lower) immunoprecipitation with anti-Flag antibody and subsequent immunoblotting with anti-GFP antibody (α-GFP). (B Upper) COS-1 cells were transfected with expression vectors encoding wt AR or the K386R/K520R mutant, and the cell extracts were analyzed by immunoblotting with anti-AR antibody (α-AR); (B Lower) immunoprecipitation with anti-Flag antibody and subsequent immunoblotting with anti-SUMO-1 antibody (α-SUMO). Arrowheads depict the slowly migrating SUMO-1-conjugated ARs.
Ubc9 Catalyzes Sumoylation of AR in Vitro.
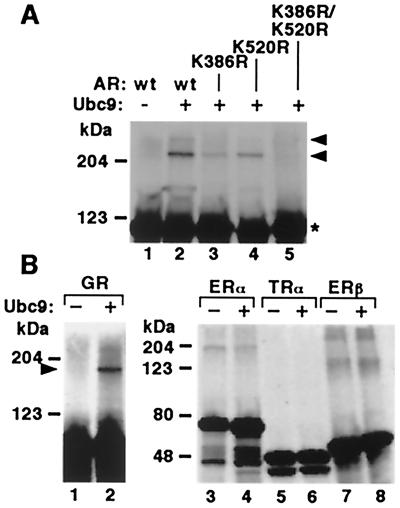
To study the catalytic role of Ubc9 in SUMO-1 modification of AR, 35S-Met-labeled AR was incubated in the absence and presence of GST-Ubc9 with GST-SUMO-1 and a HeLa cell fraction containing SUMO-1-activating enzyme (15). Ubc9 was required to produce the two slowly migrating forms of AR, corresponding to the AR SUMO-1 conjugates (Fig. 3A, lanes 1 and 2). This pattern is in line with the sumoylated AR forms detected with GFP-SUMO-1 by immunoblotting with anti-AR antibody (Fig. 2A Upper), because the size of GST-SUMO-1 is comparable to that of GFP-SUMO-1. If the HeLa cell fraction or GST-SUMO-1 was omitted from the reaction, the slowly migrating AR bands were not detectable (data not shown). With the K386R mutant of AR, only a single very weak additional band was obtained in the presence of Ubc9 (Fig. 3A, lane 3). The incubation of K520R mutant with Ubc9 yielded also only one additional major band, which was more intense than that observed with K386R (Fig. 3A, lane 4). Importantly, no sumoylated forms of AR were detected with the double mutant. Together, these results confirm that AR is modified by SUMO-1 on the amino acids K386 and K520, with the former one being the principal acceptor site.
Figure 3.
Ubc9 catalyzes attachment of SUMO-1 to AR and GR in vitro. (A) In vitro-translated 35S-Met-labeled wt and mutated AR constructs were incubated with GST-SUMO-1 and HeLa cell fraction containing SUMO-1-activating enzyme in the presence (+) or absence (−) of GST-Ubc9. The samples were resolved on 7.5% SDS/PAGE and subjected to fluorography. Arrowheads depict the slowly migrating sumoylated forms of AR; Star, unmodified AR. (B) In vitro SUMO-1 conjugation reactions with GR, ERα, ERβ, and TRα. The reactions were performed in the presence (+) or absence (−) of GST-Ubc9 as described in A.
To examine whether some other nuclear receptors could serve as substrates for Ubc9, we looked for putative sumoylation sites in the sequences of steroid receptors and thyroid hormone receptor α (TRα). N-terminal regions of AR, GR, mineralocorticoid, and progesterone receptors and ligand-binding domains of GR, mineralocorticoid receptor, and TRα contain putative SUMO-1 attachment sites, whereas estrogen receptors α (ERα) and β (ERβ) do not possess sequences matching with the consensus sequence (Table 1). After incubation of in vitro-translated GR, ERα, ERβ, and TRα under the same conditions as those for AR, a slowly migrating form of GR appeared in the presence but not in the absence of Ubc9 (Fig. 3B Left). In contrast, no Ubc9-dependent derivatives of ERα, ERβ, or TRα were detected (Fig. 3B Right), suggesting that the latter receptors are not sumoylated. The high-molecular weight bands seen with ERα and ERβ are not likely to represent SUMO-1 conjugates, because their appearance did not depend on Ubc9 that is the only SUMO-1-conjugating enzyme known thus far.
