Abstract
The evolutionarily conserved Par-3/Par-6/aPKC complex is essential for the establishment and maintenance of polarity of a wide range of cells. Both Par-3 and Par-6 are PDZ domain containing scaffold proteins capable of binding to polarity regulatory proteins. In addition to three PDZ domains, Par-3 also contains a conserved N-terminal oligomerization domain (NTD) that is essential for proper subapical membrane localization and consequently the functions of Par-3. The molecular basis of NTD-mediated Par-3 membrane localization is poorly understood. Here, we describe the structure of a monomeric form of the Par-3 NTD. Unexpectedly, the domain adopts a PB1-like fold with both type-I and type-II structural features. The Par-3 NTD oligomerizes into helical filaments via front-to-back interactions. We further demonstrate that the NTD-mediated membrane localization of Par-3 in MDCK cells is solely attributed to its oligomerization capacity. The data presented in this study suggest that the Par-3 NTD is likely to facilitate the assembly of higher-order Par-3/Par-6/aPKC complex with increased avidities in targeting the complex to the subapical membrane domain and in binding to other polarity-regulating proteins.
Keywords: cell polarity, Par-3, Par complex, PB1 domain, scaffold protein
Introduction
Polarity is a hallmark for many cell types in multicellular eukaryotes. Establishment of cell polarity is a fundamental process for asymmetric cell divisions at the early stages of development. Acquisition of polarity during maturation and maintenance of such polarity after maturation of neurons ensure that each neuronal cell contains multiple dendrites and single long axon. Establishment of apical–basal polarity is absolutely required for proper functioning of epithelial cells. Active segregation of molecular components to different subcellular domains directly orchestrates cell polarity. Genetic and molecular studies have identified distinct groups of genes that function coordinately to establish cell polarity (for reviews see (Kemphues, 2000; Macara, 2004; Suzuki and Ohno, 2006)). The products of these genes are multidomain proteins and enzymes and they often interact with each other to form large protein complexes that mediate processes including assembly of cell junctions, organization of signal transduction complexes, regulation of cytoskeletal dynamics and asymmetric trafficking of cell fate determinants.
An evolutionarily conserved tripartite protein complex composed of Par-3, Par-6 and atypical protein kinase C (aPKC) has been shown to be a central player in both establishment and maintenance of cell polarity in a number of distinct biological contexts (e.g. zygotes of Caenorhabditis elegans, neuroblasts of Drosophila, neurons and epithelia of mammals). The genes encoding Par-3 and Par-6 were originally identified in the search for genes that are required to establish anterior/posterior polarity of C. elegans zygotes (Kemphues et al, 1988; Etemad-Moghadam et al, 1995; Watts et al, 1996). Subsequently, it was shown that Par-3 and Par-6 function together with aPKC to establish embryo polarity in C. elegans (Tabuse et al, 1998). In Drosophila, the orthologous proteins control both the fate of neuroblasts and epithelial cell polarity (Wodarz et al, 1999; Huynh et al, 2001; Benton and St Johnston, 2003b; Hutterer et al, 2004). The mammalian counterparts of Par-3, Par-6 and aPKC have been implicated in the establishment of epithelial cell polarity and both axonal and dendritic development of neurons (Lin et al, 2000; Hurd et al, 2003; Plant et al, 2003; Shi et al, 2003; Chen and Macara, 2005; Nishimura et al, 2005; Zhang and Macara, 2006).
A central feature of both Par-3 and Par-6 is that both proteins are multimodular scaffold proteins capable of binding to a diverse range of polarity-regulating proteins (Macara, 2004; Margolis and Borg, 2005; Suzuki and Ohno, 2006). Par-6 contains an N-terminal PB1 domain, a C-terminal PDZ domain and a semi-CRIB motif immediately preceding the PDZ domain (see Figure 6B for a schematic diagram). Par-3 contains a conserved N-terminal domain (hereafter referred to as NTD), three central PDZ domains and a C-terminal aPKC-binding domain. The PB1 domain of Par-6 forms a heterodimer with the PB1 domain of aPKC (Hirano et al, 2005). The PDZ domain of Par-6 can bind to the first PDZ domain of Par-3. The semi-CRIB motif of Par-6 specifically binds to the GTP-bound form of Cdc42. The binding of Cdc42 to Par-6 regulates dissociation of Par-3 from the Par-6/aPKC complex (Joberty et al, 2000; Lin et al, 2000), hence Par-6 acts as a direct downstream target of Cdc42. Recently, it was shown that the conserved NTD of Par-3 can self-associate (Benton and St Johnston, 2003a; Mizuno et al, 2003). The NTD-mediated self-association of Par-3 provides a molecular basis for further oligomerization of the Par-3/Par-6/aPKC complex into a higher molecular assembly (Figure 6B). This large (Par-3/Par-6/aPKC)n complex assembly may serve as a platform for organizing other polarity-regulating proteins such as the Lgl/Scribble/DLG and Pals1/Crumbs/Patj complexes (Hurd et al, 2003; Plant et al, 2003; Wang et al, 2004). Deletion of the NTD results in mislocalization of Par-3 from the more apical side to the basal side in epithelial cells and strongly impairs cell polarity in both Drosophila and mammals (Benton and St Johnston, 2003a; Mizuno et al, 2003). Overexpression of mPar-3 NTD in neurons leads to mislocalization of endogenous mPar-3 and mPar-6 and severely compromises axon specification in developing neurons (Shi et al, 2003). Despite the critical roles of NTD in organizing the Par-3/Par-6/aPKC complex and cell polarity, the molecular mechanism of the Par-3 NTD-mediated Par complex assembly remains obscure.
Figure 6.
