Abstract
The prothrombin (F2) 3′ end formation signal is highly susceptible to thrombophilia-associated gain-of-function mutations. In its unusual architecture, the F2 3′ UTR contains an upstream sequence element (USE) that compensates for weak activities of the non-canonical cleavage site and the downstream U-rich element. Here, we address the mechanism of USE function. We show that the F2 USE contains a highly conserved nonameric core sequence, which promotes 3′ end formation in a position- and sequence-dependent manner. We identify proteins that specifically interact with the USE, and demonstrate their function as trans-acting factors that promote 3′ end formation. Interestingly, these include the splicing factors U2AF35, U2AF65 and hnRNPI. We show that these splicing factors not only modulate 3′ end formation via the USEs contained in the F2 and the complement C2 mRNAs, but also in the biocomputationally identified BCL2L2, IVNS and ACTR mRNAs, suggesting a broader functional role. These data uncover a novel mechanism that functionally links the splicing and 3′ end formation machineries of multiple cellular mRNAs in an USE-dependent manner.
Keywords: F2, mRNA processing, polyadenylation, splicing factor, upstream sequence element (USE)
Introduction
With the exception of some histone mRNAs, all eukaryotic mRNAs possess poly(A)-tails at their 3′ end, which are produced by a two-step reaction involving endonucleolytic cleavage and subsequent poly(A) tail addition (Colgan and Manley, 1997; Keller and Minvielle-Sebastia, 1997; Zhao et al, 1999; Gilmartin, 2005). The specificity and efficiency of 3′ end processing is determined by the binding of a multiprotein complex to the 3′ end processing signal. Most cellular pre-mRNAs contain two core elements. The canonical polyadenylation signal AAUAAA upstream of the cleavage site is recognized by the multimeric cleavage and polyadenylation specificity factor (CPSF). This RNA–protein interaction determines the site of cleavage 10–30 nt downstream, preferentially immediately 3′ of a CA dinucleotide. The second canonical sequence element is characterized by a high density of G/U or U residues and is located up to 30 nt downstream of the cleavage site. This downstream sequence element (DSE) is bound by the 64 kDa subunit of the heterotrimeric cleavage stimulating factor (CstF) that promotes the efficiency of 3′ end processing. Additional proteins, cleavage factors I and II (CF I and CF II), associate and the pre-mRNA is cleaved by CPSF 73 (Ryan et al, 2004; Dominski et al, 2005; Mandel et al, 2006). Subsequently, poly(A) polymerase (PAP) adds ∼250 A-nucleotides to the 3′ end in a template-independent manner. Finally, several molecules of the poly(A)-binding protein II (PABPN1) bind to the growing poly(A) tail and determine its length. These proteins remain attached to the poly(A) tail during nuclear export and enhance both, the stability and the translation of the mRNA (von der Haar et al, 2004). Therefore, defects of mRNA 3′ end formation can profoundly alter cell viability, growth and development by interfering with essential and well-coordinated cellular processes.
Although almost all pre-mRNAs are constitutively polyadenylated, alternative and regulated poly(A) site selection represents an important regulatory mechanism for spatial and temporal control of gene expression (Zhao and Manley, 1996; Edwalds-Gilbert et al, 1997; Barabino and Keller, 1999; Zhao et al, 1999). Some 49% of human mRNAs contain more than one polyadenylation site (Yan and Marr, 2005). Alternative and regulated 3′ end processing serves to direct important cellular processes such as immunoglobulin class switch (Takagaki et al, 1996) or the regulated expression of the transcription factor NF-ATc during T-cell differentiation (Chuvpilo et al, 1999).
The medical consequences of errors of 3′ end processing are exemplified by the molecular sequelae of a common prothrombotic mutation in the prothrombin (F2) mRNA (F2 20210G → A). This mutation affects the most 3′ nucleotide of the mature mRNA, where the pre-mRNA is endonucleolytically cleaved and polyadenylated; it reverts the physiologically inefficient F2 cleavage site into the most favorable CA dinucleotide context, increasing cleavage site recognition and resulting in the accumulation of correctly 3′ end processed F2 mRNA in the cytoplasm. From these studies, enhanced mRNA 3′ end formation efficiency emerged as a novel molecular principle underlying pathological gene expression and explaining the role of F2 20210G → A in the pathogenesis of thrombophilia (Gehring et al, 2001).
Subsequent analyses of the F2 mRNA 3′ end revealed an unusual architecture of non-canonical 3′ end processing signals that explain the susceptibility of the F2 3′ UTR and 3′ flanking sequence to additional, clinically relevant gain-of-function mutations (Danckwardt et al, 2004, 2006a, 2006b). The presence of a sequence element that is located upstream (upstream sequence element (USE)) of the cleavage site within the 3′ UTR stimulates F2 3′ end processing. Moreover, this 15-nucleotide spanning element is both necessary and sufficient to enhance 3′ end processing when inserted into a heterologous β-globin mRNA 3′ UTR (Danckwardt et al, 2004).
Unlike (retro-)viral RNAs (Gilmartin et al, 1995; Graveley et al, 1996), stimulatory USEs have been experimentally documented in only a few mammalian mRNAs such as the human complement C2 (Moreira et al, 1998), lamin B2 (Brackenridge and Proudfoot, 2000), cyclooxygenase-2 (Hall-Pogar et al, 2005) and the collagen genes (Natalizio et al, 2002). Biocomputational analyses now predict that USEs may represent a common and evolutionarily conserved feature of mammalian 3′ end formation signals (Legendre and Gautheret, 2003; Hu et al, 2005), suggesting a broad role of USEs in cellular 3′ end mRNA processing.
We systematically analyzed the F2 USE and determined its mechanism of function. We show that several splicing factors, CPSF and CstF components specifically bind to the highly conserved USE. The functional characterization of these RNA-binding proteins by RNAi reveals a specific stimulatory effect of known splicing factors on the 3′ end processing of the F2 and C2 USE-containing pre-mRNAs as well as the biocomputationally predicted targets BCL2L2, IVNS and ACTR mRNAs. We propose a model of USE-directed 3′ end processing that involves a novel mRNP that integrates different nuclear pre-mRNA processing steps. Our data also implicate USE-dependent RNP complex formation in the physiology of important cellular processes such as hemostasis (and other thrombin-dependent processes) and the regulation of C2 gene expression as a component of innate immunity.
Results
The F2 USE increases mRNA 3′ end processing efficiency in a position- and sequence-dependent manner
To systematically define the F2 USE and study its mechanism of function, we established an internally controlled in vivo 3′ end processing assay (Danckwardt et al, 2004) and generated constructs that contain a tandem array of 3′ end formation signals, with modifications of the F2 USE within the 5′ site (Figure 1A). In contrast, the unmodified downstream site consists of sequences originating from the wild-type F2 3′ UTR and its 3′ flanking sequences. Thus, the smaller mRNA isoform detected in the poly(A) test (PAT) analysis has been cleaved and polyadenylated at the 5′ site, whereas the longer isoform has been processed at the 3′ site. This experimental setting enabled us to directly compare the processing efficiency of the (manipulated) 5′ site in relation to the control 3′ site, providing an internal control for other potential variables such as transcription or splicing efficiency, which could influence the abundance of the mRNA encoded by the transfected constructs.
Figure 1.
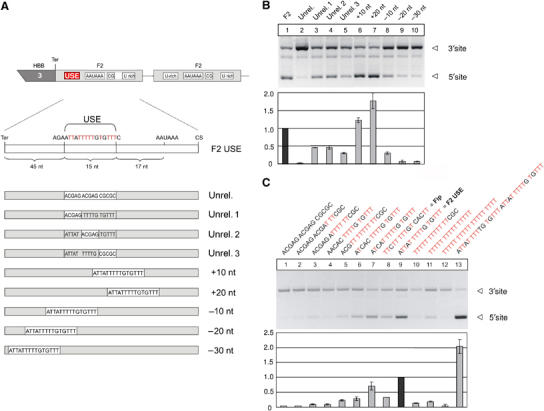
The F2 USE stimulates mRNA expression and 3′ end formation in a position- and sequence-dependent manner. (A) Schematic representation of the HBB (human β-globin)—F2 hybrid gene construct with a tandem array of two F2 3′ end formation signals used in transient transfection experiments (Ter: stop codon). The F2 USE was either completely or partially replaced by an unrelated nucleotide sequence, or displaced downstream and upstream of its original position as depicted. (B) In vivo assay carried out by transient transfection of a HBB-F2 hybrid gene construct with modifications of the USE as depicted in (A). The bar diagram shows the fold difference of the mRNA ratio processed at the 5′ or the 3′ site (5′/3′) relative to the F2 WT construct (highlighted)±s.e. (at least four independent experiments). (C) In vivo assay carried out by transient transfection of a HBB-F2 hybrid gene construct with modifications of the USE as depicted. The bar diagram shows the fold difference of the mRNA ratio processed at the 5′ or the 3′ site (5′/3′) relative to the F2 WT construct (highlighted)±s.e. (at least four independent experiments).
The in vivo assay carried out in transiently transfected HeLa cells (Figure 1B) indicates that the replacement of the entire USE (Unrel., lane 2) almost completely abolishes 3′ end formation at the affected 5′site, when compared to F2 WT (USE, lane 1). In contrast, partial replacement of the first, second or third nucleotide quintett of the USE motif by an unrelated sequence reduces the 3′ end formation capacity at the respective site by ∼2-fold (Figure 1B, lanes 3–5), although significant 3′ end formation was still observed.
Because of the critical spatial relationship of canonical 3′ end formation signals to each other, we next analyzed the positional requirements of the USE on mRNA expression and 3′ end formation. For this purpose, it is important to note that the 15-nucleotide spanning USE per se is sufficient to enhance 3′ end processing even in a heterologous 3′ UTR in a context-independent manner (Danckwardt et al, 2004). Displacing the USE, therefore, assays the positional requirements of USE function and is not expected to be compounded by a potential disruption of the surrounding mRNA architecture. The successive shift of the USE downstream towards the polyadenylation signal enhances 3′ end processing (Figure 1B, compare lane 1 with lanes 6 and 7). In contrast, shifting the USE further upstream (by 10, 20 and 30 nucleotides, respectively) resulted in a successive down-modulation of mRNA expression through loss of function of 3′ end processing (Figure 1B, compare lane 1 with lanes 8, 9 and 10). Furthermore, the (relative) changes of the efficiency of the 5′ poly(A) site upon modification (in the context of the tandem construct) were also reflected on the level of absolute mRNA abundance (Supplementary Figure S1), which indicates that the results of the PAT analysis as shown here are not compounded by the fact that the normal F2 3′ end processing site is <100% efficient. Thus, the position of the USE with respect to the canonical polyadenylation signals seems to be a quantitative determinant of its function in 3′ end processing.
Previously published data suggest that USEs might stimulate 3′ end processing, at least in part, by recruiting components of the canonical CstF complex (Moreira et al, 1998), which, under normal circumstances, critically depends on the density of U residues. We therefore analyzed whether the F2 USE activity depends on its uridine (U) content or on a more specific sequence context. To this end, we tested constructs with increasing number of U residues within the USE core region (Figure 1C). While decreasing the number of U residues caused a gradual reduction of the 3′ end processing efficiency (Figure 1C, lanes 1–7 and lane 9), increasing the number of U residues also reduced the 3′ end formation efficiency (Figure 1C, lanes 10 and 11), eventually even ablating 3′ end maturation completely (lane 12). Furthermore, the USE of the L3 mRNA that is bound by hFip1 (Kaufmann et al, 2004) was less efficient as the wild-type F2 USE (Figure 1C, lanes 8 and 9). However, duplicating the wild-type USE motif had a stimulatory impact on 3′ end formation by ∼2-fold (Figure 1C, lane 13). These effects seem to be independent of a specific cell type, as similar results were obtained both in transfected HUH-7 and HeLa cells (not shown).
These results show that USE function is sequence and position sensitive, and that its potency is not simply determined by its U content. Because CstF binding at U-rich DSEs depends on the density of U-residues, these findings suggest that the F2 USE plays a specific role and does not simply compensate for the absent DSE in the F2 pre-mRNA. In this respect, the F2 mRNA appears to differ from the otherwise similar C2 mRNA (Moreira et al, 1998).
Finally, a sequence alignment revealed that the F2 USE is highly conserved among higher eukaryotes and is located at similar positions 17–22 nucleotides upstream of the poly(A) signal (Figure 2A). It comprises two highly conserved overlapping 3′ UTR motifs (UAUUUUU and UUUUGU) belonging to the top 10 out of 106 highly conserved 3′ UTR motifs, with a cross-species conservation rate of 30 and 24%, respectively (Xie et al, 2005). Interestingly, the disruption of either motif individually and/or the presence of only one motif highly correlated with loss of function (Figure 1B and C; Supplementary Figure S2, and data not shown), which emphasizes the importance of both sequence elements. It seems likely, therefore, that the F2 USE has evolved as an optimal sequence context that includes a nonameric core sequence (Figure 2A) in a functionally important region up to 40 nucleotides upstream of the poly(A) site (previously designated as core upstream element (CUE); Hu et al, 2005) to promote 3′ end processing. It is noteworthy that the USE as identified here does not include the tetramers UGUA and UAUA that have recently been shown to account for 3′ end formation at another non-canonical poly(A) site by recruiting the human 3′ processing factor CFIm (Venkataraman et al, 2005).
Figure 2.
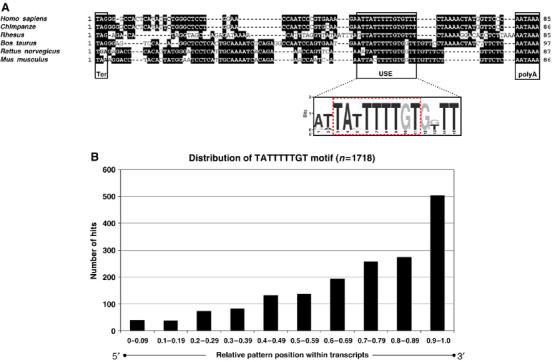
The F2 USE is highly conserved among higher eukaryotes. (A) Sequence comparison of the 3′ ends of vertebrate F2 genes (encompassing the entire 3′ UTR until the poly(A) signal). Shaded sequences denote identity. The graphical representation of the nucleic acid multiple sequence alignment (shown below) highlights the sequence conservation of the F2 USE according to the WebLogo 3 algorithm (Materials and methods), which contains a composite of two highly conserved sequence motifs (TATTTTTGT, highlighted; Xie et al, 2005). (B) Applying a sequence search algorithm that takes into consideration both the strand specificity and the typical length distribution for 3′ UTR motifs (peak >8-mers after exclusion of miRNAs target sites; Xie et al, 2005), more than 1700 human transcripts were identified to contain the nonameric USE core sequence motif. Number of hits are shown according to the localization of the sequence element within the mRNAs (bar diagram, x-axis, 5′ to 3′, left to right). Positive hits were filtered according to their relative location with respect to the poly(A) signal (AATAAA and ATTAAA, respectively). The identity of transcripts that contained the USE core sequence in close proximity to the poly(A) signal (<30 nucleotides upstream of the poly(A) signal (90 and 61 transcripts upstream of the AATAAA or ATTAAA, respectively)) is depicted in Supplementary Tables I and II.
We next analyzed if other mRNAs that contain the nonameric USE core sequence can be identified. By using a sequence search algorithm that takes into consideration both the strand specificity and the typical length distribution for 3′ UTR motifs (peak >8-mers after exclusion of miRNAs target sites; Xie et al, 2005), we identified more than 1500 human transcripts that contain the nonameric USE core sequence (Figure 2B). Remarkably, a considerable amount of positive hits were identified in human transcripts with unusually long 3′ UTRs (>1000 nucleotides, not shown). Filtering hits according to the localization of the sequence element within transcripts showed a polar distribution toward their 3′ end (Figure 2B), with more than 500 transcripts that contained the USE motif in the ultimate (tenth) part (0.9–1.0) in a 5′ to 3′ direction. Considering the critical spatial relationship of this sequence element for its function, we identified more than 150 human transcripts that contained the USE core sequence in close proximity to the poly(A) signal (less than 30 nucleotides upstream of the AATAAA and ATTAAA, respectively; see Supplementary Tables I and II). Interestingly, with the exception of four transcripts, all of them contained the USE core sequence motif in their 3′ UTRs. These finding suggest, therefore, that USE-dependent 3′ end processing plays a more general role in many transcripts.
Identification of specific nuclear USE-binding proteins
To identify trans-acting factors that specifically interact with the F2 USE to promote 3′ end processing, we next performed electromobility shift assays (EMSA) and UV crosslinking experiments. We used a 32P-5′ end-labeled 21-mer RNA oligonucleotide probe including the 15 nucleotide USE core sequence that is both necessary and sufficient to promote 3′ end processing when inserted into a heterologous β-globin gene context (Danckwardt et al, 2004). Incubation of the USE probe with nuclear extract elicits a specific shift (Figure 3A, lanes 2–5 and 6–9). A 21-mer RNA oligonucleotide in which the USE core was replaced by a non-functional unrelated sequence fails to revert the observed shift (Unrel. comp cold, lane 10), whereas an RNA oligonucleotide containing the hFip1-binding site of the L3 mRNA (see above) competes for the formation of the shifted complex (Fip comp cold, lane 11), indicating that the hFip1-binding site interacts with at least one protein that is essential for the F2 USE gel shift. However, recombinant hFip1 failed to result in a shift of the USE oligonucleotide under physiological conditions, nor could it be identified as an interacting protein on the (entire) F2 3′ UTR by RNAse H protection analysis (not shown). No significant shift was observed when the USE probe was incubated with equal amounts of cytoplasmic extract (S100, lanes 12 and 13), indicating that at least one essential protein bound by the USE is nuclear.
Figure 3.
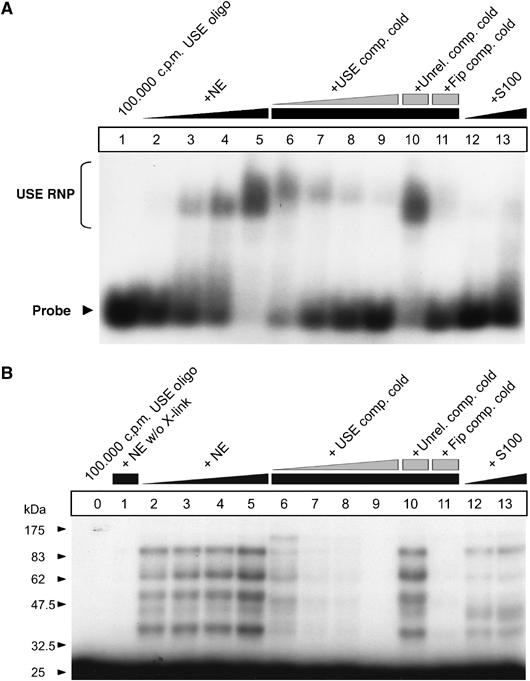
The F2 USE specifically interacts with nuclear proteins. (A) EMSA carried out with a F2 USE containing 21-mer RNA oligonucleotide (lane 1, free probe) after incubation in HeLa nuclear extract (NE, lanes 2–11) or cytoplasmatic extract (S100, lanes 12 and 13), respectively. Specificity of the RNA–protein interaction is shown by coincubation of an unlabeled F2 USE-containing 21-mer RNA oligonucleotide as cold competitor (lanes 6–9), of an unrelated competitor (lane 10) and a competitor, including the hFip1 binding site of the L3 RNA (lane 11). (B) UV crosslinking study carried out with the same USE-containing or competitor RNA oligonucleotides (lane 0, free probe) after incubation in HeLa nuclear extract (NE, lanes 1–11 (lane 1 without UV light exposure)) or cytoplasmatic extract (S100, lanes 12 and 13), respectively.
We next investigated the USE-binding proteins by UV crosslinking (Figure 3B). The USE-specific 21-mer RNA was specifically UV crosslinked to at least five proteins of ∼30–100 kDa. These crosslinks can be competed by cold USE and hFip1-binding site-specific 21-mers (lanes 6–9 and lane 11). Crosslinking studies with cytoplasmic extracts (lanes 12 and 13) showed that some of the USE-binding proteins also appear to be present in the cytoplasm, but the overall pattern of crosslinks is distinct from that generated with nuclear extracts. The affinity of the interaction between the USE and the crosslinking cytoplasmic proteins, however, does not appear to be sufficient to cause a shift in the EMSA. These results demonstrate that the F2 USE directly interacts with at least five different proteins that are predominantly located in the nuclear compartment. Furthermore, the functional significance of this interaction is highlighted by RNA–protein interaction studies using a template with a triple point mutation within the 15-nt USE core affecting the highly conserved nonamer (USEmut). This manipulation results in loss of protein binding (Supplementary Figure S2D and E), which highly correlates with loss of function of the USE (Supplementary Figure S2B, USE and USEmut; lanes 3 and 5).
Splicing factors and 3′ end processing proteins bind to the USE
We next aimed to identify the F2 USE-binding proteins by affinity purification followed by mass spectrometry. For this purpose, we first ascertained that the 3′biotin-TEG (triethylenglycol)-linker-modification used for immobilization of the RNA bait does not interfere with protein binding to the short 21-mer RNA oligonucleotides (Supplementary Figure S2A). As a specificity control, we used a template with a triple point mutation within the 15-nt USE core (USEmut), which results in similar loss of function as the replacement of the entire USE does (Supplementary Figure S2B, USE, USEmut and Unrel.; lanes 2, 3 and 5). RNA–protein interaction studies based on EMSA and UV crosslinking revealed that this loss of function correlates highly with loss of protein binding (Supplementary Figure S2D and E). The point mutated 21-mer USE sequence (USEmut) thus qualifies as an excellent specificity control for non-functional RNA–protein interactions during the affinity purification procedure. As an additional control for nonspecific RNA–protein interactions, an immobilized 21-mer RNA oligonucleotide with an unrelated sequence was used (Supplementary Figure S2C).
Affinity purification yielded several bands that were specific for the USE bait compared with the controls (Figure 4A, lanes 1–3). Comparison of the USE affinity purification-specific band pattern with the patterns of UV crosslinking experiments revealed that the size of some of the affinity-purified proteins corresponds to the size of the proteins identified by UV crosslinking; these proteins thus likely interact with the USE motif directly (Figure 4A, lanes 4 and 5, that is, PSF, p54nrb, U2AF65, hnRNPI, UFAF35). This analysis revealed that the F2 USE-binding proteins include factors known to be involved in 3′ end processing and, surprisingly, in splicing (Table I).
Figure 4.
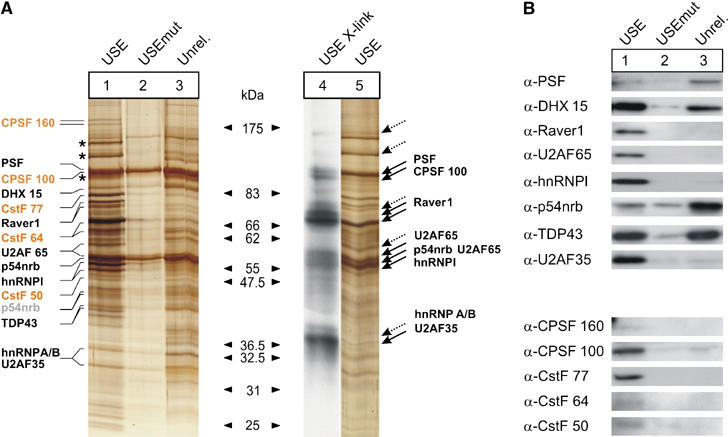
Affinity purification followed by mass spectroscopy identifies proteins that specifically interact with the F2 USE. (A) Silver-stained SDS–PAGE polyacrylamide gel of protein samples derived from affinity purification with immobilized 3′ biotinylated 21-mer RNA oligonucleotides with the F2 USE motif (USE, lanes 1 and 5), with a mutated F2 USE motif (USEmut, lane 2) or with an unrelated sequence context (Unrel., lane 3). Lanes 1–3 show protein samples eluted with up to 2000 mM NaCl (starting concentration 150 mM NaCl). Lane 4 shows the autoradiograph of a UV crosslink that highlights the size of direct interaction partners of the F2 USE (dotted arrows indicate putative direct USE-binding proteins that could not be unequivocally assigned by the mass spectrometry data) in comparison to the band pattern of directly and indirectly interacting proteins derived from affinity purification (lane 1). Silver-stained bands in lane 1 were cut out and subjected to mass spectrometry (protein names are only indicated at the respective size where the respective peptide score was maximal; canonical 3′ end processing factors are highlighted in yellow; p54nrb peptides of unexpected small size are highlighted in light gray; for mass spectrometry data, see also Table I). The analysis also revealed the presence of ATP citrate synthase, LRP 130, HSP 90- and tubulin (asterisks), which were judged to represent likely contaminants and were not included in Table I. (B) Immunoblots of eluates from affinity purifications for proteins identified by mass spectrometry (Table I). Lanes 1–3 correspond to samples as indicated in Figure 4A. Additional information on controls for unspecific RNA–protein interaction and equal loading is available in Supplementary Figure S2.
Table 1.
The F2 USE specifically interacts with splicing factors, 3′ end processing proteins and other RNA binding proteins
| Characteristics of USE-binding proteins | Accession number | Score max | Peptides | Immunoblot | |
|---|---|---|---|---|---|
| Splicing factors | |||||
| PSF/SFPQ | Splicing factor, proline and glutamine rich | P23246 | 547 | 15 | USE=Unrel. |
| p54nrb/NonO | Non-POU domain-containing octamer-binding protein | Q15233 | 414 | 15 | USE<Unrel. |
| hnRNPI/PTB | Polypyrimidine tract-binding protein 1 | P26599 | 262 | 8 | USE |
| DHX 15 | Putative pre-mRNA splicing factor RNA helicase (DEAH box protein 15) | O43143 | 190 | 7 | USE=Unrel. |
| TDP43 | TAR DNA-binding protein 43 | Q13148 | 161 | 3 | USE=Unrel. |
| DDX5 p68 | Probable RNA-dependent helicase p68 (DEAD/H box 5) | P17844 | 84 | 2 | Very weak |
| SF1 | Splicing factor 1 (zinc-finger protein 162) (transcription factor ZFM1) | Q15637 | 78 | 5 | Not detectable |
| U2AF65 | Splicing factor U2AF 65-kDa subunit | P26368 | 74 | 2 | USE |
| U2AF35 | Splicing factor U2AF 35-kDa subunit | Q01081 | 50 | 1 | USE |
| SF3B | Splicing factor 3B subunit 3 | Q15393 | 36 | 2 | Not determined |
| 3′ End processing | |||||
| CstF 50 | Cleavage stimulation factor, 50-kDa subunit | Q05048 | 182 | 8 | USE |
| CPSF 160 | Cleavage and polyadenylation specificity factor, 160-kDa subunit | Q10570 | 168 | 8 | USE |
| CstF 77 | Cleavage stimulation factor subunit 3 | gi∣4557495 | 134 | 6 | USE |
| CstF 64 | Cleavage stimulation factor, 64-kDa subunit | P33240 | 102 | 3 | USE |
| CPSF 100 | Cleavage and polyadenylation specificity factor, 100-kDa subunit | Q9P2I0 | 89 | 3 | USE |
| Other | |||||
| Similar to Raver1? | Unknown (protein for IMAGE:5113697) | gi∣22902182 | 91 | 2 | USE |
| hnRNP C1/C2 | Heterogeneous nuclear ribonucleoproteins C1/C2 | P07910 | 83 | 2 | USE=USEmut=Unrel. |
| hnRNP A2/B1 | Heterogeneous nuclear ribonucleoproteins A2/B1 | P22626 | 50 | 2 | Not detectable |
| Nuclear DNA helicase II | ATP-dependent RNA helicase A | Q08211 | 43 | 2 | Not determined |
| USE-binding proteins identified by affinity purification and subsequent mass spectrometry of USE-specific bands shown in Figure 4A. The analysis also revealed the presence of ATP citrate synthase, LRP 130, HSP 90-alpha and tubulin (Figure 4A, lane 1, black asterisks), which were judged to represent likely contaminants and were not included in the table. Data interpretation was performed with the MASCOT V2.1 search engine for mass spectrometric data (see Materials and methods section). The maximal scores obtained for each individual protein and the number of aligned peptides are shown. The immunoblot data shown in Figure 4B are summarized. | |||||
We next confirmed the identity of the proteins found by mass spectrometry. Immunoblots of eluates derived from affinity purification with the USE bait and the respective controls (Figure 4B; Table I) show that six out of 13 proteins identified by mass spectrometry were specifically present in the eluates of the USE columns. For three proteins (CPSF 160, CstF 64 and CstF 50) the signal is weak, which may indicate the low abundance of these proteins in the eluates or reflect a lower affinity of the antibodies. Four of the proteins (PSF, DHX 15, p54nrb, TDP43) are also present in the eluates of the USEunrel and/or USEmut controls, which indicates nonspecific RNA-binding and/or indirect RNA-interaction properties via yet unidentified proteins bound to the RNA baits in lanes 1, 2 and 3. Taken together, these results reveal that the USE interacts specifically with two predominant classes of proteins with known roles in splicing and 3′ end processing (Table I).
RNAi demonstrates a role of splicing factors in USE-dependent F2 3′ end processing
We next analyzed the functional importance of the identified proteins on USE-dependent 3′ end processing. We first established the siRNA-mediated, target-specific depletion of the splicing factors shown in the upper panel of Figure 4B, and subsequently performed an in vivo 3′ end formation assay by transfecting suitable constructs that contain a tandem array of 3′ end formation signals, with and without an F2 USE within the 5′ site (Figure 5B).
Figure 5.
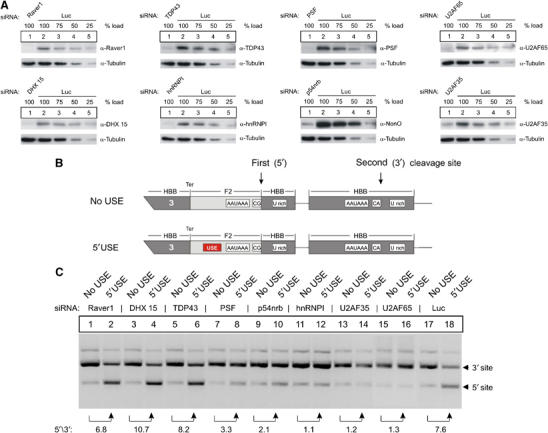
USE-binding splicing factors specifically promote the expression of USE-containing mRNAs. (A) Representative immunoblots of protein lysates obtained from cells treated with siRNAs directed against USE-binding proteins (lane 1, each panel). Lanes 2–5 in each panel show a serial reduction of the load of protein lysates obtained from cells treated with a luciferase siRNAs as control for nonspecific RNAi effects and for quantification of the RNAi efficiency. (B) Schematic representation of the HBB-F2 hybrid gene construct with a tandem array of two 3′ end formation signals. The F2 USE of the 5′ located 3′ end processing signal was either maintained (‘5′USE' construct) or completely replaced by an unrelated nucleotide sequence (‘no USE' construct). (C) In vivo assay carried out by transient transfection of a HBB-F2 hybrid gene construct, with or without a USE (B), after depletion of USE-binding proteins (A). Each number represents the fold difference of the mRNA ratio processed at the 5′ or the 3′ site (5′/3′) relative to the respective ratio in the odd numbered lanes (representative figure of three independent experiments).
Functional RNAi resulted in efficient protein depletion of each target protein to below 25% of control levels (Figure 5A, each panel, lanes 1 and 2–5). Importantly, the protein abundance of the other seven proteins was unaffected by the target-specific depletions (not shown).
Expectedly, the analysis of transfected F2 mRNA reporter abundance revealed a significant upmodulation of 3′ end processing at the 5′ site in the presence of a functional F2 USE of ∼7.6-fold, when compared with the respective mRNA counterpart derived from constructs without an USE (Figure 5C, lanes 17 and 18). This USE-dependent stimulatory effect on 3′ end processing was slightly reduced in cells upon depletion of PSF and p54nrb (to 3.3- and 2.1-fold, lanes 7 and 8, and 9 and 10), and almost completely abolished after depletion of the splicing factors hnRNPI, U2AF35 and U2AF65 (lanes 11 and 12, 13 and 14, 15 and 16). In contrast, depletion of Raver1, DHX 15 and TDP did not reduce USE-mediated 3′ end processing (lanes 1–6). These data indicate that USE function on F2 3′ end processing critically depends on the splicing factors hnRNPI, U2AF35 and U2AF65. It should be noted that we did not observe a significant proportion of unspliced reporter mRNAs upon depletion of these splicing factors in the PAT assay. Efficient depletion of PSF, U2AF35 and U2AF65, however, strongly affected cell morphology and plasmid transfection efficiencies.
hnRNPI and U2AF65 interact with the F2 USE directly
To identify the functionally relevant splicing factors that directly interact with the F2 mRNA in vivo, we next performed an RNP immunoprecipitation (IP) assay and monitored the endogenous F2 mRNA contained in the IPs. For this purpose, IPs were carried out with cell lysates after UV or formaldehyde (FA) crosslinking to specifically assay for direct RNA–protein interactions (Niranjanakumari et al, 2002), with antibodies directed against hnRNPI, U2AF35, p54nrb, U2AF65 and PSF. Nonspecific association of mRNAs with IP reagents was controlled by parallel incubations with anti-mouse antibodies (Figure 6A).
Figure 6.
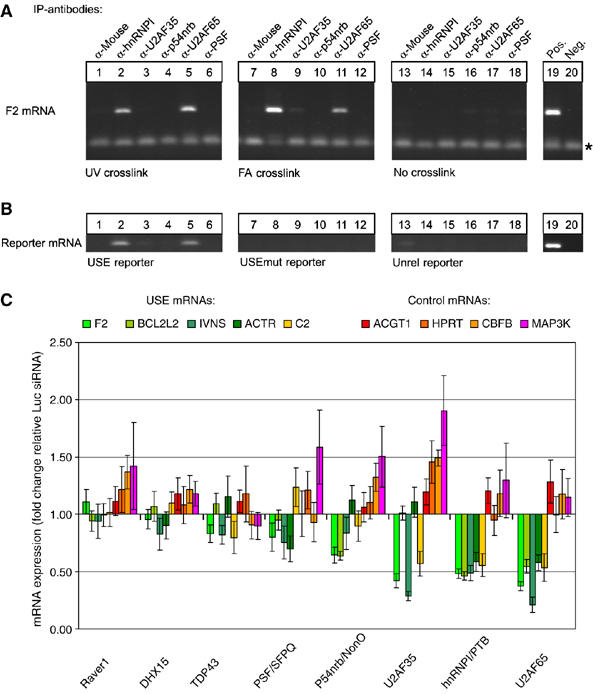
Depletion of the USE-binding protein hnRNPI and U2AF65 specifically reduces the mRNA expression of USE-containing endogenous transcripts. (A) In vivo RNA–protein interaction assay based on mRNA analyses in RNP IPs carried out with antibodies as indicated. IPs were carried out with HUH-7 cell lysates after UV crosslinking (lanes 1–6), FA crosslinking (lanes 7–12) or without crosslinking (lanes 13–18; for further information see Materials and methods). The F2 mRNA was analyzed by RT–PCR (primer dimers are indicated by asterisks); lanes 19 and 20 represent positive and negative controls, respectively. (B) In vivo RNA–protein interaction assay based on reporter mRNA analysis in IP samples derived from FA crosslinked HeLa cells transfected with reporter constructs containing either a F2 USE (USE reporter), a mutated F2 USE (USEmut reporter) or an unrelated sequence context (Unrel. reporter, see Supplementary Figure S2). (C) Endogenous F2, BCL2L2, IVNS, ACTR and C2 mRNA, and ACTG1, HPRT, CBFB and MAP3K mRNA abundance of HUH-7 cells after RNAi directed against USE-binding proteins as indicated (x-axis). The fold change of mRNA expression upon candidate siRNA treatment (y-axis) is quantified relative to the mRNA expression of cells treated with luciferase siRNAs after normalization against endogenous ACTB mRNA. Each bar represents values of at least five independent RNAi experiments determined by quantitative RT–PCR in duplicates (±s.e.).
In IPs carried out with antibodies directed against hnRNPI and U2AF65, the endogenous F2 mRNA was specifically enriched in samples derived from cells after UV and FA crosslinking (Figure 6A, lanes 2 and 5, 8 and 11), whereas the F2 mRNA could not be detected in IPs with other antibodies or in IPs with cell lysates that were not crosslinked, respectively. These results thus indicate that hnRNPI and U2AF65 interact with the F2 mRNA directly. In contrast, U2AF35, p54nrb and PSF either do not directly interact with the F2 mRNA or a direct interaction is masked or otherwise undetectable.
We next investigated whether hnRNPI and U2AF65 interact with the F2 mRNA in a USE-dependent manner (Figure 6B). For this purpose, we extended the in vivo RNA–protein interaction study to cells transfected with reporter constructs either with or without a functional F2 USE (compare Supplementary Figure S2B, USE, USEmut and Unrel.), followed by assaying the FA-crosslinked reporter-specific mRNA in the IP material. In order to compensate for the ∼5-fold difference of mRNA expression that depends on the functionality/presence of the USE (Figure 1B and data not shown), the amount of transfected reporter plasmid DNA was adjusted accordingly.
As shown in the left panel in Figure 6B, the USE-containing reporter mRNA was specifically detected in IPs carried out with antibodies directed against hnRNPI and U2AF65 (lanes 2 and 5). In contrast, reporter mRNAs with a non-functional USE (USEmut, middle panel) or without a USE (Unrel., right panel) were not detectable in IPs carried out with lysates of cells transfected with the USEmut or Unrel. constructs, respectively. The intact pyrimidine-rich F2 USE thus represents a direct binding site for hnRNPI and U2AF65 (Singh et al, 1995).
RNAi demonstrates a role of splicing factors in USE-dependent mRNA expression of several endogenous transcripts
We next analyzed the functional importance of the identified proteins in USE-dependent mRNA expression of endogenous transcripts. For this purpose, we first established the siRNA-mediated, target-specific depletion of the splicing factors in HUH-7 cells and monitored endogenous mRNA abundance by RT–PCR (Figure 6C). The quantification of endogenous F2 mRNA abundance revealed a significant down-modulation to approximately 80% upon depletion of TDP43 and PSF. The quantitatively most profound reduction (to below 60%) resulted from depletion of the splicing factors p54nrb, U2AF35, hnRNPI and U2AF65 (Figure 6C, quantified after normalization against the ACTB mRNA abundance, which lacks the nonameric USE core sequence motif). Finally, we analyzed whether the functional effects on F2 mRNA abundance could be extended to other USE-containing mRNAs. For this analysis, we selected the USE core sequence-containing BCL2L2, IVNS1ABP and ACTR3B mRNAs (see Supplementary Tables I and II), and the C2 mRNA that has previously been shown to be processed in an USE- and hnRNPI-dependent manner (Moreira et al, 1998). Whereas depletion of U2AF35 resulted in a down-modulation of the F2, IVNS1ABP and C2 mRNAs, depletion of hnRNPI and U2AF65 strikingly affected all tested USE-containing mRNAs (Figure 6C, compare green and yellow bars).
In order to demonstrate that the functional effects of RNAi of the USE-binding proteins was specific for the USE core sequence-containing genes, we also monitored the expression of actin (ACTG1), the hypoxanthine guanine phosphoribosyltransferase 1 (HPRT1) and the polyomavirus enhancer-binding protein 2 (CBFB) mRNAs as representative examples of spliced mRNAs with a conventional 3′ end formation signal. Furthermore, we analyzed the expression of the mitogen-activated protein kinase kinase kinase 1 (MAP3K1) that contains the nonameric USE core sequence within the ORF far upstream of a potential downstream poly(A) signal: The quantification of the ACTG1, HPRT1, CBFB and MAP3K1 mRNAs shows that these controls are not down-modulated upon depletion of the USE-binding proteins (Figure 6C, red bars). Thus, the depletion of the USE-binding splicing factors hnRNPI, U2AF65 and—in part also U2AF35—reduces the expression of the nonameric USE core sequence-containing F2, BCL2L2, IVNS1ABP and ACTR3B mRNAs, whereas the USE core sequence-containing MAP3K1 mRNA that lacks a downstream poly(A) signal in close proximity was unaffected by these manipulations.
These results thus recapitulate the positional effect of USE function (Figure 1) and highlight the specific stimulatory effect of these splicing factors on USE-mediated mRNA expression for several endogenous transcripts.
Discussion
USE-dependent 3′ end processing has been experimentally documented for a number of genes that are involved in important physiological processes such as hemostasis (prothrombin; Danckwardt et al, 2004), innate immunity (complement C2; Moreira et al, 1998), inflammation (cyclooxygenase-2; Hall-Pogar et al, 2005) and in the maintenance of cell structure (lamin B2; Brackenridge and Proudfoot, 2000) and collagen (Natalizio et al, 2002). Biocomputational analyses predict that USE-dependent 3′ end processing may be quite common among cellular mRNAs (Legendre and Gautheret, 2003; Hu et al, 2005). USEs thus represent one of the important regulatory sequence elements contained in 3′ UTRs.
Sequence comparisons of the entire USE did not reveal a clear consensus in other genes, including those with functionally defined USEs. However, when recently identified conserved 3′ UTR sequence motifs (Hu et al, 2005; Xie et al, 2005) were considered, it became apparent that the F2 USE includes two overlapping motifs (UAUUUUU and UUUUGU) belonging to the top 10 out of 106 highly conserved 3′ UTR motifs with a cross-species conservation rate of 30 and 24%, respectively (Xie et al, 2005). Of note, the conservation rate of the highly conserved polyadenylation signal is 46%, whereas control random motifs show a conservation rate of only 10%. Importantly, the UAUUUUU motif is destroyed in the non-functional USE motif (USEmut), which highly correlates with loss of protein binding (Supplementary Figure S2). However, the presence of the UAUUUUU motif alone yielded only moderate 3′ end processing efficiencies, whereas full activity was observed in the presence of both elements (Figure 1). This indicates that the F2 USE has evolved as an optimal sequence context consisting of a composite of two highly conserved 3′ UTR motifs.
Interestingly, the complement C2 mRNA contains a suboptimal match of another conserved top 10 hexamers (UGUUUU; Hu et al, 2005), which can also be found at the 3′ end of the F2 USE. The biocomputational predictions and the functional analyses reported here thus implicate that different, highly conserved U-rich (Legendre and Gautheret, 2003) 3′ UTR sequence motifs can function as USEs and enhance 3′ end processing in a position-dependent manner, showing highest activities when located in a region recently designated as core upstream element (CUE; Hu et al, 2005). In that respect, the sequence element identified here differs from tetrameric sequence elements that have recently been identified to account for 3′ end processing at the non-canonical poly(A) site of the PAPOLA and PAPOLG mRNA by recruiting CFIm (Venkataraman et al, 2005).
Different steps of gene expression pathways are thought to be coupled (Hirose and Manley, 2000; Proudfoot et al, 2002). In this context, the important finding reported here is that two different classes of RNA processing proteins bind to the F2 USE (Table I) and thus provide further evidence for an extensive molecular network that effectively coordinates gene expression (Maniatis and Reed, 2002). In line with the notion that processing factors involved in pre-mRNA splicing and 3′ end formation can influence each other positively (Niwa et al, 1990; Wassarman and Steitz, 1993; Gunderson et al, 1994; Lutz et al, 1996; Vagner et al, 2000; Li et al, 2001; McCracken et al, 2002, 2003; Millevoi et al, 2002, 2006; Awasthi and Alwine, 2003; Kyburz et al, 2006), we identify here that the splicing factors hnRNPI, U2AF65 and, in part, U2AF35, represent components of a functionally relevant USE-dependent RNP. The requirement of these splicing factors for 3′ end processing seemed to be USE specific, because the depletion of these factors did not influence the expression of those tested mRNAs that do not contain the nonameric USE core sequence motif. Furthermore, the USE stimulates polyadenylation (Supplementary Figure S3) and its function critically depends on a tight spatial relationship to the canonical 3′ end formation signals. Although it is formally possible that the USE and the 3′ terminal splice site may potentiate polyadenylation through common means, our findings indicate that the functional effect of the F2 USE does not result from a nonspecific coupling of splicing or 3′ end terminal exon definition and 3′ end processing.
Previously, p54nrb and PSF have been shown to interact with the carboxy-terminal domain (CTD) of RNA polymerase II to link transcriptional activities with splicing (Emili et al, 2002; Kameoka et al, 2004), whereas 3′ end processing has been suggested to be more indirectly affected by these proteins (Rosonina et al, 2005). p54nrb has recently been shown to be a component of the snRNP-free U1A (SF-A) complex that appears to directly promote pre-mRNA cleavage (Liang and Lutz, 2006). Moreover, hnRNPI, U2AF35, U2AF65 and PSF have previously been identified in CF II preparations (de Vries et al, 2000). With respect to our findings presented here, it is particularly interesting that U2AF65 has previously been shown to directly interact with the CTD of the PAP (Vagner et al, 2000) and cleavage factor CF I (Millevoi et al, 2006). U2AF65 has also been reported to stimulate the 3′ end cleavage reaction when tethered more than 150 nucleotides upstream of the AAUAAA hexanucleotide (Millevoi et al, 2002). However, this finding is unlikely to be related to the USE effect, because we show that the USE is virtually non-functional when shifted more than 50 nucleotides upstream of the AAUAAA (Figure 1).
The second class of proteins interacting with the USE are canonical 3′ end processing factors (Table I). Therefore, the USE-dependent RNP complex may promote 3′ end processing by serving as an additional anchor for the (canonical) 3′ end processing machinery or by stabilizing the RNA interaction of both CPSF and CstF components (Brackenridge and Proudfoot, 2000). Importantly, cooperative binding has also been implicated to account for CstF binding to the DSE, which greatly enhances the affinity of CPSF to the AAUAAA hexamer and vice versa (Colgan and Manley, 1997; Zhao et al, 1999). We therefore propose the existence of a novel and complex RNP, most likely consisting of at least two components of two distinct complexes (U2AF65 of the heterodimeric complex U2AF35/U2AF65, and hnRNPI known to interact with p54nrb and PSF) that cooperatively promote 3′ end processing in a USE-dependent manner (Figure 7). However, it should be noted that USE-dependent 3′ end processing of some of the USE core sequence-containing transcripts analyzed here seems to require different cofactors (Figure 6C), opening the perspective of transcript specific regulation of 3′ end processing. Despite previous reports implying a more general function of p54nrb and PSF in 3′ end processing (Lutz et al, 1998; Liang and Lutz, 2006), these splicing factors are likely dispensable for a USE-specific function in 3′ end processing (Figures 4B and 6C).
Figure 7.
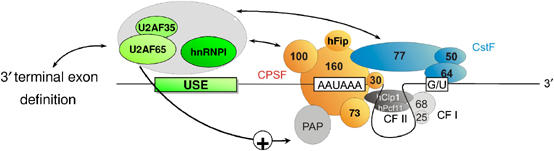
Model for USE-dependent RNA processing at (non-canonical) 3′ end formation signals. 3′ End processing of USE-containing mRNAs is proposed to depend on the formation of USE-dependent RNP complexes that participate in an extensive molecular network coordinating gene expression. USE-binding splicing factors are depicted as positive effectors (green complex), potentially involving a direct stimulation of PAP via U2AF65. The USE-binding protein complex (Table I) is indicated by gray shading. The USE-binding proteins are proposed to bridge and promote splicing and 3′ end processing. Specifically, the 65 kDa subunit of the U2AF dimer may link 3′terminal exon definition with efficient 3′ end processing. On the other hand, the USE-dependent RNP complexes establish the link to the 3′ end processing machinery by interaction with CPSF 100 and CstF 77. The polypyrimidine tract-binding protein PTB/hnRNPI and U2AF65 likely represent direct USE-interacting proteins and may thus nucleate the USE-dependent RNP complexes for efficient stimulation of 3′ end formation.
Taken together, the data presented here functionally link the splicing and 3′ end processing machineries in a USE-dependent manner. It will be interesting to dissect the different cofactor requirements and to analyze whether this novel mechanism is subject to specific regulatory steps that may respond to external stimuli.
Materials and methods
Detailed information on Materials and methods is available in Supplementary data.
Supplementary Material
Supplementary Figures
Supplementary Figure Legends, Tables and Information
Acknowledgments
We thank Margit Happich for excellent technical assistance, Pavel Ivanov, Stephen Breit, Marcelo Viegas and other members of the Molecular Medicine Partnership Unit for advice and helpful discussions. We also acknowledge Brigitte Jockusch for kindly providing the anti-Raver1 antibody. This work was funded by grants from the Deutsche Forschungsgemeinschaft (KU563/7-1 and KU563/8-1 to AEK), the Fritz-Thyssen Stiftung (grant 1999-1076 to AEK) and by the ‘Young Investigator Award' fellowship from the University of Heidelberg (to SD). This work was supported by the DFG Forschergruppe (FOR 426): Complex RNA–protein interactions in the maturation and function of eukaryotic mRNA. Work in the laboratory of WK was supported by the University of Basel and the Swiss National Science Foundation.
References
- Awasthi S, Alwine JC (2003) Association of polyadenylation cleavage factor I with U1 snRNP. RNA 9: 1400–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabino SM, Keller W (1999) Last but not least: regulated poly(A) tail formation. Cell 99: 9–11 [DOI] [PubMed] [Google Scholar]
- Brackenridge S, Proudfoot NJ (2000) Recruitment of a basal polyadenylation factor by the upstream sequence element of the human lamin B2 polyadenylation signal. Mol Cell Biol 20: 2660–2669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuvpilo S, Zimmer M, Kerstan A, Glockner J, Avots A, Escher C, Fischer C, Inashkina I, Jankevics E, Berberich-Siebelt F, Schmitt E, Serfling E (1999) Alternative polyadenylation events contribute to the induction of NF-ATc in effector T cells. Immunity 10: 261–269 [DOI] [PubMed] [Google Scholar]
- Colgan DF, Manley JL (1997) Mechanism and regulation of mRNA polyadenylation. Genes Dev 11: 2755–2766 [DOI] [PubMed] [Google Scholar]
- Danckwardt S, Gehring NH, Neu-Yilik G, Hundsdoerfer P, Pforsich M, Frede U, Hentze MW, Kulozik AE (2004) The prothrombin 3′end formation signal reveals a unique architecture that is sensitive to thrombophilic gain-of-function mutations. Blood 104: 428–435 [DOI] [PubMed] [Google Scholar]
- Danckwardt S, Hartmann K, Gehring NH, Hentze MW, Kulozik AE (2006a) 3′ End processing of the prothrombin mRNA in thrombophilia. Acta Haematol 115: 192–197 [DOI] [PubMed] [Google Scholar]
- Danckwardt S, Hartmann K, Katz B, Hentze M, Levy Y, Eichele R, Deutsch V, Kulozik A, Ben-Tal O (2006b) The prothrombin 20209 C>T mutation in Jewish–Moroccan Caucasians: molecular analysis of gain-of-function of 3′end processing. J Thromb Haemost 4: 1078–1085 [DOI] [PubMed] [Google Scholar]
- de Vries H, Ruegsegger U, Hubner W, Friedlein A, Langen H, Keller W (2000) Human pre-mRNA cleavage factor II(m) contains homologs of yeast proteins and bridges two other cleavage factors. EMBO J 19: 5895–5904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z, Yang XC, Marzluff WF (2005) The polyadenylation factor CPSF 73 is involved in histone-pre-mRNA processing. Cell 123: 37–48 [DOI] [PubMed] [Google Scholar]
- Edwalds-Gilbert G, Veraldi KL, Milcarek C (1997) Alternative poly(A) site selection in complex transcription units: means to an end? Nucleic Acids Res 25: 2547–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emili A, Shales M, McCracken S, Xie W, Tucker PW, Kobayashi R, Blencowe BJ, Ingles CJ (2002) Splicing and transcription-associated proteins PSF and p54nrb/nonO bind to the RNA polymerase II CTD. RNA 8: 1102–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring NH, Frede U, Neu-Yilik G, Hundsdoerfer P, Vetter B, Hentze MW, Kulozik AE (2001) Increased efficiency of mRNA 3′ end formation: a new genetic mechanism contributing to hereditary thrombophilia. Nat Genet 28: 389–392 [DOI] [PubMed] [Google Scholar]
- Gilmartin GM (2005) Eukaryotic mRNA 3′ processing: a common means to different ends. Genes Dev 19: 2517–2521 [DOI] [PubMed] [Google Scholar]
- Gilmartin GM, Fleming ES, Oetjen J, Graveley BR (1995) CPSF recognition of an HIV-1 mRNA 3′-processing enhancer: multiple sequence contacts involved in poly(A) site definition. Genes Dev 9: 72–83 [DOI] [PubMed] [Google Scholar]
- Graveley BR, Fleming ES, Gilmartin GM (1996) RNA structure is a critical determinant of poly(A) site recognition by cleavage and polyadenylation specificity factor. Mol Cell Biol 16: 4942–4951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunderson SI, Beyer K, Martin G, Keller W, Boelens WC, Mattaj LW (1994) The human U1A snRNP protein regulates polyadenylation via a direct interaction with poly(A) polymerase. Cell 76: 531–541 [DOI] [PubMed] [Google Scholar]
- Hall-Pogar T, Zhang H, Tian B, Lutz CS (2005) Alternative polyadenylation of cyclooxygenase-2. Nucleic Acids Res 33: 2565–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose Y, Manley JL (2000) RNA polymerase II and the integration of nuclear events. Genes Dev 14: 1415–1429 [PubMed] [Google Scholar]
- Hu J, Lutz CS, Wilusz J, Tian B (2005) Bioinformatic identification of candidate cis-regulatory elements involved in human mRNA polyadenylation. RNA 11: 1485–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kameoka S, Duque P, Konarska MM (2004) p54(nrb) associates with the 5′ splice site within large transcription/splicing complexes. EMBO J 23: 1782–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann I, Martin G, Friedlein A, Langen H, Keller W (2004) Human Fip1 is a subunit of CPSF that binds to U-rich RNA elements and stimulates poly(A) polymerase. EMBO J 23: 616–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller W, Minvielle-Sebastia L (1997) A comparison of mammalian and yeast pre-mRNA 3′-end processing. Curr Opin Cell Biol 9: 329–336 [DOI] [PubMed] [Google Scholar]
- Kyburz A, Friedlein A, Langen H, Keller W (2006) Direct interactions between subunits of CPSF and the U2 snRNP contribute to the coupling of pre-mRNA 3′ end processing and splicing. Mol Cell 23: 195–205 [DOI] [PubMed] [Google Scholar]
- Legendre M, Gautheret D (2003) Sequence determinants in human polyadenylation site selection. BMC Genomics 4: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Chen ZY, Wang W, Baker CC, Krug RM (2001) The 3′-end-processing factor CPSF is required for the splicing of single-intron pre-mRNAs in vivo. RNA 7: 920–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S, Lutz CS (2006) p54nrb is a component of the snRNP-free U1A (SF-A) complex that promotes pre-mRNA cleavage during polyadenylation. RNA 12: 111–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz CS, Cooke C, O'Connor JP, Kobayashi R, Alwine JC (1998) The snRNP-free U1A (SF-A) complex(es): identification of the largest subunit as PSF, the polypyrimidine-tract binding protein-associated splicing factor. RNA 4: 1493–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz CS, Murthy KG, Schek N, O'Connor JP, Manley JL, Alwine JC (1996) Interaction between the U1 snRNP-A protein and the 160-kDa subunit of cleavage-polyadenylation specificity factor increases polyadenylation efficiency in vitro. Genes Dev 10: 325–337 [DOI] [PubMed] [Google Scholar]
- Mandel C, Kaneko S, Zhang H, Gebauer D, Vethantham V, Manley J, Tong L (2006) Polyadenylation factor CPSF 73 is the pre-mRNA 3′-end-processing endonuclease. Nature 444: 953–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Reed R (2002) An extensive network of coupling among gene expression machines. Nature 416: 499–506 [DOI] [PubMed] [Google Scholar]
- McCracken S, Lambermon M, Blencowe BJ (2002) SRm160 splicing coactivator promotes transcript 3′-end cleavage. Mol Cell Biol 22: 148–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S, Longman D, Johnstone IL, Caceres JF, Blencowe BJ (2003) An evolutionarily conserved role for SRm160 in 3′-end processing that functions independently of exon junction complex formation. J Biol Chem 278: 44153–44160 [DOI] [PubMed] [Google Scholar]
- Millevoi S, Geraghty F, Idowu B, Tam JL, Antoniou M, Vagner S (2002) A novel function for the U2AF 65 splicing factor in promoting pre-mRNA 3′-end processing. EMBO Rep 3: 869–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millevoi S, Loulergue C, Dettwiler S, Karaa SZ, Keller W, Antoniou M, Vagner S (2006) An interaction between U2AF 65 and CF I(m) links the splicing and 3′ end processing machineries. EMBO J 25: 4854–4864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira A, Takagaki Y, Brackenridge S, Wollerton M, Manley JL, Proudfoot NJ (1998) The upstream sequence element of the C2 complement poly(A) signal activates mRNA 3′ end formation by two distinct mechanisms. Genes Dev 12: 2522–2534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natalizio BJ, Muniz LC, Arhin GK, Wilusz J, Lutz CS (2002) Upstream elements present in the 3′-untranslated region of collagen genes influence the processing efficiency of overlapping polyadenylation signals. J Biol Chem 277: 42733–42740 [DOI] [PubMed] [Google Scholar]
- Niranjanakumari S, Lasda E, Brazas R, Garcia-Blanco MA (2002) Reversible cross-linking combined with immunoprecipitation to study RNA–protein interactions in vivo. Methods 26: 182–190 [DOI] [PubMed] [Google Scholar]
- Niwa M, Rose SD, Berget SM (1990) In vitro polyadenylation is stimulated by the presence of an upstream intron. Genes Dev 4: 1552–1559 [DOI] [PubMed] [Google Scholar]
- Proudfoot NJ, Furger A, Dye MJ (2002) Integrating mRNA Processing with transcription. Cell 108: 501–512 [DOI] [PubMed] [Google Scholar]
- Rosonina E, Ip JYY, Calarco JA, Bakowski MA, Emili A, McCracken S, Tucker P, Ingles CJ, Blencowe BJ (2005) Role for PSF in mediating transcriptional activator-dependent stimulation of pre-mRNA processing in vivo. Mol Cell Biol 25: 6734–6746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K, Calvo O, Manley JL (2004) Evidence that polyadenylation factor CPSF 73 is the mRNA 3′ processing endonuclease. RNA 10: 565–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Valcarcel J, Green MR (1995) Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science 268: 1173–1176 [DOI] [PubMed] [Google Scholar]
- Takagaki Y, Seipelt RL, Peterson ML, Manley JL (1996) The polyadenylation factor CstF 64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell 87:941–952 [DOI] [PubMed] [Google Scholar]
- Vagner S, Vagner C, Mattaj IW (2000) The carboxyl terminus of vertebrate poly(A) polymerase interacts with U2AF 65 to couple 3′-end processing and splicing. Genes Dev 14: 403–413 [PMC free article] [PubMed] [Google Scholar]
- Venkataraman K, Brown KM, Gilmartin GM (2005) Analysis of a noncanonical poly(A) site reveals a tripartite mechanism for vertebrate poly(A) site recognition. Genes Dev 19: 1315–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Haar T, Gross JD, Wagner G, McCarthy JE (2004) The mRNA cap-binding protein eIF4E in post-transcriptional gene expression. Nat Struct Mol Biol 11: 503–511 [DOI] [PubMed] [Google Scholar]
- Wassarman KM, Steitz JA (1993) Association with terminal exons in pre-mRNAs: a new role for the U1 snRNP? Genes Dev 7: 647–659 [DOI] [PubMed] [Google Scholar]
- Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M (2005) Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature 434: 338–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Marr TG (2005) Computational analysis of 3′-ends of ESTs shows four classes of alternative polyadenylation in human, mouse, and rat. Genome Res 15: 369–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Hyman L, Moore C (1999) Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev 63: 405–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Manley JL (1996) Complex alternative RNA processing generates an unexpected diversity of poly(A) polymerase isoforms. Mol Cell Biol 16: 2378–2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures
Supplementary Figure Legends, Tables and Information