Table 1.
Potential acceptor sites for SUMO-1 in selected nuclear receptors
| Receptor | Residue | Domain | Sequence | Swiss-Prot no. |
|---|---|---|---|---|
| hAR | K386 | NT | PHARIKLENPLDY | P10275 |
| K520 | NT | SPTCVKSEMGPWD | ||
| hGR | K277 | NT | TLPQVKTEKEDFI | P04150 |
| K293 | NT | TPGVIKQEKLGTV | ||
| K703 | LBD | GKAIVKREGNSSQ | ||
| hMR | K89 | NT | LTSDIKTELESKE | P08235 |
| K399 | NT | IVQYIKPEPDGAF | ||
| K427 | NT | FSVPIKQESTKHS | ||
| K494 | NT | FPVGIKQEPDDGS | ||
| K953 | LBD | ESHALKVEFPAML | ||
| hPR | K388 | NT | PALKIKEEEEGAE | P06401 |
| hERα | P03372 | |||
| hERβ | Q92731 | |||
| hTRα | K283 | LBD | GEMAVKREQLKNG | P21205 |
NT, amino-terminal region; LBD, ligand-binding domain.
Disruption of the SUMO-1 Acceptor Sites Enhances Transcriptional Activity of AR.
Transactivation assays using minimal ARE2TATA-LUC reporter containing two hormone response elements were used to examine whether substitutions in the SUMO-1 attachment sites of AR influence the transcriptional activity of the receptor. The K520R mutant that is modified by SUMO-1 almost as well as wt AR had transcriptional activity in COS-1 cells indistinguishable from that of wt AR (Fig. 4A). In contrast, K386R and K386R/K520R mutants, which were poor targets for SUMO-1 conjugation, had activities 1.9- and 2.4-fold higher than that of wt AR, respectively. More importantly, the compound mutant was constantly 2.4- to 3.3-fold more active than the wt receptor over a wide range of expression plasmid amounts (1–100 ng/well) and testosterone concentrations (0.1–100 nM) (Fig. 4B and data not shown). The higher transcriptional activity of K386R/K520R mutant cannot be explained by receptor protein levels because, if anything, the expression level of the double mutant was lower than that of wt AR as assessed by immunoblotting (Fig. 4B Inset). In addition to COS-1 cells, K386R and K386R/K520R mutants were 2- and 2.4-fold more active than wt AR in HeLa cells (data not shown).
Figure 4.
Substitutions of the sumoylation sites activate AR. (A) Transactivation by 10 ng of wt Flag-AR and the mutants K386R, K520R, and K386R/K520R was studied on minimal pARE2-TATA-LUC reporter (150 ng) in COS-1 cells. The cells received 100 nM testosterone (+T) or vehicle (−T) 18 h after transfection. LUC activities in cell extracts were adjusted to the transfection efficiency by using β-galactosidase activity. The activity of AR in the presence of testosterone is set as 100, and the means ± SD from at least six independent experiments are shown. (B) As in A, except that cells were transfected with 1–100 ng of AR expression plasmids as indicated. The amount of transfected DNA was balanced by the addition of empty expression vector. (Inset) Expression levels of wt AR and K386R/K520R mutant in an experiment corresponding to data shown in B. AR was immunoblotted with AR antibody from the same lysates (pooled from triplicate wells) that were used for reporter gene assays. (C) Effect of coexpressed Ubc9 on transactivation by AR and K386R/K520R. The conditions were as in A, except that indicated amounts of pFLAG-Ubc9 (in μg) were cotransfected and the total amount of DNA was balanced by adding empty pFLAG-CMV2. Black bars depict wt AR and gray ones K386R/K520R mutant.