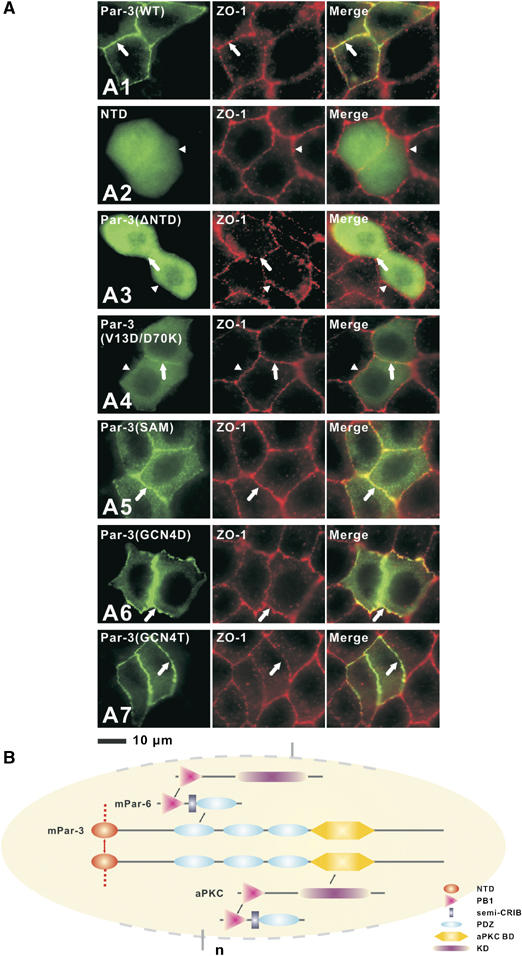
Multimerization of NTD is essential for Par-3 membrane targeting. (A) MDCK II cells were transiently transfected with pEGFP-Par-3(WT) (subpanel A1), pEGFP-NTD (subpanel A2), pEGFP-Par-3(ΔNTD) (subpanel A3), pEGFP-Par-3(V13D/D70K) (subpanel A4), pEGFP-Par-3(SAM) (subpanel A5), pEGFP-Par-3(GCN4D) (subpanel A6) and pEGFP-Par-3(GCN4T) (subpanel A7), respectively. After transfection, the cells were maintained for 72 h to form a confluent monolayer. Cells were then fixed and stained with antibody for ZO-1. Cellular localizations of various forms of Par-3 were visualized by fluorescence signals from EGFP. Colocalization of the Par-3 signal with the tight junction maker ZO-1 is indicated by arrows (subpanels A1–A7). Arrowheads point to the tight junction with no Par-3 enrichment (A2 and A3). (B) Schematic diagram showing the high-order Par-3/Par-6/aPKC complex assembly. Formation of the PB1 heterodimer links aPKC with Par-6. Formation of the PDZ domain heterodimer connects Par-6 to Par-3. The direct interaction between the kinase domain (KD) of aPKC and with the C-terminal kinase binding domain (aPKC-BD) of Par-3 not only links the two proteins but also provides a potential regulatory mechanism of aPKC's kinase activity. Finally, NTD domain-mediated oligomerization of Par-3 provides a mechanism for the Par-3/Par-6/aPKC to further polymerize into supramolecular assemblies.
We report here that the Par-3 NTD is capable of forming high-order aggregates that can be dissociated by high concentrations of salt. The three-dimensional (3D) structure of an NTD monomeric subunit was solved by NMR spectroscopy. Unexpectedly, we found that the Par-3 NTD adopts an atypical PB1-domain-like structure capable of homo-oligomerization. We further demonstrate that NTD-mediated Par-3 membrane localization is solely attributed to the oligomerization capacity of the domain.
Results and discussion
Par-3 NTD forms homo-oligomers
To uncover the molecular basis governing the self-association of Par-3 NTD, we characterized biochemical properties of the purified recombinant NTD. Purified NTD was eluted from an analytical gel filtration column at a molecular mass much greater than its theoretical mass of 11.2 kDa, indicating that NTD forms homo-oligomers (Figure 1A). Gel filtration chromatographic analysis further showed that NTD tends to aggregate into larger homo-oligomers at high protein concentrations (e.g. the majority of the protein was eluted at a void volume in the gel filtration column used when the NTD concentration reached 15 mg/ml). The formation of NTD homo-oligomers was readily confirmed by a chemical crosslinking assay (Figure 1B). We then probed the driving force that mediates the self-association of NTD by assaying the association status of the domain in 50 mM Tris, pH 7.5 containing various additives including different detergents and/or salts. We found that high concentrations of salt in the buffer were sufficient to convert the NTD homo-oligomers into monomers (Figure 1C), indicating that electrostatic interactions play key roles in the oligomerization of NTD. Consistent with this observation, the HSQC (heteronuclear single quantum coherence) spectrum of 15N-labeled NTD in the presence of 2 M KCl is indicative of a relatively small protein with sharp linewidth and good spectrum homogeneity. In contrast, no signals could be detected when the salt concentration of the NTD solution was lowered (Supplementary Figure 1A and B). We next searched for single amino-acid residue point mutations that can disrupt self-oligomerization of NTD. We chose to alter the electrostatic property of NTD (i.e. by changing the charged residues directly or converting the surrounding uncharged residues to charged ones) and assayed the gel filtration profiles of each of these point mutants. Among the more than 30 different mutants of NTD that we characterized (Supplementary Table 1), two monomeric mutants, V13D and D70K NTD, were selected for detailed structural characterization (see below for the rationales of the selection). Both of these mutants were eluted as monomer in low-salt concentration buffer with a wide protein concentration range (Figure 1D). Furthermore, neither of the two mutants displayed any tendency of forming high-molecular mass aggregates in the chemical crosslinking assay (Figure 1B).
Figure 1.
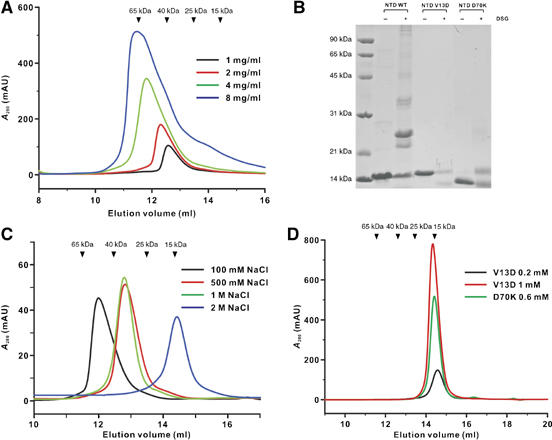
Biochemical properties of Par-3 NTD. (A) Analytical gel filtration profiles of the wild-type Par-3 NTD at different loading concentrations. The elution volumes of the molecular mass standards are marked at the top of the panel. (B) Chemical crosslinking of the wild-type Par-3 NTD and the two point-substituion mutants (V13D NTD and D70K NTD). The wild-type Par-3 NTD formed various forms of oligomers after chemical crosslinking. In contrast, no multimeric proteins could be detected for either of the mutants in the presence of the crosslinker. (C) Gel filtration profiles of the wild-type Par-3 NTD in a column buffer containing various concentrations of salt. (D) Gel filtration profiles of the two monomeric mutants of Par-3 NTD mutants. Two different concentrations of the V13D mutant were analyzed to show that the elution profile of the protein does not depend on the sample concentration.
Par-3 NTD adopts a PB1 domain-like fold
The V13D monomeric mutant of NTD displays excellent HSQC spectrum (Supplementary Figure 1C). We further showed that substitution of Val13 with Asp only resulted in minimal chemical shift changes to the residues in close vicinity to Val13, indicating that the mutation did not alter the overall conformation of NTD (Supplementary Figure 2). The 3D structure of the V13D NTD mutant was solved using NMR spectroscopy. Except for a few residues in the N-terminus and one residue in the C-terminus, the entire V13D NTD is well defined (Figure 2A; Table I). V13D NTD adopts a ubiquitin-like fold comprising five β strands (β1–β5) and two α helices (α1–α2) (Figures 2B and 3A). The five-stranded β-sheet forms a half β-barrel, and the barrel is completed by packing of the long amphipathic α1 helix. One of the two opening sides is capped by the very short α2 helix that is almost perpendicular to the α1 helix (Figure 2B). The overall structure of V13D NTD is very similar to that of ubiquitin, with a root mean square deviation (r.m.s.d.) of ∼1.4 Å for the secondary structured regions, although the two proteins share no amino-acid sequence homology.
Figure 2.

Structure of the Par-3 V13D NTD. (A) Stereoview showing the backbone of 20 superimposed NMR-derived structures of the Par-3 V13D NTD. The N- and C-termini of the protein are labeled. (B) Ribbon diagram of a representative NMR structure of the Par-3 V13D NTD. The secondary structure elements of the protein are labeled with the same scheme used for ubiquitin and PB1 domains.
Table 1.
Structural statistics for the family of 20 structures of Par-3 V13D NTD
| Distance restraints | |
| Intra-residue(i−j=0) | 597 |
| Sequential(∣i−j∣=1) | 341 |
| Medium range (2<∣i−j∣<4) | 221 |
| Long range(∣I−j∣>5) | 430 |
| Hydrogen bonds | 82 |
| Total | 1671 |
| Dihedral angle restraints | |
| Φ | 37 |
| Ψ | 38 |
| Total | 75 |
| Mean r.m.s. deviations from the experimental restraints | |
| Distance (Å) | 0.010±0.000 |
| Dihedral angle (Å) | 0.200±0.036 |
| Mean r.m.s. deviations from idealized covalent geometry | |
| Bond (Å) | 0.001±0.000 |
| Angle (deg) | 0.259±0.004 |
| Improper (deg) | 0.133±0.006 |
| Mean energies (kcal/mol)a | |
| ENOE | 7.41±1.32 |
| Ecdih | 0.19±0.07 |
| EL−J | −317.49±6.10 |
| Ramachandran plotb | |
| Residues 2–83 in Par-3 V13D NTD | |
| % Residues in the most favorable regions | 76.3 |
| Additional allowed regions | 22.9 |
| Generously allowed regions | 0.8 |
| Atomic r.m.s. differences (Å) | |
| Full molecule (residues 2–82 in Par-3 V13D NTD) | |
| Backbone heavy atoms (N, Cα and C′) | 0.33 |
| Heavy atoms | 0.90 |
| None of the structures exhibits distance violations greater than 0.3 Å or dihedral angle violations greater than 4°. | |
| The final values of the square-well NOE and dihedral angle potentials were calculated with force constants of 50 kcal/mol/Å and 200 kcal/mol/rad. | |
| The program PROCHECK (Laskowski et al, 1996) was used to assess the overall quality of the structures. | |
Figure 3.
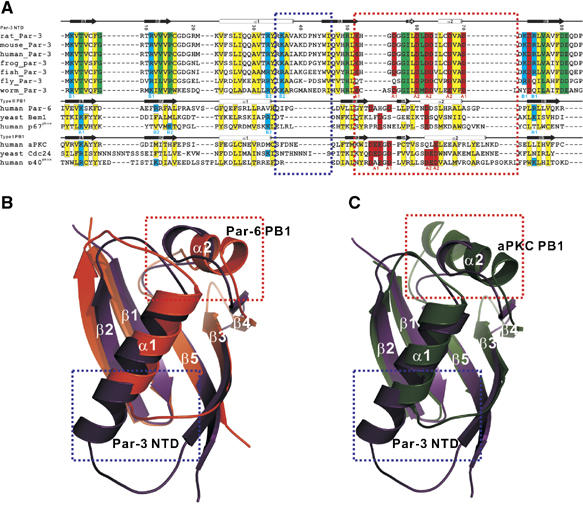
Par-3 NTD adopts a PB1-like fold. (A) Structure-based sequence alignment of Par-3 NTD from different species. The amino-acid sequences of selected PB1 domains are also included in the alignment. In this alignment, the hydrophobic residues forming the core of the domains are in yellow; the negatively charged resides forming the ‘A1' and ‘A2' regions are in red; the positively charged residues forming the ‘B1' and ‘B2' patches are in cyan and the rest of the highly conserved residues are in green. The specific amino-acid residues forming the ‘A1 and A2' and ‘B1 and B2' regions are marked at the bottom of the sequences. The secondary structures of the Par-3 NTD and the PB1 domains are labeled on the top of the respective sequences. The amino-acid residue number of the Par-3 NTD is also indicated in the figure. The blue and the red boxes highlight the two regions of the Par-3 NTD that are distinctly different from the corresponding regions in PB1 domains. (B, C) Structural comparison of the Par-3 NTD with two representative types, type I (from Par-6) and type II (from aPKC), of PB1 domains. The two distinct regions of Par-3 and the PB1 domains indicated in the sequence alignment are also indicated in the structural comparison.
The overall structural similarity of V13D NTD with ubiquitin immediately suggested a potential functional similarity of Par-3 NTD with the PB1 domains, given that two proteins in the Par complex (Par-6 and aPKC) contain a PB1 domain in each (Ito et al, 2001; Terasawa et al, 2001; Wilson et al, 2003; Hirano et al, 2005). The overall structures of the PB1 domains from Par-6 and aPKC are very similar to the structure of V13D NTD (pairwise r.m.s.d. value of ∼1.5 Å for the secondary structured regions; Figure 3). The hallmark of PB1 domains is their charge–charge interaction-mediated heterodimerization (Ito et al, 2001; Terasawa et al, 2001; Wilson et al, 2003; Hirano et al, 2005). The intermolecular interaction between the wild-type Par-3 NTD is also dominated by charge–charge interactions (Figure 1), although the Par-3 NTD forms homo-oligomers instead of the heterodimers observed in canonical PB1 domains. Structure-based amino-acid sequence alignment of Par-3 NTD with different types of PB1 domains indicates their overall structural similarities (Figure 3A). This sequence alignment analysis also reveals a number of critically important differences of the Par-3 NTD with PB1 domains. Specifically, the Par-3 NTD contains an elongated α1 helix followed by a rigid α1/β3-loop (the blue box in Figure 3A and B). The structural features of the region spanning β3, β4 and α2 in the Par-3 NTD (the red box in Figure 3) are also distinctly different from that of the same region in PB1 domains. This region in the type-I PB1 domains (e.g. the aPKC PB1) encodes a so-called ‘OPCA motif' with a stretch of negatively charged residues in the β3/β4-loop, which is largely responsible for binding to the conserved positively charged residues at the back (i.e. β1, β2 and β5) of the type-II PB1 domains (e.g. the Par-6 PB1) (Figure 3 and 4). In the Par-3 NTD, the β3/β4-loop is shortened (hence lacking the negatively charged residues found in the type-I PB1 domains); the slightly lengthened β4/α2-loop contains three conserved Asp residues. Additionally, the Par-3 NTD also contains two negatively charged residues that are near to each other and accessible to the solvent in the α2 helix (Figures 3 and 4). We further note that the α2 helix in the Par-3 NTD is shorter than the corresponding helix in the PB1 domains. These negatively charged residues are also located on one side of the Par-3 NTD and can be divided into two patches (‘A1' and ‘A2') as seen in the type-I PB1 domains such as aPKC PB1 (Figure 4A and C). In addition to these negatively charged residues, the Par-3 NTD also contains several conserved and positively charged residues at positions corresponding to those in type-II PB1 domains (β1, β2, β5 and the C-terminal end of α1; Figure 3B). These positively charged residues are located on the opposite side of the acidic surface described above and can also be divided into two patches (‘B1' and ‘B2') as observed in the type-II PB1 domains (Figure 4B and D). Therefore, the Par-3 NTD contains structural features found in both type-I and type-II PB1 domains, and we thus define the Par-3 NTD as a PB1-like domain. Currently available domain prediction computer algorithms (e.g. SMART, Pfam, Prosite and 3D-PSSM) cannot predict Par-3 NTD as an ubiquitin fold protein, possibly due to the significant amino-acid sequence variations in the two boxed regions as shown in Figure 3. Finally, Par-3 NTD shares no surface charge similarity with ubiquitin, even though the backbone structures of the two proteins are very similar.
Figure 4.
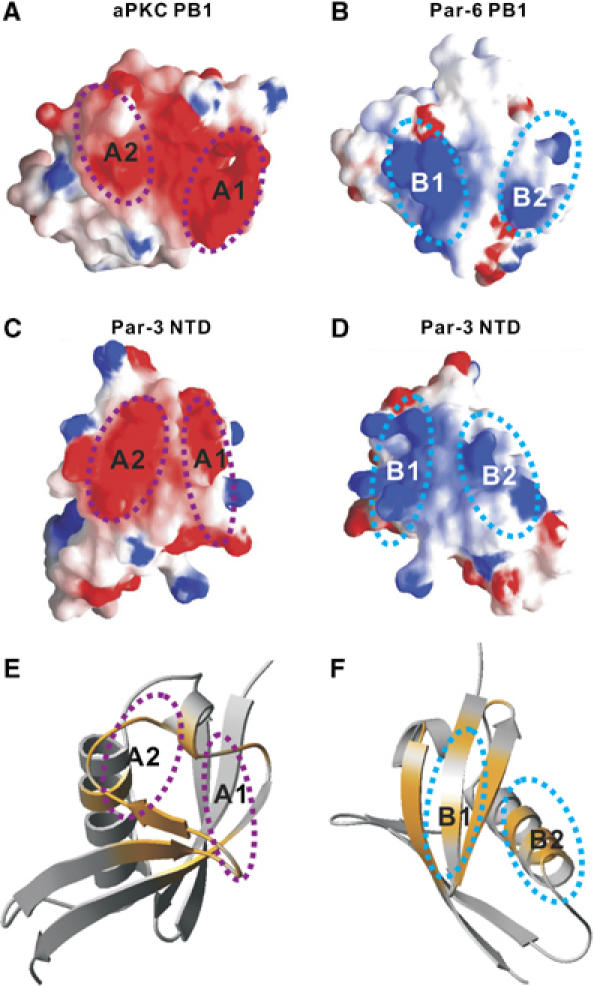
Interaction interface between the Par-3 NTD dimers. (A) Surface representation of the type-I PB1 domain from aPKC showing the negatively charged ‘A1' and ‘A2' regions. (B) Surface diagram representation showing the positively charged ‘B1' and ‘B2' regions in the type-II PB1 domain from Par-6. (C, D) Surface representation of the Par-3 NTD ‘A1 and A2' and ‘B1 and B2' regions, respectively. In the electrostatic potential surface diagrams, the positively charged surface is in blue and the negatively charged surface is in red. (E) Summary of the chemical shift changes of V13D NTD induced by binding of D70K NTD. The chemical shift changes of the backbone amides of V13D NTD (colored in orange) are mapped onto the ribbon diagram of the monomeric NTD structure. The orientations of NTD shown in panels C and E are the same. (F) Summary of the chemical shift changes D70K NTD resulting from binding to V13D NTD. The orientations of NTD shown in the panels D and F are identical.
Molecular basis of the Par-3 NTD self-association
The existence of both positively charged and negatively charged surfaces at the two opposite sides of Par-3 NTD indicates that multiple NTD are likely to be able to interact with each other via charge–charge interactions in a front-to-back manner (Figure 4C and D). Mutation of Val13 to Asp introduced a negative charge at the center of the positively charged surface, thereby blocking V13D NTD from associating with each other (Supplementary Figure 3). Similarly, substitution of Asp70 at the end of the α2 helix with Lys introduced a positive charge to the negatively charged surface of NTD, and the D70K NTD behaved as a monomer in solution (Supplementary Figure 3). We predicted that the V13D NTD should be able to interact with the D70K NTD if the wild-type NTD indeed interacts with each other in a front-to-back manner. A fluorescence polarization assay demonstrated that the two NTD mutants specifically interacted with each other, with a dissociation constant of ∼100 μM (Supplementary Figure 4). The interaction between these two mutants was disrupted by inclusion of 0.5 M NaCl (Supplementary Figure 4). We then mapped the interaction sites of the two mutants by NMR spectroscopy. We first titrated 15N-labeled V13D NTD with unlabeled D70K NTD. Saturation of V13D NTD with the D70K mutant led to disappearance of a selected set of peaks (Supplementary Figure 5), and these peaks corresponded to the negatively charged residues in the ‘A1' and ‘A2' patches of NTD (Figure 4E). Reciprocally, we titrated 15N-labeled D70K NTD with the unlabeled V13D mutant. Binding of V13D NTD specifically induced chemical shift changes to the residues in the ‘B1' and ‘B2' regions of the D70K mutant (Figure 4F; Supplementary Figure 6). Taken together, the above biochemical and NMR spectroscopic data indicate that Par-3 NTD can interact with each other via the two oppositely charged surfaces in a front-to-back manner, thereby forming homo-oligomers. Finally, based on the well-established equilibrium polymerization model and the measured dissociation constant between the two monomeric forms of Par-3 NTD (Greer, 1998; Dudowicz et al, 2000), Par-3 is expected to be able to oligomerizes efficiently via its NTD domain in Par-3-enriched regions (e.g. near the tight junctions of epithelial cells).
The above NMR titration-based study cannot differentiate pairwise interaction sites (e.g. A1-B1/A2-B2 or A1-B2/A2-B1) between the two NTD monomers. To obtain this information, we resorted to the paramagnetic pseudo-contact shift approach (Ikegami et al, 2004; Pintacuda et al, 2004; Su et al, 2006). In this experiment, two out of three Cys residues in D70K NTD were mutated (Cys16 to Ala, Cys66 to Val). The remaining Cys6 (solvent accessible and located at the bottom of the B1 site of D70K NTD) was then labeled with S-(2-pyridylthione)-cysteaminyl–EDTA (Supplementary Figure 7). Addition of paramagnetic lanthanide ion Dy3+ induced chemical shift changes to the residues limited within the A1 region of V13D NTD (Supplementary Figure 7A–D), supporting that B1 in D70K is in direct contact with A1 in V13D.
Par-3 NTD cannot interact with Par-6 PB1 or aPKC PB1
Since the PB1-like Par-3 NTD contains two positively and negatively charged surfaces located at the opposite sides of the protein, we wondered whether Par-3 NTD might be able to interact with the type-I PB1 domain using its positively charged surface and bind to the type-II PB1 domain via its negatively charged residues. We modeled Par-3 NTD onto the aPKC/Par-6 PB1 heterodimer (Hirano et al, 2005) by superimposing the structure of the Par-3 NTD with that of Par-6 PB1. The elongated α1 helix abducted from Par-3 NTD spatially crashed with the β3 and β4/α2-loop of aPKC PB1 (Figure 3B; Supplementary Figure 8A), suggesting that Par-3 NTD is unlikely capable of binding to type-I PB1 domains. Reciprocally, we modeled Par-3 NTD onto the Par-6 PB1 domain in the aPKC/Par-6 PB1 heterodimer. The shortened β3/β4-loop, the α2 helix and the lack of the negatively charged residues in the loop led to the lack of necessary surfaces as well as charge complementation between the Par-3 NTD and the Par-6 PB1 domain (Figure 3C; Supplementary Figure 8B), indicating that there is little chance for potential interaction between Par-3 NTD and the type-II PB1 domains. We tested potential interactions between Par-3 NTD and the PB1 domains from aPKC or Par-6 using an in vitro pull-down assay. Consistent with the structure-based predictions, no direct interactions could be observed between Par-3 NTD and PB1 domains either from aPKC or Par-6 (data not shown).
A structural model of the Par-3 NTD oligomer
Having mapped the interaction surface between two Par-3 NTD domains, we tried to build an NTD oligomer model by determining the structure of the V13D NTD/D70K NTD complex. Unfortunately, the poor quality of the NMR spectra precluded us from solving the high-resolution structure of the V13D NTD/D70K NTD complex (Supplementary Figures 5 and 6). Extensive search for various mutants hoping to obtain stable NTD dimer was not successful either. In an alternative approach, we decided to build a Par-3 NTD dimer structural model based on the chemical shift perturbation data derived from the NMR titration experiments and other biochemical data. During this model building process, we included one ambiguous distance restraint (10 Å) between B1/B2 and A1/A2, and calculated the Par-3 NTD dimer structure model using the program HADDOCK (Dominguez et al, 2003). Only one converged dimer structure ensemble was derived and this dimer structure model matches the pairwise interactions involving A1–B1 and A2–B2 pairs (Figure 5A and B). Importantly, the NTD dimer model calculated from the HADDOCK-based calculation agrees well with the contact site information derived from the paramagnetic pseudo-contact shift experiment shown in the Supplementary Figure 7. Additional biochemical data further supported the B1–A1 and B2–A2 pairwise interactions seen in the NTD dimer model (Supplementary Figure 7E). Finally, the A1–B1 and A2–B2 pairwise interaction pattern in the NTD dimer model is also observed in known PB1–PB1 heterodimers (Ito et al, 2001; Terasawa et al, 2001; Wilson et al, 2003; Hirano et al, 2005).
Figure 5.
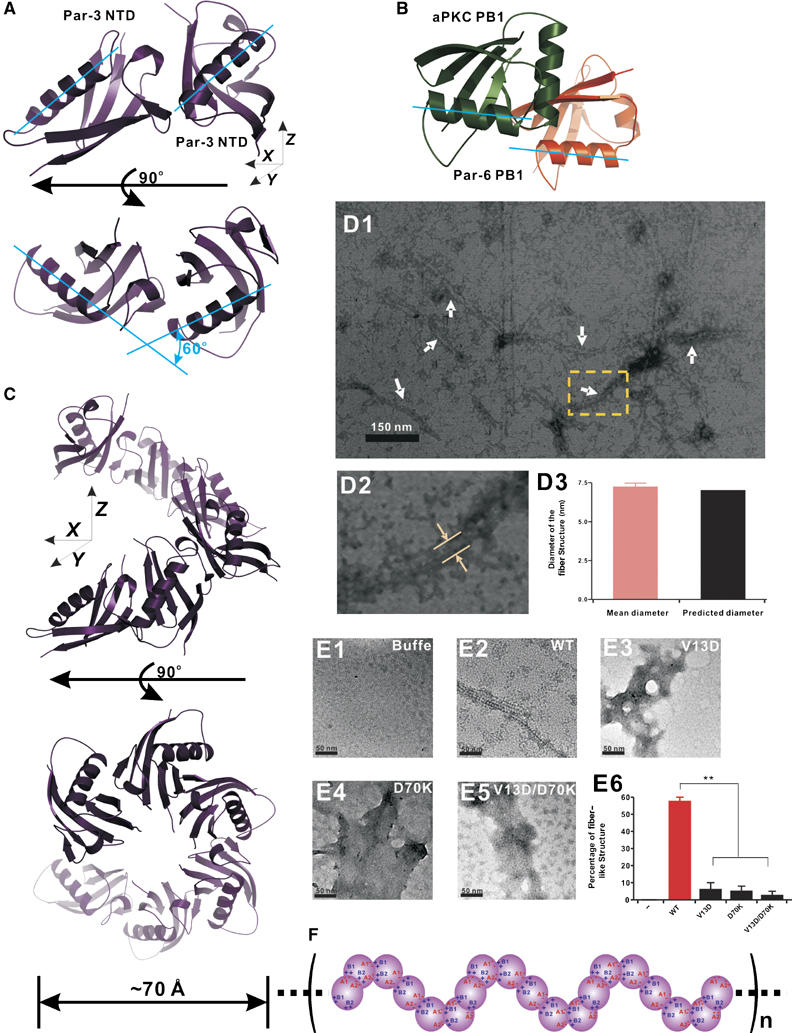
The oligomeric structural model of Par-3 NTD oligomers. (A) Ribbon representation of the Par-3 NTD dimer model. The two monomers are related by a 60° rotation in the x–y plane. (B) Ribbon representation of aPKC/Par-6 PB1 heterodimer complex. In this diagram, the aPKC PB1 is in green and the Par-6 PB1 is in orange. Two PB1 domains in the complex are related with each other with a simple lateral translation in the x–y plane. (C) Horizontal and top views of the Par-3 NTD oligomer model. The Par-3 NTD forms a helical filament with six monomeric units in one complete helical turn. The diameter of the filament is ∼70 Å. (D1–D3) Electron micrographs of the negatively stained Par-3 NTD showing that NTD forms fiber-like structures in low-salt solution. In panel D1, the arrows indicate the negatively stained fiber-like structures formed by the wild-type NTD. Panel D2 shows a magnified micrograph of the NTD fiber indicated by the yellow box in panel D1. The measured diameter of the fiber structure is indicated by arrows. The average diameter of the fiber-like structure obtained from 50 fibers is indicated in panel D3. (E1–E5) Monomeric NTD mutants show radically different negative staining patterns from the wild-type NTD. In contrast to the wild-type NTD (panel E2), V13D NTD (panel E3), D70K NTD (panel E4) and V13D/D70K NTD (panel E5) form amorphous deposits on carbon-coated copper grids. Panel E6 shows quantification of fiber-like images observed for the wild-type NTD and its mutants under negative staining electron microscopy. (F) Schematic diagram of the Par-3 NTD oligomer model. The formation of elongated, helical filamentous structures of the Par-3 NTD oligomer is mediated mainly by the electrostatic interactions via A1/A2 and B/B2 charged surfaces.
Although similar, the two monomers in the Par-3 NTD are packed with each other in a significantly different manner when compared with the two PB1 domains in PB1 heterodimers (Wilson et al, 2003; Hirano et al, 2005). Instead of forming the nearly perfect front-to-back dimers seen in PB1 heterodimers (i.e. the two units are related to each other by simple lateral translation), the two monomers in the Par-3 dimer are related to each other by ∼60° rotation in the x–y plane in addition to the lateral translation (Figure 5A and B). Based on this Par-3 NTD dimer model, we were able to build a 3D polymer model of the Par-3 NTD (Figure 5C). In this model, the Par-3 NTD is capable of forming helical filaments. One complete helix in the filament contains six units of Par-3 NTD (6 × 60°). The calculated diameter of the Par-3 NTD helical filament is ∼70 Å. Electron micrographs of the negatively stained protein showed that the wild-type Par-3 NTD eluted from a low-salt buffer forms filaments with a diameter of ∼70 Å (Figure 5D1–D3), correlating well with the width of the filament calculated from the 3D NTD polymer model. In line with our structural characterization, several monomeric mutants of NTD (V13D, D70K and the V13D/D70K double mutant) do not form filamentous structure under the same conditions used for the wild-type protein in the negative staining electron microscopy studies (Figure 5E1–E5). These monomeric mutants form predominantly amorphous deposits on the supporting matrix used in the electron microscopic studies (Figure 5E6).
Although the NTD alone is capable of forming oligomers, it is not known whether Par-3 can also form oligomer. To test this possibility, we purified a much longer form of Par-3 containing N-terminal 704 amino-acid residues (Supplementary Figure 9). This Par-3 fragment includes the NTD domain and all three PDZ domains of the protein (the full-length Par-3 could not be expressed in soluble form in bacterial cells). Electron microscopic studies showed that this 704-residue Par-3 fragment also forms fiber-like structure. Consistent with the observation in NTD alone, V13D Par-3(1–704) and V13D/D70K Par-3(1–704) do not form fiber structure under EM (Supplementary Figure 9).
The oligomerization of the N-terminal domain is critical for proper cellular localization of Par-3
Although it has been shown that NTD is essential for proper functioning of Par-3 in polarity regulation (Benton and St Johnston, 2003a; Fang et al, 2003; Mizuno et al, 2003), the molecular basis of NTD's function is not clear. Our detailed structural and biochemical studies provide an opportunity for us to test the role of the NTD oligomerization in the function of Par-3. Consistent with previous work (Mizuno et al, 2003), both NTD alone and Par-3 with its NTD deleted (ΔNTD) showed diffused cellular distribution patterns when expressed in MDCK cells (Figure 6A2 and A3). As expected, the wild-type Par-3 colocalized well with endogenous ZO-1 (Figure 6A1). Since both V13D and D70K single-point mutants of Par-3 are still capable of binding to the wild-type Par-3 and could in turn interfere with endogenous Par-3, we created a V13D/D70K double mutant of Par-3. The Par-3 NTD bearing the V13D/D70K substitutions showed no detectable binding to the wild-type NTD, and the mutant protein properly folded (data not shown). When expressed in MDCK cells, the V13D/D70K Par-3 mutant showed predominantly diffused cellular localization (Figure 6A4), indicating that proper membrane localization of Par-3 requires NTD-mediated oligomerization. To further confirm this conclusion, we generated several chimeric Par-3 mutants by replacing its NTD with protein domains with various oligomerization capacities. Replacing the Par-3 NTD with a weak oligomerization SAM domain of neuronal scaffold protein Shank1 (EGFP-Par-3(SAM); our unpublished data) led to a partial rescue of membrane localization of Par-3 (Figure 6A5). Replacing the NTD with a stable dimerization GCN4 leucine zipper (EGFP-Par-3(GCN4D)) also partially rescued membrane localization of Par-3 (Figure 6A6). When the NTD was replaced with a GCN4 construct capable of forming tight tetramers (EGFP-Par-3(GCN4T)) (Harbury et al, 1993), the resulting Par-3 chimera was completely membrane localized and the chimera essentially recapitulated the localization pattern of the wild-type Par-3, indicating that stable oligomerization enhances Par-3 membrane localization (Figure 6A7). The data observed for the chimeric Par-3 further indicated that the NTD-mediated apical membrane localization of Par-3 does not occur by binding of the NTD to apical anchor(s), as otherwise the substitution of the NTD with the SAM domain and the GCN4 leucine zippers would unlikely rescue the function of the NTD. Taken together, the data presented in Figure 6A firmly establish that the NTD is absolutely required for the membrane localization of Par-3 and NTD-mediated Par-3 apical membrane localization is solely caused by the oligomerization of the domain.
In summary, we discovered that the conserved NTD of Par-3 adopts a PB1-like fold with two charged binding surfaces located on the opposite sides of the protein. The Par-3 NTD is capable of forming high-order homo-oligomers in solution via these two oppositely charged surfaces in a front-to-back manner (Figure 5F). The NTD-mediated oligomerization of Par-3 is essential for the apical membrane localization of this key cell polarity determinant. The NTD-mediated homo-multimerization of Par-3 provides a molecular basis for further oligomerization of the Par-3/Par-6/aPKC complex (Figure 6B). One can envision that a high-order (Par-3/Par-6/aPKC)n complex may increase the avidity of this polarity determining complex in interacting with apical membrane components. The oligomerization of the Par-3/Par-6/aPKC complex may also increase the local concentration of aPKC in the apical membrane domain, thereby preventing the basal–lateral determinants such as Lgl from entering the apical membrane domain. This large (Par-3/Par-6/aPKC)n complex assembly can also serve as a platform for organizing other polarity-regulating proteins such as the Lgl/Scribble/DLG and Pals1/Crumbs/Patj complexes.
Materials and methods
Protein expression and purification
The coding sequence for the Par-3 NTD (residues 2–83) was amplified by PCR from full-length Rat Par-3 (Asip) and cloned into a modified pET32a vector (Long et al, 2003). The resulting NTD contained a His6 tag at its N-terminus. All point mutants of NTD were prepared using PCR-based methods. Proteins were expressed in BL21 (DE3) Escherichia coli cells at 37°C. The His6-tagged NTD proteins were purified by Ni-nitrilotriacetic acid (Ni-NTA) agarose (Qiagen) affinity chromatography followed by size-exclusion chromatography. Uniformly isotope-labeled NTD were prepared by growing bacteria in M9 minimal medium using 15NH4Cl as the sole nitrogen source or 15NH4Cl and 13C6-glucose (Cambridge Isotope Laboratories Inc.) as the sole nitrogen and carbon sources, respectively. The NMR samples were concentrated to ∼0.1 mM (for HSQC-based sample screening and titration experiments) or ∼1 mM (for structural determinations) in 50 mM Tris–HCl, pH 7.0 in the presence of 100 mM NaCl, 1 mM DTT and 1 mM EDTA. For in vitro biochemical analysis, the wild-type NTD and mutants were expressed as GST-fusion proteins using pGEX-4T-1 vector and purified by glutathione–Sepharose-4B affinity chromatography (Amersham Biosciences).
NMR spectroscopy
NMR spectra were acquired at 30°C on Varian Inova 500 or 750 MHz spectrometers, each equipped with a z-axis-shielded triple resonance probehead. Sequential backbone and non-aromatic, non-exchangeable side chain resonance assignments of Par-3 V13D NTD were achieved by standard heteronuclear correlation experiments including HNCO, HNCACB, CBCA(CO)NH and HCCH-TOCSY using 15N-/13C-labeled samples, and confirmed by a 3D 15N-seperated NOESY experiment using an 15N-labeled sample (Bax and Grzesiek, 1993; Kay and Gardner, 1997). The side chains of aromatics were assigned by 1H 2D TOCSY and NOESY experiments using unlabeled samples in D2O (Wüthrich, 1986).
Structure calculations
Approximate interproton distance restraints were derived from 2D 1H-NOESY, 3D 15N-seperated NOESY and 3D 13C-seperated NOESY spectra. NOE restrains were grouped into three distance ranges: 1.8–2.7 Å (1.8–2.9 Å for NOEs involving NH protons), 1.8–3.3 Å (1.8–3.5 Å for NOEs involving NH protons) and 1.8–5.0 Å, corresponding to strong, medium and weak NOEs, respectively. Hydrogen bonding restraints were generated from the standard secondary structure of the protein based on the NOE patterns and backbone secondary chemical shifts. Backbone dihedral angle restraints (φ and ψ angles) were derived from the secondary structure of the protein and backbone chemical shift analysis program TALOS (Cornilescu et al, 1999). Structures were calculated using the program CNS (Brunger et al, 1998).
Model building of Par-3 NTD dimers and oligomers
One ambiguous distance restraint (with a distance up-limit of 10 Å) between residues in the ‘B1, B2' and the ‘A1, A2' regions of two NTD monomers was included to calculate the Par-3 NTD dimer model using the program HADDOCK. The selection of amino-acid residues was based on the NMR titration experiments and interaction studies using a large array of mutants of NTD (Supplementary Table 1). The Par-3 NTD oligomer model was built by stepwise assembly of dimer models using the same ambiguous distance restraints used for the dimer model construction, and a homo-hexamer model was calculated in this study.
Gel filtration chromatography
Analytical gel filtration chromatography was carried out on an AKTA FPLC system using a Suprose-12 10/30 column (Amersham Pharmacia Biotech). Protein samples were dissolved in 50 mM Tris–HCl buffer (pH 7.5) with various concentrations of salts and 1 mM DTT. The column was calibrated with the low-molecular-mass column calibration kit from Amersham Pharmacia Biotech.
Fluorescence assays
Fluorescence polarization assays were performed on a PerkinElmer LS-55 fluorimeter equipped with an automated polarizer at 20°C. Purified D70K NTD was chemically modified by conjugating a fluorescent naphthalene derivative 1,5-IAEDANS (5-((((2-iodoacetyl)amino)ethyl)amino)naphthalene-1-sulfonic acid (Molecular Probes) to the Cys residues of the protein. Fluorescence titration was performed by adding increasing amounts of unlabeled V13D NTD to a constant amount of IAEDANS-labeled D70K NTD (1 μM) in 50 mM sodium phosphate buffer, pH 7.5, containing 1 mM EDTA and 1 mM DTT, with or without 500 mM NaCl.
Chemical crosslinking assay
Protein samples (0.5 mg/ml) were dissolved in 50 mM sodium phosphate buffer (pH 7.0) containing 100 mM NaCl, 1 mM DTT, 1 mM EDTA and 1 mM crosslinker disuccinimidyl glutarate (DSG, from Pierce, crosslinker arm length of 7.72 Å). The reaction typically lasted for 15–30 min at room temperature and was then quenched by addition of excess amounts of 1 M Tris–HCl stock solution.
GST pull-down assays
GST- or GST-tagged proteins (50 μl from 1 mg/ml stock solutions) were incubated with 20 μl of glutathione–Sepharose-4B slurry beads in an assay buffer (50 mM Tris–HCl, pH 7.5, 100 mM NaCl, 1 mM DTT and 0.5% NP-40) for 1 h at 4°C. The beads were washed three times using 500 μl assay buffer. The GST-fusion protein loaded beads were then mixed with each potential binding protein and the mixtures were incubated for another 2 h at 4°C. The beads were further washed with 500 μl assay buffer three times. The proteins captured by affinity beads were eluted by boiling, resolved by 15% SDS–PAGE and detected by Coomassie blue staining.
Electron microscopy
Wild-type NTD and its mutants (1 mg/ml) were dissolved in 50 mM Tris–HCl buffer (pH 7.5) containing 100 mM NaCl and 1 mM DTT. Samples were applied to parlodion–carbon films mounted on 400 × 400 copper grids (from SPI Supplies). The NTD-loaded copper grids were first washed with distilled water and then incubated with 1% uranyl acetate for 10 min at room temperature to stain the proteins. Excess staining dye was removed by rinsing the copper grids with distilled water. After air drying, the sample specimens were examined using a JEOL 2010 transmission electron microscope.
Cell culture, transfection and immunofluorescence
NTD-EGFP was constructed by inserting the NTD coding sequence into the HindIII and EcoRI sites of pEGFP N1 vector. EGFP-Par-3(V13D/D70K) and EGFP-Par-3(ΔNTD) were created using PCR-based mutagenesis and cloned into the pEGFP C2 vector. Various EGFP-Par-3 chimeras, EGFP-Par-3(SAM), EGFP-Par-3(GCN4D) and EGFP-Par-3(GCN4 T), were generated by substituting NTD with the Shank1a SAM domain (residues 2023–2082), a dimeric form of GCN4 leucine zipper, and a tetrameric form of GCN4 (provided by Professor M Yuzaki, Keio University School of Medicine), respectively. MDCK II cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS, Invitrogen) at 37°C and 5% CO2. The cells were transiently transfected with 0.7 μg plasmids per well using a lipofectamine PLUS kit (Invitrogen). The expression level of the EGFP-tagged proteins were checked by Western blot assay. The anti-ZO-1 monoclonal antibody was provided by Professor B Peng (HKUST). Immunofluorescence labeling was performed using Rhodamine Red-X-conjugated secondary antibodies and the images were acquired using a Nikon TE2000E inverted fluorescent microscope.
Illustrations
The figures were prepared using the programs MOLMOL (Koradi et al, 1996), MOLSCRIPT (Kraulis, 1991), PyMOL (http://pymol.sourceforge.net/) and GRASP (Nicholls, 1992).
Coordinates
The atomic coordinates of Par-3 V13D NTD have been deposited in the Protein Data Bank (accession code 2NS5).
Supplementary Material
Supplementary Figures and Legends
Acknowledgments
We thank Professor M Yuzaki, Keio University School of Medicine, Japan for GCN4 constructs with various multimerization capacities used in this study and Professor B Peng in HKUST for the anti-ZO-1 antibody. The electron microscopic images were acquired using the JEOL 2010 transmission electron microscope in the MCPF, HKUST. This work was supported by grants from the Research Grants Council of Hong Kong, and the Human Frontier Science Program to MZ The NMR spectrometer used in this work was purchased with funds donated to the Biotechnology Research Institute by the Hong Kong Jockey Club. MZ was a recipient of the Croucher Foundation Senior Research Fellow Award.
References
- Bax A, Grzesiek S (1993) Methodological advances in protein NMR. Acc Chem Res 26: 131–138 [Google Scholar]
- Benton R, St Johnston D (2003a) A conserved oligomerization domain in Drosophila Bazooka/PAR-3 is important for apical localization and epithelial polarity. Curr Biol 13: 1330–1334 [DOI] [PubMed] [Google Scholar]
- Benton R, St Johnston D (2003b) Drosophila PAR-1 and 14-3-3 inhibit Bazooka/PAR-3 to establish complementary cortical domains in polarized cells. Cell 115: 691–704 [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Chen X, Macara IG (2005) Par-3 controls tight junction assembly through the Rac exchange factor Tiam1. Nat Cell Biol 7: 262–269 [DOI] [PubMed] [Google Scholar]
- Cornilescu G, Delaglio F, Bax A (1999) Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR 13: 289–302 [DOI] [PubMed] [Google Scholar]
- Dominguez C, Boelens R, Bonvin AM (2003) HADDOCK: a protein–protein docking approach based on biochemical or biophysical information. J Am Chem Soc 125: 1731–1737 [DOI] [PubMed] [Google Scholar]
- Dudowicz J, Freed KF, Douglas JF (2000) Lattice model of living polymerization. III. Evidence for particle clustering from phase separation properties and ‘rounding' of the dynamical clustering transition. J Chem Phy 113: 434–446 [Google Scholar]
- Etemad-Moghadam B, Guo S, Kemphues KJ (1995) Asymmetrically distributed PAR-3 protein contributes to cell polarity and spindle alignment in early C. elegans embryos. Cell 83: 743–752 [DOI] [PubMed] [Google Scholar]
- Fang M, Tao YX, He F, Zhang M, Levine CF, Mao P, Tao F, Chou CL, Sadegh-Nasseri S, Johns RA (2003) Synaptic PDZ domain-mediated protein interactions are disrupted by inhalational anesthetics. J Biol Chem 278: 36669–36675 [DOI] [PubMed] [Google Scholar]
- Greer SC (1998) Physical chemistry of equilibrium polymerization. J Phys Chem B 102: 5413–5422 [Google Scholar]
- Harbury PB, Zhang T, Kim PS, Alber T (1993) A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science 262: 1401–1407 [DOI] [PubMed] [Google Scholar]
- Hirano Y, Yoshinaga S, Takeya R, Suzuki NN, Horiuchi M, Kohjima M, Sumimoto H, Inagaki F (2005) Structure of a cell polarity regulator, a complex between atypical PKC and Par6 PB1 domains. J Biol Chem 280: 9653–9661 [DOI] [PubMed] [Google Scholar]
- Hurd TW, Gao L, Roh MH, Macara IG, Margolis B (2003) Direct interaction of two polarity complexes implicated in epithelial tight junction assembly. Nat Cell Biol 5: 137–142 [DOI] [PubMed] [Google Scholar]
- Hutterer A, Betschinger J, Petronczki M, Knoblich JA (2004) Sequential roles of Cdc42, Par-6, aPKC, and Lgl in the establishment of epithelial polarity during Drosophila embryogenesis. Dev Cell 6: 845–854 [DOI] [PubMed] [Google Scholar]
- Huynh JR, Petronczki M, Knoblich JA, St Johnston D (2001) Bazooka and PAR-6 are required with PAR-1 for the maintenance of oocyte fate in Drosophila. Curr Biol 11: 901–906 [DOI] [PubMed] [Google Scholar]
- Ikegami T, Verdier L, Sakhaii P, Grimme S, Pescatore B, Saxena K, Fiebig KM, Griesinger C (2004) Novel techniques for weak alignment of proteins in solution using chemical tags coordinating lanthanide ions. J Biomol NMR 29: 339–349 [DOI] [PubMed] [Google Scholar]
- Ito T, Matsui Y, Ago T, Ota K, Sumimoto H (2001) Novel modular domain PB1 recognizes PC motif to mediate functional protein–protein interactions. EMBO J 20: 3938–3946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joberty G, Petersen C, Gao L, Macara IG (2000) The cell-polarity protein Par6 links Par3 and atypical protein kinase C to Cdc42. Nat Cell Biol 2: 531–539 [DOI] [PubMed] [Google Scholar]
- Kay LE, Gardner KH (1997) Solution NMR spectroscopy beyond 25 kDa. Curr Opin Struct Biol 7: 722–731 [DOI] [PubMed] [Google Scholar]
- Kemphues K (2000) PARsing embryonic polarity. Cell 101: 345–348 [DOI] [PubMed] [Google Scholar]
- Kemphues KJ, Priess JR, Morton DG, Cheng NS (1988) Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell 52: 311–320 [DOI] [PubMed] [Google Scholar]
- Koradi R, Billeter M, Wuthrich K (1996) MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph 14: 51–55 [DOI] [PubMed] [Google Scholar]
- Kraulis PJ (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr 24: 946–950 [Google Scholar]
- Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM (1996) AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR 8: 477–486 [DOI] [PubMed] [Google Scholar]
- Lin D, Edwards AS, Fawcett JP, Mbamalu G, Scott JD, Pawson T (2000) A mammalian PAR-3–PAR-6 complex implicated in Cdc42/Rac1 and aPKC signalling and cell polarity. Nat Cell Biol 2: 540–547 [DOI] [PubMed] [Google Scholar]
- Long JF, Tochio H, Wang P, Fan JS, