Abstract
GABAergic dysfunction is present in the hippocampus in schizophrenia (SZ) and bipolar disorder (BD). The trisynaptic pathway was “deconstructed” into various layers of sectors CA3/2 and CA1 and gene expression profiling performed. Network association analysis was used to uncover genes that may be related to regulation of glutamate decarboxylase 67 (GAD67), a marker for this system that has been found by many studies to show decreased expression in SZs and BDs. The most striking change was a down-regulation of GAD67 in the stratum oriens (SO) of CA2/3 in both groups; CA1 only showed changes in the SO of schizophrenics. The network generated for GAD67 contained 25 genes involved in the regulation of kainate receptors, TGF-β and Wnt signaling, as well as transcription factors involved in cell growth and differentiation. In SZs, IL-1β, (GRIK2/3), TGF-β2, TGF-βR1, histone deacetylase 1 (HDAC1), death associated protein (DAXX), and cyclin D2 (CCND2) were all significantly up-regulated, whereas in BDs, PAX5, Runx2, LEF1, TLE1, and CCND2 were significantly down-regulated. In the SO of CA1 of BDs, where GAD67 showed no expression change, TGF-β and Wnt signaling genes were all up-regulated, but other transcription factors showed no change in expression. In other layers/sectors, BDs showed no expression changes in these GAD67 network genes. Overall, these results are consistent with the hypothesis that decreased expression of GAD67 may be associated with an epigenetic mechanism in SZ. In BD, however, a suppression of transcription factors involved in cell differentiation may contribute to GABA dysfunction.
Keywords: epigenetics, network association analysis, PAX5, Runx2, HDAC1
In recent years, the study of psychotic disorders has been seeking to define endophenotypes at the regional, cellular, molecular, and genetic levels. The term endophenotype has been defined as “measurable components unseen by the unaided eye” that occur along the continuum of proximal phenotype and distal genotype (1). The quantifiable aspects of an endophenotype can be identified by using many different forms of technology, including brain imaging, linkage analysis, high density haplotyping, SNP analysis, and gene expression profiling (GEP) studies in postmortem studies. The research described herein has attempted to elucidate the complex network of gene interactions that underlie the susceptibility for these two disorders. To accomplish this, it is necessary to examine large numbers of genes simultaneously in an attempt to understand how their complex interactions may contribute to normal and abnormal functioning (2).
There is now compelling evidence that a GABA defect occurs in corticolimbic regions of patients with either schizophrenia (SZ) or bipolar disorder (BD) and involves decreased expression of transcripts for GAD67. Evidence for decreased GABAergic activity has been obtained by many different laboratories by using a variety of techniques (for a review, see ref. 3). In the anterior cingulate cortex, such changes are selectively found in layer II (4), whereas in the hippocampus, they are particularly robust in sectors CA2/3 (5). The trisynaptic pathway consists of a complex relay of extrinsic and intrinsic inputs operating at various steps along the way toward CA1. This pathway includes mossy fiber projections from the granule cell layer that terminate on the dendrites of pyramidal neurons within the stratum radiatum (SR) of sectors CA3/2. These latter neurons, in turn, send Schaffer collaterals to the SR of sector CA1 where they terminate on the dendrites of pyramidal neuronss. These projection neurons send projections to the subiculum, entorhinal cortex, and dorsolateral prefrontal area (for a review, see ref. 6). Although GABA cells are found throughout the trisynaptic pathway, the stratum oriens (SO) and SR are almost exclusively populated by these inhibitory interneurons (7). It is believed that fast-field oscillations in the CA1 region reflect summed inhibitory postsynaptic currents in pyramidal cells as a result of high-frequency barrages from interneurons (8). It seems likely that the regulation of GAD67 expression and, by inference, the GABA cell phenotype, might involve variable extrinsic and intrinsic inputs at different points along the circuit.
To explore this question, the current study has used a combination of laser-capture microdissection and microarray-based GEP to identify whether there are networks of functionally related genes that show changes in expression in GABA cells of the trisynaptic pathway. This strategy has been used to assess whether such changes in expression vary according to specific locations within different layers and sectors of the hippocampus. Toward this end, the trisynaptic pathway has been partially deconstructed in a way that sectors CA3/2 and CA1, as well as their component layers, SR, stratum pyramidale (SP) and SO, have been separately examined. Neurons in SR and SO are almost exclusively GABAergic in nature, and this segregation has been exploited so that genes related to the regulation of GAD67 may be separately examined. Network association analysis has been used to establish relationships of GAD67 with other potential genes in signaling, metabolism, and transcriptional clusters.
Results
The percent present calls for SO, SP, and SR of sectors CA3/2 and CA1 was compared in BDs and SZs. Similar to a previous postmortem study from this laboratory (9), the number of genes satisfying the inclusion criterion of P = 0.05 was much greater in BDs than in SZs and was more pronounced in SO of CA3/2 and CA1 than in the other layers. In SP and SR, the number of genes meeting the inclusion criterion was much lower in both groups.
GAD67 and GAD65 Expression.
As shown in Table 1, expression data for GAD67 mRNA in whole hippocampal extracts were much lower and statistically less robust than those from the LMD preparations (9). In the laser microdissection study, the most striking decrease in GAD67 expression occurred in the SO of BDs with a fold change of −9.59 and a P = 0.0000048. The SZs also showed similarly robust P values in this layer, although the magnitude of the respective fold change was similar to that seen in SP and SR. In CA1, the only significant change in GAD67 expression was observed in SO in the SZ group, and the fold change was −3.27 (P = 0.043).
Table 1.
Expression of GAD67 in hippocampus of SZs and BDs
| Fold change | P | Fold change | P | ||
|---|---|---|---|---|---|
| Whole hippocampus extracts* | |||||
| BD vs. CON | −1.84 | 0.013 | |||
| SZ vs. CON | NC | NC | |||
| LMD dissections | CA2/3 | CA2/3 | CA1 | CA1 | |
| Stratum radiatum | BD vs. CON | NC | NC | NC | NC |
| SZ vs. CON | −2.81 | 0.000051 | NC | NC | |
| Stratum pyramidale | BD vs. CON | −2.74 | 0.048 | NC | NC |
| SZ vs. CON | −2.94 | 0.038 | NC | NC | |
| Stratum oriens | BD vs. CON | −9.59 | 0.0000048 | NC | NC |
| SZ vs. CON | −2.81 | 0.000051 | −3.27 | 0.043 | |
*From Konradi et al. (9); P, probability of significance; NC, no change.