We have previously shown that overexpression of Ubc9 can result in enhancement of AR-mediated transcription. As shown in Fig. 4C, ligand-dependent activity of wt AR was clearly enhanced (3.2-fold) when 100 ng/well of Ubc9 was coexpressed, whereas the compound mutant showed practically no response to Ubc9 (1.2-fold). However, K386R/K520R mutant was stimulated by a higher Ubc9 dose (300 ng/well), even though the induction (1.9-fold) was clearly lower than that with wt receptor (5.1-fold). Thus, effects of Ubc9 overexpression on AR-dependent transcription are complex and not merely a direct consequence of enhanced sumoylation of AR. This is in line with the finding that sumoylation-negative Ubc9 mutant can also stimulate AR-dependent transactivation (9). Overexpression of Ubc9 may, in fact, stall the sumoylation machinery because other components of the conjugation system are likely to become limiting. This should lead to enhancement rather than inhibition of AR-dependent transcription.
In contrast to transcriptional activation, the transrepressing function of AR was not influenced by substitution of SUMO-1 acceptor sites, because both wt AR and the K386R/K520R mutant repressed a NF-κB/RelA-activated promoter to the same extent (Fig. 5A). Furthermore, the effects of the point mutations on ligand binding and DNA binding by AR were investigated in intact cells. Consistent with the lower expression level of the compound mutant, cells transfected with the mutant construct showed ≈30% less androgen-binding sites than cells transfected wt AR, as assessed by whole cell-binding assays (data not shown). DNA-binding capacity of compound mutant, as examined by promoter interference assays using the same double hormone response elements as in ARE2TATA-LUC, was somewhat lower (10–20%) than that of wt AR (30) (Fig. 5B), which is in line with androgen-binding data.
Figure 5.
Effect of K386R/K520R mutation on transrepressing activity of AR and DNA binding in intact cells. (A) Repression of RelA-dependent transactivation by wt AR and K386R/K520R mutant. COS-1 cells were transfected with pκB6tk-LUC (150 ng), pCMV-RelA (30 ng), and AR expression vectors (150 ng). The relative LUC activity in the absence of cotransfected AR is set as 100. Values are means ± SD from six independent experiments. (B) Binding of the wt and mutant AR to androgen response elements in intact cells as determined by a promoter interference assay (30). COS-1 cells were transfected with pCMV-ARE2-LUC reporter (100 ng) and increasing amounts (10, 50, and 100 ng) of wt Flag-AR and K386R/K520R mutant expression vectors. The total amount of DNA was kept constant by adding empty pcDNA3.1(+) when necessary. Reporter gene activity in the absence of AR expression vector is set as 100, and the means ± SD from three independent experiments are shown. Black and gray bars depict wt AR and K386R/K520R mutant, respectively.
Discussion
In the present study, we have shown that a ligand-activated transcription factor, AR, is covalently modified by SUMO-1 in intact cells and under cell-free conditions. Androgen promoted sumoylation in cells, probably by facilitating the nuclear transfer of cytoplasmic AR. The sumoylation sites of human AR are K386 and K520. The former appears to act as a master switch of sumoylation, because the K386 mutant was modified only poorly, whereas the K520 mutation impaired the extent of SUMO-1 conjugation much less. The overall cellular distribution of the K386R/K520R mutant and its hormone-induced nuclear translocation were very similar to those of the wt AR (data not shown). Both sumoylation sites are fully conserved in mammalian AR sequences, and a site corresponding to human K386 is also present in the Xenopus AR.
In addition to AR, several other, but not all, members of the steroid receptor family contain potential (I/L/V)KXE attachment sites for SUMO-1 (Table 1). In this study, we show that GR is modified by SUMO-1 in vitro under the same Ubc9-dependent conditions as those used for AR. Intriguingly, the sumoylated sites in AR and the two potential modification sites in the N-terminal domain of GR are identical with the protein motifs that have recently been demonstrated to restrict the transcriptional synergy of the two receptors (29). In line with our data on AR, disruption of these sites by replacing the central SUMO-1 acceptor Lys with Arg led to enhancement of AR-dependent transcription on promoters with more than one hormone response elements (29). The synergy control motifs are identical with the sumoylation consensus sequence, and they can be found in negative regulatory regions of many, otherwise unrelated, transcription factors (29), suggesting that sumoylation can act as a general mechanism controlling their activities.
In contrast to AR, sumoylation of p53 has recently been implicated in the activation of p53-dependent transcription (27, 28). However, similar to AR, the basal transcriptional activity of the sumoylation-defective p53 mutant was either equal to or higher than that of wt p53, depending on the promoter studied. In the case of AR, the extent to which SUMO-1 acceptor sites were mutated correlated with increased transcriptional activity, indicating that sumoylation can, in fact, attenuate AR function. This is in agreement with a recent report showing that c-Jun is negatively regulated by sumoylation (42). The result that the K386R/K520R mutant was coactivated by GR-interacting protein 1 (GRIP1) in a fashion similar to wt AR (data not shown) is in keeping with the fact that the sumoylation sites do not overlap with the core transactivation domain of AR (43). The sumoylation sites localize to the N-terminal region of AR that is involved in interactions with the hormone-bound ligand-binding domain, and therefore, attachment of bulky “side chains” to this region is likely to perturb with the ability of AR to make intramolecular contacts (33). NF-κB-transrepressing activity of AR (44) is not influenced by point mutations substituting the sumoylation sites. This suggests that sumoylation modulates interactions specific for the transactivation process. These results are in agreement with those obtained by using the synergy control motif-disrupted GR (29).
Only a relatively small proportion of total AR can be detected as conjugated to SUMO-1 in transfected cells. This is not completely in line with data from transactivation experiments, in which substitutions of the AR residues responsible for sumoylation clearly enhanced the transcriptional activity of AR. These results suggest that the modification is transient and that there is a dynamic equilibrium between SUMO-1-conjugated and unconjugated receptor forms. Thus, sumoylation is likely to represent a mechanism for a rapid and reversible attenuation of AR function in distinct promoter contexts.
In conclusion, the importance of SUMO-1 modifications in transcriptional regulation is emerging. Current results demonstrate the existence of a new modulatory system to restrict steroid receptor activity. Further characterization of sumoylation of AR has implications in various physiological processes and in pathological states, including prostate cancer.
Acknowledgments
We thank Kati Saastamoinen, Leena Pietilä, and Seija Mäki for excellent technical assistance; and Jan-Åke Gustafsson, Benita Katzenellenbogen, Eckardt Treuter, Paul M. Yen, and Patrick Baeuerle for plasmids. This work was supported by grants from the Medical Research Council (Academy of Finland), the Finnish Foundation for Cancer Research, the Sigrid Jusélius Foundation, Biocentrum Helsinki, the Helsinki University Central Hospital, CaP CURE, the Finnish Medical Society Duodecim, the Finnish Cultural Foundation, and the Research and Science Foundation of Farmos.
Abbreviations
- AR
androgen receptor
- GR
glucocorticoid receptor
- LUC
luciferase
- SUMO-1
small ubiquitin-like modifier 1
- GFP
green fluorescent protein
- GST
glutathione S-transferase
- CMV
cytomegalovirus
- EGFP
enhanced GFP
- TRα
thyroid hormone receptor α
- PR
progesterone receptor
- ERα
estrogen receptor α
- ERβ
estrogen receptor β
- DBD
DNA-binding domain
- wt
wild type
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Beato M, Herrlich P, Schütz G. Cell. 1995;83:851–857. doi: 10.1016/0092-8674(95)90201-5. [DOI] [PubMed] [Google Scholar]
- 2.McKenna N J, Lanz R B, O'Malley B W. Endocr Rev. 1999;20:321–344. doi: 10.1210/edrv.20.3.0366. [DOI] [PubMed] [Google Scholar]
- 3.Freedman L P. Cell. 1999;97:5–8. doi: 10.1016/s0092-8674(00)80708-4. [DOI] [PubMed] [Google Scholar]
- 4.Weigel N L, Zhang Y. J Mol Med. 1997;76:469–479. doi: 10.1007/s001090050241. [DOI] [PubMed] [Google Scholar]
- 5.Lange C A, Shen T, Horwitz K B. Proc Natl Acad Sci USA. 2000;97:1032–1037. doi: 10.1073/pnas.97.3.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li X-Y, Boudjelal M, Xiao J-H, Peng Z-H, Asuru A, Kang S, Fisher G J, Voorhees J J. Mol Endocrinol. 1999;13:1686–1699. doi: 10.1210/mend.13.10.0362. [DOI] [PubMed] [Google Scholar]
- 7.Nawaz Z, Lonard D M, Dennis A P, Smith C L, O'Malley B W. Proc Natl Acad Sci USA. 1999;96:1858–1862. doi: 10.1073/pnas.96.5.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Göttlicher M, Heck S, Doucas V, Wade E, Kullmann M, Cato A C B, Evans R M, Herrlich P. Steroids. 1996;61:257–262. doi: 10.1016/0039-128x(96)00032-3. [DOI] [PubMed] [Google Scholar]
- 9.Poukka H, Aarnisalo P, Karvonen U, Palvimo J J, Jänne O A. J Biol Chem. 1999;274:19441–19446. doi: 10.1074/jbc.274.27.19441. [DOI] [PubMed] [Google Scholar]
- 10.Melchior F. Annu Rev Dev Biol. 2000;16:591–626. doi: 10.1146/annurev.cellbio.16.1.591. [DOI] [PubMed] [Google Scholar]
- 11.Desterro J M P, Rodriguez M S, Kemp G D, Hay R T. J Biol Chem. 1999;274:10618–10624. doi: 10.1074/jbc.274.15.10618. [DOI] [PubMed] [Google Scholar]
- 12.Okuma T, Honda R, Ichikawa G, Tsumagari N, Yasuda H. Biochem Biophys Res Commun. 1999;254:693–698. doi: 10.1006/bbrc.1998.9995. [DOI] [PubMed] [Google Scholar]
- 13.Johnson E S, Schwienhorst I, Dohmen R J, Blobel G. EMBO J. 1997;16:5509–5519. doi: 10.1093/emboj/16.18.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson E S, Blobel G. J Biol Chem. 1997;272:26799–26802. doi: 10.1074/jbc.272.43.26799. [DOI] [PubMed] [Google Scholar]
- 15.Schwarz S E, Matuschewski K, Liakopoulos D, Scheffner M, Jentsch S. Proc Natl Acad Sci USA. 1998;95:560–564. doi: 10.1073/pnas.95.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li S-J, Hochstrasser M. Nature (London) 1999;398:246–251. doi: 10.1038/18457. [DOI] [PubMed] [Google Scholar]
- 17.Suzuki T, Ichiyama A, Saitoh H, Kawakami T, Omata M, Chung C H, Kimura M, Shimbara N, Tanaka K. J Biol Chem. 1999;274:31131–31134. doi: 10.1074/jbc.274.44.31131. [DOI] [PubMed] [Google Scholar]
- 18.Gong L, Millas S, Maul G G, Yeh E T H. J Biol Chem. 2000;275:3355–3359. doi: 10.1074/jbc.275.5.3355. [DOI] [PubMed] [Google Scholar]
- 19.Seufert W, Futcher B, Jentsch S. Nature (London) 1995;373:78–81. doi: 10.1038/373078a0. [DOI] [PubMed] [Google Scholar]
- 20.Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- 21.Mahajan R, Gerace L, Melchior F. J Cell Biol. 1998;140:259–270. doi: 10.1083/jcb.140.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matunis M J, Wu J, Blobel G. J Cell Biol. 1998;140:499–509. doi: 10.1083/jcb.140.3.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Müller S, Matunis M J, Dejean A. EMBO J. 1998;17:61–70. doi: 10.1093/emboj/17.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duprez E, Saurin A J, Desterro J M, Lallemand-Breitenbach V, Howe K, Boddy M N, Solomon E, de Thé H, Hay R T, Freemont P S. J Cell Sci. 1999;112:381–393. doi: 10.1242/jcs.112.3.381. [DOI] [PubMed] [Google Scholar]
- 25.Ishov A M, Sotnikov A G, Vladimirova O V, Neff N, Kamitani T, Yeh E T H, Strauss J F, III, Maul G G. J Cell Biol. 1999;147:221–233. doi: 10.1083/jcb.147.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Desterro J M P, Rodriguez M S, Hay R T. Mol Cell. 1998;2:233–239. doi: 10.1016/s1097-2765(00)80133-1. [DOI] [PubMed] [Google Scholar]
- 27.Gostissa M, Hengstermann A, Fogalm V, Sandy P, Schwarz S E, Scheffner M, Del Sal G. EMBO J. 1999;18:6462–6471. doi: 10.1093/emboj/18.22.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodriguez M S, Desterro J M P, Lain S, Midgley C A, Lane D P, Hay R T. EMBO J. 1999;18:6455–6461. doi: 10.1093/emboj/18.22.6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iñiguez-Lluhi J A, Pearce D. Mol Cell Biol. 2000;20:6040–6050. doi: 10.1128/mcb.20.16.6040-6050.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karvonen U, Kallio P J, Jänne O A, Palvimo J J. J Biol Chem. 1997;272:15973–15979. doi: 10.1074/jbc.272.25.15973. [DOI] [PubMed] [Google Scholar]
- 31.Moilanen A-M, Poukka H, Karvonen U, Häkli M, Jänne O A, Palvimo J J. Mol Cell Biol. 1998;18:5128–5139. doi: 10.1128/mcb.18.9.5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmitz M L, Baeuerle P A. EMBO J. 1991;10:3805–3815. doi: 10.1002/j.1460-2075.1991.tb04950.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ikonen T, Palvimo J J, Jänne O A. J Biol Chem. 1997;272:29821–29828. doi: 10.1074/jbc.272.47.29821. [DOI] [PubMed] [Google Scholar]
- 34.Rosenthal N. Methods Enzymol. 1987;152:704–720. doi: 10.1016/0076-6879(87)52075-4. [DOI] [PubMed] [Google Scholar]
- 35.Poukka H, Aarnisalo P, Santti H, Jänne O A, Palvimo J J. J Biol Chem. 2000;275:571–579. doi: 10.1074/jbc.275.1.571. [DOI] [PubMed] [Google Scholar]
- 36.Kamitani T M, Nguyen H P, Yeh E T H. J Biol Chem. 1997;272:14001–14004. doi: 10.1074/jbc.272.22.14001. [DOI] [PubMed] [Google Scholar]
- 37.Jenster G, Trapman J, Brinkmann A O. Biochem J. 1993;293:761–768. doi: 10.1042/bj2930761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou Z, Sar M, Simental J A, Lane M V, Wilson E M. J Biol Chem. 1994;269:13115–13123. [PubMed] [Google Scholar]
- 39.Poukka H, Karvonen U, Yoshikawa N, Tanaka H, Palvimo J J, Jänne O A. J Cell Sci. 2000;113:2991–3001. doi: 10.1242/jcs.113.17.2991. [DOI] [PubMed] [Google Scholar]
- 40.Sternsdorf T, Jensen K, Reich B, Will H. J Biol Chem. 1999;274:12555–12566. doi: 10.1074/jbc.274.18.12555. [DOI] [PubMed] [Google Scholar]
- 41.Johnson E S, Blobel G. J Cell Biol. 1999;147:981–993. doi: 10.1083/jcb.147.5.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Müller S, Berger M, Lehembre F, Seeler J-S, Haupt Y, Dejean A. J Biol Chem. 2000;275:13321–13329. doi: 10.1074/jbc.275.18.13321. [DOI] [PubMed] [Google Scholar]
- 43.Jenster G, van der Korput H A, Trapman J, Brinkmann A O. J Biol Chem. 1995;270:7341–7346. doi: 10.1074/jbc.270.13.7341. [DOI] [PubMed] [Google Scholar]
- 44.Palvimo J J, Reinikainen P, Ikonen T, Kallio P J, Moilanen A, Jänne O A. J Biol Chem. 1996;271:24151–24156. doi: 10.1074/jbc.271.39.24151. [DOI] [PubMed] [Google Scholar]