As shown in Table 2, GAD65 also showed significant negative fold changes (i.e., −2.29 and −3.0, respectively) in the SO of CA2/3 of both the SZ and BD groups. The magnitude of these changes compares well with those for GAD67 (Table 1). Unlike the latter, however, no other changes in expression were observed for GAD65 in any of the other layers or sectors. As shown in Table 2, 10 of 13 genes showing significant changes in expression showed fold changes lower than for GAD67 (i.e., less than −2.0) in the SZs. For the BDs, virtually all of the genes showing significant changes had fold changes (i.e., −1.2 to −2.8) much lower than those for GAD67.
Table 2.
Genes involved in the regulation of GAD67 in SO of CA3/2
| Genes | SZ |
BD |
||
|---|---|---|---|---|
| Fold change | P | Fold change | P | |
| GAD65 | −2.29 | 0.008 | −3.00 | 0.004 |
| GRIK1 | −1.39 | 0.049 | −1.36 | 0.047 |
| GRIK2 | 1.50 | 0.038 | −1.20 | 0.022 |
| GRIK3 | 1.60 | 0.012 | — | — |
| GRIK4 | — | — | — | — |
| GRIK5 | — | — | — | — |
| TGFB2 | 1.43 | 0.016 | ||
| TGFBR1 | 1.31 | 0.011 | ||
| SMURF1 | — | — | −1.30 | 0.046 |
| SMAD1 | — | — | 2.30 | 0.029 |
| GSK-3β | — | — | — | — |
| CTNNB1 | — | — | — | — |
| PAX5 | — | — | −1.50 | 0.027 |
| LEF1 | — | — | −2.10 | 0.027 |
| TLE1 | −1.30 | 0.023 | −1.60 | 0.024 |
| IL1B | 1.28 | 0.037 | — | — |
| RUNX2 | — | — | −2.80 | 0.002 |
| FOXG1B | −2.47 | 0.001 | — | — |
| MSX1 | — | — | — | — |
| LHX2 | −2.10 | 0.026 | −2.30 | 0.018 |
| DLX2 | — | — | — | — |
| DAXX | 1.30 | 0.032 | — | — |
| HDAC1 | 1.50 | 0.022 | — | — |
| ID3 | — | — | — | — |
| CCND1 | — | — | — | — |
| CCND2 | 1.40 | 0.027 | −2.20 | 0.001 |
Target Genes for GAD67 Regulation.
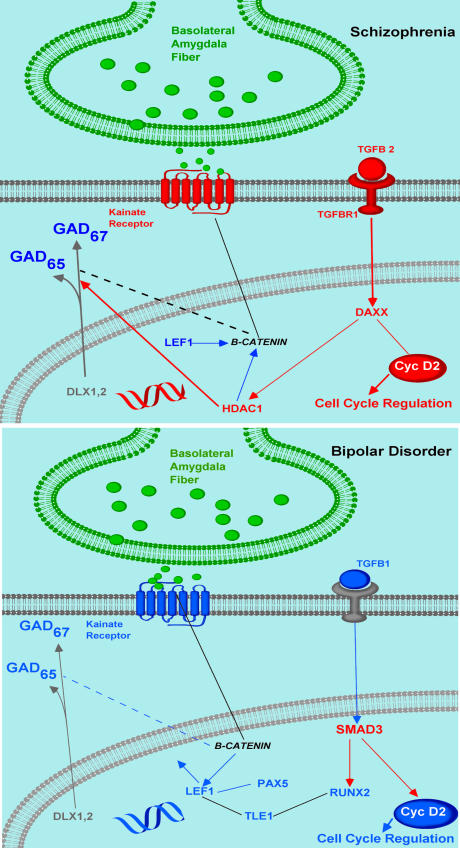
The network association analysis yielded a cluster consisting of several genes potentially involved in the regulation of GAD67 expression. These were configured into a schematic diagram indicating in what cellular compartments the respective gene products might be found (Fig. 1 and Table 2). Genes not showing significant changes were included in the diagram because they were constitutively expressed and, as such, could interact in a functionally meaningful way with the genes showing significant changes. In addition to kainate receptor subunits [glutamate receptor subunits 1–5 of the kainate receptor (GRIK1–5)], IL-1β, TGF-β signaling (i.e., TGF-β1, TGF-βR1, SMURF1, and SMAD1), Wnt signaling (i.e., GSK-3β and CTNNB1), cell cycle regulation (i.e., CCND1), and neurogenesis (i.e., PAX5), several transcription factors involved in cell differentiation (i.e., DLX1 and -2, LHX1, TLE1, Runx2, LEF1, and PAX5) were also represented in the network.
Fig. 1.
Schematic diagram based on ingenuity association analysis depicting the respective changes in the GAD67 regulatory network shown in Fig. 2 of SZs vs. BDs. The kainate receptors, which are up- and down-regulated, respectively, in SZ and BDs, are depicted as playing a central role in mediating the effect of basolateral amygdalar afferents on GABA cells in the SO of CA2/3. Cyclin D2 may be a pivotal gene that interfaces this network with that for cell cycle regulation.
SO of CA2/3.
The most striking changes occurred in SO of CA3/2 [supporting information (SI) Fig. 3A]. In SZs, IL-β1, GRIK2/3, TGF-β2, TGF-βR1, DAXX, HDAC1, and CCND2 were all significantly up-regulated, whereas FOXG1B and LHX1 were both down-regulated. In BDs, GRIK1/2, SMURF1, LHX1, Runx2, PAX5, LEF1, and CCND2 were all significantly down-regulated, whereas SMAD3 was the only up-regulated gene in SO of CA2/3 in BDs (Fig. 1 and Table 2).
SO-CA1.
In CA1 (SI Fig. 3B), the GAD67 regulatory network showed changes in gene expression in both BDs and SZs, but there were notable differences to respective findings in SO-CA2/3. In SZs, IL-1β, LEF1 and SMAD3 were all down-regulated; however, there were no changes in regulation for HDAC1 or DAXX. For the BDs, where there were no changes in the regulation of GAD67 in this locus, TGF-β2, GSK3β, CTNNB1, and FOXG1B were all up-regulated, whereas GRIK1/5, LEF1, SMAD3, and DLX2 were down-regulated. In both groups, CCND2 did not show changes in expression.
SR and SP of CA2/3 and CA1.
There were very few changes in the regulation of genes in the SR and SP of either sector in the SZ or BD groups (not shown).
Quantitative RT-PCR (QRT-PCR).
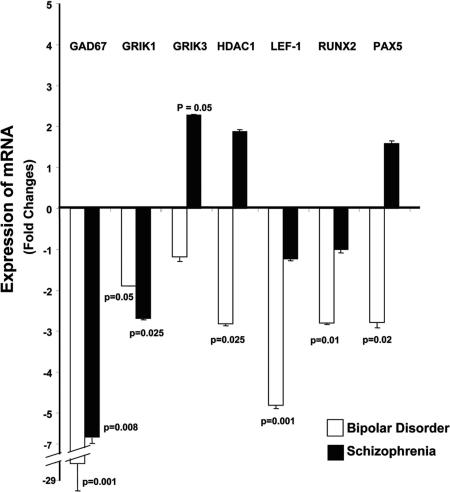
As shown in Fig. 2, the changes in gene expression observed in SO-CA2/3 were validated by using QRT-PCR. In all cases, the changes in expression occurred in the same direction as that seen with the microarrays. For GAD67 expression, the BDs showed a −27-fold change and the SZs a −6.5- fold change. The expression of HDAC1 showed a 2-fold increase in the SZs, but in BDs, it was significantly decreased. As observed with GEP, LEF1, Runx2, and PAX5 were all down-regulated in the BDs.
Fig. 2.
QRT-PCR validation of microarray-based GEP data. The results for GAD67, HDAC1, LEF1, Runx2, PAX5, and GRIK1/3 show changes in the same direction as those observed with microarray. The error bars are very small; however, the P value of 0.001 is robust for the REST analysis that was used. The error bars for GAD67 appear to be larger than those of the other genes because of the proportions of the graph needed to accommodate the data for all of the genes. On a percent basis, the error bars for GAD67 when expressed as a percent of the mean are relatively similar to those for the other genes.
Potential Confounding Effects.
As shown in SI Table 3, the normal control (CON), SZ, and BD groups were group-matched with respect to age, postmortem interval (PMI), gender, and laterality. Except for gender and hemispheric ratios, which were different in the BDs, age, PMI, pH, and 18S/28S ratios all showed complete overlap across the three groups. Hence, it is unlikely that any of these potential confounds influenced the data. In the case of the gender ratio, estrogen could have influenced the values obtained in the BDs because the SZs and CONs were matched with respect to this variable. The individual values for male vs. female groups were not appreciably different. Additionally, estrogens do not appear to exert a direct effect on GABAergic neurotransmission (10), making it unlikely that a higher ratio of females in the BD group could account for the GAD67 expression results. All of the SZs and six of the BDs were treated with antipsychotic medication during the year before death, making it unlikely that these drugs were responsible for the pattern of findings described in this report. Both SZs and BDs showed a significant reduction in the expression of GAD67, whether they were treated with “high” or “low” doses of neuroleptic. All of the BDs and many of the SZs were treated with one or more mood-stabilizing agents. The subjects receiving lithium alone, lithium plus other mood-stabilizing anticonvulsants, and mood-stabilizing anticonvulsants alone did not show meaningful differences in gene expression. No specific changes in the expression of genes associated with the network appeared to be related to the cause of death of the subjects.
Discussion
The results of this study have demonstrated a unique network consisting of 25 different genes that may be involved in the regulation of GAD67 expression in human hippocampus. That there were 10 genes (40% of total) in the BDs and 12 genes (48% of the total) in the SZs showing significant changes in expression argues against the possibility that the inclusion of these genes in the network occurred as a result of random effects. It also seems unlikely that the changes in expression seen in these various genes may be attributable to psychotropic medication effects. It is important to emphasize that the pattern of gene expression changes noted within this network varies on the basis of hippocampal layer, sector, and psychiatric diagnosis. The nature of these changes was largely consistent with the reduction in the expression of GAD67, suggesting there may be a potential cause–effect relationship among the various components of the network. Additionally, the most pronounced decreases of GAD67 expression, as well as other genes within this network, occurred preferentially in the SO of CA3/2 in both the SZ and BD groups. This is consistent with other work showing a preponderance of abnormalities in this same locus in SZ using a variety of markers and methodological approaches (for a review, see ref. 7).
It is important to note that at least some of the genes in the GAD67 network discussed below might be expressed by glial cells rather than neurons. In adult cortex and hippocampus, glial cells are defined in part by the absence of a nucleolus where mRNA is found and Nissl-positive material (i.e., RNA) in the cytoplasm. Consistent with this, ultrastructural studies demonstrate a paucity of ribosomes in the cytoplasm of glial cells, suggesting that transcription and translation may not occur robustly in these nonneuronal cells under normal in vivo circumstances. Contrariwise, neurons show a very distinctive nucleolus, as well as a robust Nissl staining in the cytoplasm. In the current study, only cells with identifiable Nissl-positive staining in their cytoplasm were microdissected, suggesting that the RNA samples were almost exclusively obtained from neurons, rather than glial cells. Nevertheless, there is a possibility that some of the mRNA included in this study was glial-derived. To minimize contamination with glial RNA, future studies will use Nissl-staining characteristics and/or immunolocalizations of marker proteins, such as calcium-binding peptides (GABA cells), glial fibrillary protein (astrocytes), and myelin basic protein oligodendrocytes to more effectively distinguish neurons and glia.
The genes showing changes in expression are associated with different tissue compartments within the cell. For example, IL1β is located in the extracellular space and is associated with multiple functions, including suppression of Wnt-dependent development in hippocampal embryonic tissue (11), protection against oxidative stress in adult tissue (12), and regulation of cell cycle indirectly by cyclin D2 (13). Also found extracellularly is fibroblast growth factor 8 that contributes to the differentiation of mesenchymal derivatives and to a proximal to distal growth of limbs (14). Among the genes found in the plasma membrane are the GRIK1–5. As discussed below, studies from other laboratories have demonstrated that these genes play a critical role in the early development of the hippocampus, influencing the differentiation of GABA cells and their functional integrity within the trisynaptic pathway (15).
TGF-β2, like IL-1β, is associated with the extracellular space; however, it gives rise to a critical signaling pathway that includes TGF-βR2 (plasma member), SMURF1 (cytoplasm), and SMAD3 (nucleus). The TGF-β signaling pathway is associated with many different early developmental processes (16), but disruptions of its function have also been detected in several adult disease states (17). Other genes included in the GAD67 regulatory network include CNNTB1 or β-catenin, GSK-3β, and LEF1, three key components of the Wnt signaling pathway. The latter, like the TGF-β pathway, also plays a role in modulating early developmental events, such as the formation of the neural tube (18) and forebrain (19). Neurogenesis during adulthood and, by inference, cell cycle regulation, has been postulated to occur in the hippocampus of SZs. Thus far, in studies of SZ, this has primarily been associated with the granule cell layer in the dentate gyrus (20–22). Perhaps relevant to SZ is the fact that Wnt signaling contributes to neurogenesis in the adult hippocampus (23). Similarly, cyclin D2 is the only D-type cyclin expressed in dividing cells derived from neuronal precursors in the adult hippocampus (24). Additionally, cyclins D1 and -2 (CCDN1/2) are also known to play a major role in the regulation of cell cycle. In this context, it is relevant to note that robust changes in both cell cycle regulation and neurogenesis gene clusters have been observed in the current databases; however, these findings will be reported elsewhere.
Several other transcription factors that appeared as key components of the GAD67 regulatory network have been specifically implicated in the development of GABAergic cells. These include DLX1/2, that play a central role in the migration of GABA cells in the hippocampus (25) and retina during embryogenesis (26). The transcription factor LHX2 is known to specifically contribute to the differentiation of GABAergic and cholinergic neurons during embryogenesis (27). The PAX family of paired homeobox genes are transcription factors expressed in several regions of the developing murine cerebellum (28). PAX2 is expressed by many different populations of GABA neurons in the cerebellar cortex, including Golgi II, basket, and stellate cells, as well as the deep cerebellar nuclei during embryogenesis. In the current study, it is the PAX5 isoform that is expressed in human hippocampus and included in the GAD67 regulatory network.
Transcription factors involved in embryogenesis are also present in the network. For example, PAX5 binds to ID3 and is known to influence the regulation of cell cycle in B lymphocytes (29). Additionally, TLE1, a corepressor of transcription, works together with FOXG1B (see below) to promote neuronal differentiation in cerebral cortex during the embryonic period (30). FOX1G is a major regulator of telencephalic neurogenesis in many regions, including the hippocampus (31); it also helps to control the proliferation of precursor cells by regulation of fibroblast growth factor 8 signaling and differentiation (32, 33). The Runx family of transcription factors is generally involved in the regulation of cell fate (34); the Runx1 and -3 isoforms contribute to the differentiation of sensory neurons in the dorsal root ganglion (35, 36). Runx2, the isoform included in the GAD67 network, plays a specific role in the differentiation of osteoblasts during embryonic development (37). It receives a positive regulation through the Wnt signaling pathway, although its interaction with β-catenin is an indirect one.
Also found in the GAD67 regulatory network is HDAC1 that forms a complex with DNA methyl transferase 1; both genes serve as repressors of promoter complexes (38), including that of the GAD67 gene (39). HDAC1 is also essential for the maintenance of neurogenesis in zebrafish during the embryonic period (40), and it is thought to be a potentially useful target for a chromatin-remodeling deficit suspected to occur in SZ (39). Closely aligned with HDAC1 is DAXX, a transcriptional corepressor that also acts at promoter sites (41).
One other gene known to play a pivotal role in determining the GABAergic phenotype during early development is LBX1 (42), which did not appear in the GAD67 regulatory network; however, its absence can be explained by the fact that its transcript was not detectable on the microarrays of any of the groups.
Taking together what is known about the genes comprising the GAD67 regulatory network, it seems possible they may have the ability to contribute to the differentiation and functional integrity of GABAergic interneurons in the human hippocampus during the adult period. Interestingly, several of the GAD67 network genes e.g., TLE1, FOX1GB, and Runx2, are transcription factors that are associated with cell differentiation during embryogenesis and/or neurogenesis during the adult period. Indeed some have even been specifically implicated in the differentiation of the GABA cell phenotype during the early prenatal period (e.g., GRIK1–3, DLX1/2, LHX, and PAX5). Taking these observations with the fact that a high proportion of genes showed significant changes in the SZ and BD groups, it seems reasonable to postulate that the GAD67 network described herein could theoretically play a key role in the maintenance of the GABA cell phenotype in the adult hippocampus.
As described in Results, there are two distinct patterns of expression changes that were observed in SZ vs. BDs. In SO of sector CA2/3 of SZs, HDAC1 and its epigenetic corepressor, DAXX, were both up-regulated, a pattern consistent with the hypothesis that epigenetic mechanisms play a selective role in the pathophysiology of SZ (43–45). Previous work in this area has pointed to DNA methyl transferase 1 as playing the central role in the regulation of the GAD67 promoter in the dorsolateral prefrontal cortex (46). In BDs, on the other hand, HDAC1 and DAXX did not show significant changes in expression; however, several transcription factor genes, including PAX 5, Runx2, LHX2, TLE1, and LEF1, were all down-regulated. The latter pattern suggests that an entirely different molecular mechanism might be involved in the decreased expression of GAD67 in BD. Given the association between these latter genes and cellular differentiation during embryogenesis, it is tempting to speculate that the decreased expression of GAD67 in the SO of CA2/3 of BD subjects might involve a shift toward a relatively less differentiated state. A dedifferentiation of normally segregated cortico–subcortical sensorimotor maps in the putamen has been described in dystonia (47).
In the SO of sector CA1, the pattern of gene expression changes was quite different from that seen in CA2/3. In SZs, only three genes showed significant changes in expression. In BDs, however, there were several genes associated with the TGF-β and Wnt signaling pathways that were significantly up-regulated. In the absence of changes in the regulation of GAD67, these latter findings support the view that this regulatory network might be capable of shifting GABA cells toward increased expression of GAD67 and possibly even toward an enhanced degree of functional differentiation.
Why would different genes within this GAD67 regulating network show different patterns of expression on a layer-by-layer and subregional basis? If these changes were related solely to the genetic susceptibility for the respective disorders, one would expect to see similar changes in GAD67 gene expression in GABA cells throughout the hippocampus. It is well known that neurons within the trisynaptic pathway receive different sets of intrinsic and extrinsic inputs and that these influence the feed forward excitation that normally occurs along this circuit (48). For example, pyramidal neurons in CA3/2 receive a major input from the mossy fiber system originating in the dentate area and terminating in the stratum lucidum (6), whereas pyramidal neurons in sector CA1 receive the Schaffer collaterals from CA3 that terminate the SR and entorhinal projections that terminate in the SM of this sector. On the other hand, the SO of CA3/2 receives abundant projections from subcortical regions, such as the basolateral amygdala (BLA) and septal nuclei. Those fibers originating from the BLA are probably glutamatergic in nature (49), whereas those originating from the septum can be either cholinergic (50) or GABAergic (51). The latter septal inputs exert a disinhibitory influence on pyramidal cell activity and may contribute to the generation of oscillations, such as the theta rhythm (51).
Given the complex organization of the trisynaptic pathway, it seems reasonable to assume that the profile of gene expression changes found in specific subpopulations of GABAergic interneurons and at various points along the circuit might also vary from one layer to another and from one sector to another. Indeed, the results reported here are consistent with this idea and further suggest that the patterns of gene expression observed in GABA cells of the SO in CA3/2 vs. those in CA1 may be related to activity-driven influences derived from various intrinsic and extrinsic inputs. That kainate receptor subunits form an integral part of the GAD67 regulating network is consistent with the observation that glutamate exerts a potent modulatory influence on hippocampal GABA cell activity (52) and contributes to the patterns of gene expression reported above (53). Interestingly, recent genetic association studies have implicated polymorphisms of GRIK2 (54) and GRIK3 (55) in the susceptibility for SZ.
In summary, the results of this study suggest that a common cellular phenotype in SZ and BD, the decreased expression of GAD67 in GABAergic interneurons, may involve different underlying molecular mechanisms (Fig. 1) that are in part related to susceptibility genes for the respective two disorders, as well as activity-dependent changes arising from specific afferent inputs to these interneurons. Overall, this study suggests that epigenetic regulation may play a contributory role in GABA cell dysfunction in SZ, whereas disturbances in the regulation of GABA cell differentiation may be present in BD.
Methods
Subjects.
The subjects consisted of seven normal CONs, seven SZs, and seven BDs from the Harvard Brain Tissue Resource Center at McLean Hospital matched for age, postmortem disorder, hemisphere, gender, and tissue pH (SI Table 4). Psychiatric diagnoses have been established by using a retrospective review of medical records and an extensive family questionnaire that includes the medical, psychiatric, and social history of the subjects. A gross and microscopic assessment of each case did not reveal any significant neuropathological changes. For the diagnosis of SZ, the criteria of Feighner et al. (56) were used, and the diagnoses of schizoaffective disorder and BD were made according to DMS-III-R criteria. Among the SZs, some cases have the diagnosis of schizoaffective disorder. All of the subjects were taking antipsychotic medications or mood-stabilizing agents during the year before death. At the time of processing, each brain was bisected, one hemisphere was fixed, and the other was frozen by using a combination of liquid nitrogen and dry ice (57).
Tissue Preparation.
Frozen tissue blocks from each of the fresh human cases were removed from the hippocampus at the level of the lateral geniculate nucleus. A total of seven frozen tissue sections were cut from each block on a Microm HM 560 CryoStar cryostat (8 μm), mounted on LEICA Frame Slides with a PET-membrane (1.4 μm), and fixed in Streck Tissue Fixative (Streck Laboratories, Omaha, NB). The Nissl-stained sections were examined microscopically to ensure that each was cut in a transverse plane through the hippocampus and that all of the typical cytoarchitectonic features (i.e., area dentate and sectors CA4, -3, -2, and -1), as well as their associated layers, were present. The frame slides were mounted on a LEICA AS LMD apparatus, and tissue samples from SO, SP, and SR of CA2/3 and CA1 were microdissected. Each vial into which the laser-dissected specimens fell by gravity contained a small volume of a lysis/denaturing solution to inhibit RNase activity.
RNA extraction was undertaken with a Qiagen Rneasy Micro Kit (Qiagen, Valencia, CA), and the RNA yield was in the range of 20–30 ng. RNA quality was assessed by using an Agilent 2100 bioanalyser (Agilent Technologies, Palo Alto, CA). Following the manufacturer's instructions, three rounds of linear amplification of the target were carried out by using the MessageAmp aRNA Amplification kit (Ambion, Austin, TX). The use of three rounds of amplification could induce degradation of RNA and potentially bias the microarray data; however, all of the samples across the three groups were processed in an identical fashion, making it unlikely that such bias occurred in one group to a greater degree than another.
Subsequently, target labeling was performed with the MessageAMP Biotin Enhanced Kit (Ambion). Fifteen micrograms of biotinylated target RNA was fragmented and individually hybridized to the HU-133A arrays (Affymetrix, Santa Clara, CA). The microarrays were then stained with two rounds of streptavidin–phycoerythrin (Molecular Probes, Eugene, OR) and one round of biotinylated antistreptavidin antibody (Vector Laboratories, Burlingame, CA), scanned twice, and visually inspected for evidence of artifacts.
In addition to their demographic factors, the cases included in this study (Table 1) were chosen on the basis of their RNA quality. This was assessed by using tissue pH, the 18S/28S ratio, and the Percent Present Calls for each case. Although the pH and 18S/28S ratio are generally considered good indicators of RNA quality in a postmortem specimen, there are notable instances in which this is not the case. In some cases, the Percent Present Call is a more meaningful indicator of RNA integrity because it reflects the number of genes that have been detected with the microarray analysis. There were cases in which the pH and 18S/28S ratios were excellent but the Percent Present calls were very low. In contrast, there were other cases for which pH and 18S/28S ratios were poor but the Percent Present Calls were close to the average for the respective group. The cases shown in Fig. 1 were those with the most robust Percent Present Calls. Because the Affymetrix system used in this study is biased toward the detection of the 3′ end of transcripts, this system can tolerate significant RNA degradation, while yielding high-quality expression data (58, 59).
Data Analyses.
The DNA Chip Analyzer (dChip) version 1.3 software package (60) was used to evaluate the Percent Present Calls and the significance of differences between the normal CONs and the SZs or BDs. As previously reported (61), the variance obtained with the Perfect Match Model of dChip (R2 = 0.001) was considerably lower than that seen with any of the other models and was used throughout the first-stage analysis of the microarray data.
Biologically relevant clusters of genes were identified by using GenMapp algorithms (www.genmapp.org). As described (62, 63), a metric, called the composite probability, Pc, was computed for each GenMapp biopathway or cluster based on (i) the P value for each gene showing significant differences, (ii) the number of genes showing significant changes, and (iii) the total number of genes within each cluster. The P value for inclusion in this study was set at P = 0.05. The magnitude of Pc depends upon the number of genes per biological cluster and the robustness of the P values for individual genes.
For the network association analyses, a total of 12 databases representing layers SO, SP, and SR in sectors CA3/2 and CA1 from CONs vs. SZ and CONs vs. BDs were evaluated. To ensure biological relevancy, the data input for this Bayesian network modeling consisted only of the genes showing significant differences detected with the GenMapp analysis (see above). A network association algorithm from Ingenuity Systems Pathways Analysis was used to interrogate the 12 databases for a network of genes possibly involved in the regulation of GAD67 (64). The gene expression data are related to the Ingenuity Knowledge Base, which is constantly updated with new scientific reports. It contains curated relationships among proteins, genes, complexes, cells, tissues, drugs, and diseases, so that comprehensive biologically relevant networks of genes can be derived.
QRT-PCR.
One microgram of three-round amplified unbiotinylated RNA from the microarray studies was reverse-transcribed with SuperScript II (Invitrogen, Carlsbad, CA) by using an oligo(dT) primer. For these QRT-PCR studies, single-round amplification could theoretically have been used to minimize degradation-related biases in the data; however, this would have required different reagents and conditions that would have resulted in RNA yields quite different from those used for the microarray portion of the study. Primers for target genes were designed by using the Primer3 Web-based software (http://frodo.wi.mit.edu/primer3) and synthesized by Operon Biotechnologies (Huntsville, AL). The sequence information for each of the primer pairs is shown in SI Table 4.
The genes above were chosen for the QRT-PCR analyses because they were considered most important for the study. All genes analyzed by using QRT-PCR have been included in Fig. 2. PCR was performed with the iQ SYBR Green Supermix from Bio-Rad (Hercules, CA). The 2X premix contained 100 mM KCl, 40 mM Tris·HCI (pH 8.4), 0.4 mM each dNTP (dATP, dCTP, dGTP, and dTTP), as well as 50 units/ml iTaq DNA polymerase, 6 mM MgCl2, SYBR green I, 20 nM fluoresein, and stabilizers. Primers were added for a final concentration of 300 nM. Twenty nanograms of ssDNA template was run for each sample. The PCR amplifications were performed on an MJR Chromo4 (Bio-Rad) using a 2-min 30-sec hot start at 95°C, followed by 45 cycles of 15 sec at 95°C, 30 sec at 57°C, and 30 sec at 72°C. After 45 cycles, a melting curve was created by stepping the temperature from 72°C to 95°C. The fluorescence was read over 0.2°C increments.
Three potential “housekeeping” genes were considered for the purpose of normalizing the QRT-PCR data and included glycerophosphate dehydrogenase (G3PD), β-actin, and β2-microglobulin. Differences in expression across the groups were evaluated in sectors CA2/3 and CA1, and the data were expressed as a mean and standard error of the mean for the subjects in each group. β2-Microglobulin was the only gene that showed little or no change in expression in the layers and sectors examined and was designated as the normalizing gene for subsequent QRT-PCR studies. The Relative Expression Software Tool (REST), version 2, was used to analyze QRT-PCR data for the normalized target genes (65).
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants MH42261, MH31862, and MH62822.
Abbreviations
- SR
stratum radiatum
- SP
stratum pyramidale
- SO
stratum oriens
- GEP
gene expression profiling
- BD
bipolar disorder
- SZ
schizophrenic
- CON
control
- GRIK
glutamate receptor subunit of the kainate receptor
- QRT-PCR
quantitative RT-PCR.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0703806104/DC1.
References
- 1.Gottesman II, Gould TD. Am J Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- 2.Schadt EE, Lamb J, Yang X, Zhu J, Edwards S, Guhathakurta D, Sieberts SK, Monks S, Reitman M, Zhang C, et al. Nat Genet. 2005;37:710–717. doi: 10.1038/ng1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benes FM. Brain Res Rev. 2000;31:251–269. doi: 10.1016/s0165-0173(99)00041-7. [DOI] [PubMed] [Google Scholar]
- 4.Vawter MP, Evans S, Choudary P, Tomita H, Meador-Woodruff J, Molnar M, Li J, Lopez JF, Myers R, Cox D, et al. Neuropsychopharmacology. 2004;29:373–384. doi: 10.1038/sj.npp.1300337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Todtenkopf MS, Benes FM. Synapse. 1998;29:323–332. doi: 10.1002/(SICI)1098-2396(199808)29:4<323::AID-SYN4>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 6.Rosene DL, Van Hoesen GW. In: Cerebral Cortex. Peters A, Jones EG, editors. Plenum, New York: 1987. pp. 345–456. [Google Scholar]
- 7.Benes FM, Berretta S. Neuropsychopharmacology. 2001;25:1–27. doi: 10.1016/S0893-133X(01)00225-1. [DOI] [PubMed] [Google Scholar]
- 8.Ylinen A, Bragin A, Nadasdy Z, Jando G, Szabo I, Sik A, Buzsaki G. J Neurosci. 1995;15:30–46. doi: 10.1523/JNEUROSCI.15-01-00030.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Konradi C, Eaton M, MacDonald ML, Walsh J, Benes FM, Heckers S. Arch Gen Psychiatry. 2004;61:300–308. doi: 10.1001/archpsyc.61.3.300. [DOI] [PubMed] [Google Scholar]
- 10.Wilson MA. Crit Rev Neurobiol. 1996;10:1–37. doi: 10.1615/critrevneurobiol.v10.i1.10. [DOI] [PubMed] [Google Scholar]
- 11.Shimogori T, Banuchi V, Ng HY, Strauss JB, Grove EA. Development (Cambridge, UK) 2004;131:5639–5647. doi: 10.1242/dev.01428. [DOI] [PubMed] [Google Scholar]
- 12.Mark RJ, Fuson KS, Keane-Lazar K, May PC. Brain Res. 1999;830:88–93. doi: 10.1016/s0006-8993(99)01390-6. [DOI] [PubMed] [Google Scholar]
- 13.Roy D, Sarkar S, Felty Q. Front Biosci. 2006;11:889–898. doi: 10.2741/1845. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y, Liu C, Yamada Y, Fan CM. Development (Cambridge, UK) 2002;129:5289–5300. doi: 10.1242/dev.129.22.5289. [DOI] [PubMed] [Google Scholar]
- 15.Maingret F, Lauri SE, Taira T, Isaac JT. J Physiol (London) 2005;567:131–142. doi: 10.1113/jphysiol.2005.089474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knepper JL, James AC, Ming JE. Dev Dyn. 2006;235:1482–1490. doi: 10.1002/dvdy.20725. [DOI] [PubMed] [Google Scholar]
- 17.Attisano L, Wrana JL. Science. 2002;296:1646–1647. doi: 10.1126/science.1071809. [DOI] [PubMed] [Google Scholar]
- 18.Backman M, Machon O, Mygland L, van den Bout CJ, Zhong W, Taketo MM, Krauss S. Dev Biol. 2005;279:155–168. doi: 10.1016/j.ydbio.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 19.Galceran J, Miyashita-Lin EM, Devaney E, Rubenstein JL, Grosschedl R. Development (Cambridge, UK) 2000;127:469–482. doi: 10.1242/dev.127.3.469. [DOI] [PubMed] [Google Scholar]
- 20.Lipska BK. J Psychiatry Neurosci. 2004;29:282–286. [PMC free article] [PubMed] [Google Scholar]
- 21.Reif A, Fritzen S, Finger M, Strobel A, Lauer M, Schmitt A, Lesch KP. Mol Psychiatry. 2006;11:514–522. doi: 10.1038/sj.mp.4001791. [DOI] [PubMed] [Google Scholar]
- 22.Toro CT, Deakin JF. Schizophr Res. 2007;90:1–14. doi: 10.1016/j.schres.2006.09.030. [DOI] [PubMed] [Google Scholar]
- 23.Lie DC, Colamarino SA, Song HJ, Desire L, Mira H, Consiglio A, Lein ES, Jessberger S, Lansford H, Dearie AR, et al. Nature. 2005;437:1370–1375. doi: 10.1038/nature04108. [DOI] [PubMed] [Google Scholar]
- 24.Kowalczyk A, Filipkowski RK, Rylski M, Wilczynski GM, Konopacki FA, Jaworski J, Ciemerych MA, Sicinski P, Kaczmarek L. J Cell Biol. 2004;167:209–213. doi: 10.1083/jcb.200404181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pleasure SJ, Anderson S, Hevner R, Bagri A, Marin O, Lowenstein DH, Rubenstein JL. Neuron. 2000;28:727–740. doi: 10.1016/s0896-6273(00)00149-5. [DOI] [PubMed] [Google Scholar]
- 26.de Melo J, Qiu X, Du G, Cristante L, Eisenstat DD. J Comp Neurol. 2003;461:187–204. doi: 10.1002/cne.10674. [DOI] [PubMed] [Google Scholar]
- 27.Manabe T, Tatsumi K, Inoue M, Matsuyoshi H, Makinodan M, Yokoyama S, Wanaka A. J Neurochem. 2005;94:723–730. doi: 10.1111/j.1471-4159.2005.03261.x. [DOI] [PubMed] [Google Scholar]
- 28.Maricich SM, Herrup K. J Neurobiol. 1999;41:281–294. doi: 10.1002/(sici)1097-4695(19991105)41:2<281::aid-neu10>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 29.Sugai M, Gonda H, Nambu Y, Yokota Y, Shimizu A. J Mol Med. 2004;82:592–599. doi: 10.1007/s00109-004-0562-z. [DOI] [PubMed] [Google Scholar]
- 30.Marcal N, Patel H, Dong Z, Belanger-Jasmin S, Hoffman B, Helgason CD, Dang J, Stifani S. Mol Cell Biol. 2005;25:10916–10929. doi: 10.1128/MCB.25.24.10916-10929.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muzio L, Mallamaci A. J Neurosci. 2005;25:4435–4441. doi: 10.1523/JNEUROSCI.4804-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen Q, Wang Y, Dimos JT, Fasano CA, Phoenix TN, Lemischka IR, Ivanova NB, Stifani S, Morrisey EE, Temple S. Nat Neurosci. 2006;9:743–751. doi: 10.1038/nn1694. [DOI] [PubMed] [Google Scholar]
- 33.Martynoga B, Morrison H, Price DJ, Mason JO. Dev Biol. 2005;283:113–127. doi: 10.1016/j.ydbio.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 34.Blyth K, Cameron ER, Neil JC. Nat Rev Cancer. 2005;5:376–387. doi: 10.1038/nrc1607. [DOI] [PubMed] [Google Scholar]
- 35.Kramer I, Sigrist M, de Nooij JC, Taniuchi I, Jessell TM, Arber S. Neuron. 2006;49:379–393. doi: 10.1016/j.neuron.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 36.Zhong J, Pevny L, Snider WD. Neuron. 2006;49:325–327. doi: 10.1016/j.neuron.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 37.Westendorf JJ, Zaidi SK, Cascino JE, Kahler R, van Wijnen AJ, Lian JB, Yoshida M, Stein GS, Li X. Mol Cell Biol. 2002;22:7982–7992. doi: 10.1128/MCB.22.22.7982-7992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fuks F, Burgers WA, Brehm A, Hughes-Davies L, Kouzarides T. Nat Genet. 2000;24:88–91. doi: 10.1038/71750. [DOI] [PubMed] [Google Scholar]
- 39.Costa E, Grayson DR, Mitchell CP, Tremolizzo L, Veldic M, Guidotti A. Crit Rev Neurobiol. 2003;15:121–142. doi: 10.1615/critrevneurobiol.v15.i2.20. [DOI] [PubMed] [Google Scholar]
- 40.Cunliffe VT. Development (Cambridge, UK) 2004;131:2983–2995. doi: 10.1242/dev.01166. [DOI] [PubMed] [Google Scholar]
- 41.Ecsedy JA, Michaelson JS, Leder P. Mol Cell Biol. 2003;23:950–960. doi: 10.1128/MCB.23.3.950-960.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng L, Samad OA, Xu Y, Mizuguchi R, Luo P, Shirasawa S, Goulding M, Ma Q. Nat Neurosci. 2005;8:1510–1515. doi: 10.1038/nn1569. [DOI] [PubMed] [Google Scholar]
- 43.Costa E, Chen Y, Davis J, Dong E, Noh JS, Tremolizzo L, Veldic M, Grayson DR, Guidotti A. Mol Interv. 2002;2:47–57. doi: 10.1124/mi.2.1.47. [DOI] [PubMed] [Google Scholar]
- 44.Costa E, Grayson DR, Veldic M, Guidotti A. Int Rev Neurobiol. 2004;59:73–91. doi: 10.1016/S0074-7742(04)59004-9. [DOI] [PubMed] [Google Scholar]
- 45.Costa E, Grayson DR, Guidotti A. Mol Interv. 2003;3:220–229. doi: 10.1124/mi.3.4.220. [DOI] [PubMed] [Google Scholar]
- 46.Noh JS, Sharma RP, Veldic M, Salvacion AA, Jia X, Chen Y, Costa E, Guidotti A, Grayson DR. Proc Natl Acad Sci USA. 2005;102:1749–1754. doi: 10.1073/pnas.0409648102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Delmaire C, Krainik A, Tezenas du Montcel S, Gerardin E, Meunier S, Mangin J-F, Sangla S, Garnero L, Vidailht M, et al. Neurology. 2005;64:1391–1396. doi: 10.1212/01.WNL.0000158424.01299.76. [DOI] [PubMed] [Google Scholar]
- 48.Benes FM, Berretta S. Ann NY Acad Sci. 2000;911:293–304. doi: 10.1111/j.1749-6632.2000.tb06733.x. [DOI] [PubMed] [Google Scholar]
- 49.Swanson LW, Petrovich GD. Trends Neurosci. 1998;21:323–331. doi: 10.1016/s0166-2236(98)01265-x. [DOI] [PubMed] [Google Scholar]
- 50.Wu M, Shanabrough M, Leranth C, Alreja M. J Neurosci. 2000;20:3900–3908. doi: 10.1523/JNEUROSCI.20-10-03900.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Toth K, Freund TF, Miles R. J Physiol (London) 1997;500:463–474. doi: 10.1113/jphysiol.1997.sp022033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kullmann DM, Semyanov A. Epilepsia. 2002;43(Suppl 5):174–178. doi: 10.1046/j.1528-1157.43.s.5.12.x. [DOI] [PubMed] [Google Scholar]
- 53.Martin DL, Rimvall K. J Neurochem. 1993;60:395–407. doi: 10.1111/j.1471-4159.1993.tb03165.x. [DOI] [PubMed] [Google Scholar]
- 54.Bah J, Quach H, Ebstein RP, Segman RH, Melke J, Jamain S, Rietschel M, Modai I, Kanas K, Karni O, et al. NeuroReport. 2004;15:1987–1991. doi: 10.1097/00001756-200408260-00031. [DOI] [PubMed] [Google Scholar]
- 55.Begni S, Popoli M, Moraschi S, Bignotti S, Tura GB, Gennarelli M. Mol Psychiatry. 2002;7:416–418. doi: 10.1038/sj.mp.4000987. [DOI] [PubMed] [Google Scholar]
- 56.Feighner JP, Robins E, Guze SB, Woodruff RA, Jr, Winokur G, Munoz R. Arch Gen Psychiatry. 1972;26:57–63. doi: 10.1001/archpsyc.1972.01750190059011. [DOI] [PubMed] [Google Scholar]
- 57.Vonsattel JP, Aizawa H, Ge P, DiFiglia M, McKee AC, MacDonald M, Gusella JF, Landwehrmeyer GB, Bird ED, Richardson EPJ. J Neuropathol Exp Neurol. 1995;54:42–56. doi: 10.1097/00005072-199501000-00006. [DOI] [PubMed] [Google Scholar]
- 58.Luzzi V, Mahadevappa M, Raja R, Warrington JA, Watson MA. J Mol Diagn. 2003;5:9–14. doi: 10.1016/S1525-1578(10)60445-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee J, Aniko H, Willhite D, Zlotnik A, Hevezi P. FASEB J. 2005;19:1359–1361. doi: 10.1096/fj.04-3552fje. [DOI] [PubMed] [Google Scholar]
- 60.Li CX, Wong WH. Proc Natl Acad Sci USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Benes FM, Burke RE, Walsh J, Berretta S, Matzilevich D, Minns M, Konradi C. Mol Psychiatry. 2006;11:158–171. doi: 10.1038/sj.mp.4001524. [DOI] [PubMed] [Google Scholar]
- 62.Benes FM, Matzilevich D, Burke RE, Walsh J. Mol Psychiatry. 2006;11:241–251. doi: 10.1038/sj.mp.4001758. [DOI] [PubMed] [Google Scholar]
- 63.Benes FM, Burke RE, Walsh J, Matzilevich D, Berretta S, Minns M, Konradi C. Mol Psychiatry. 2004;9:932–945. doi: 10.1038/sj.mp.4001524. [DOI] [PubMed] [Google Scholar]
- 64.Butte AJ, Kohane IS. Pac Symp Biocomput. 2000:418–29. doi: 10.1142/9789814447331_0040. [DOI] [PubMed] [Google Scholar]
- 65.Pfaffl MW, Horgan GW, Dempfle L. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.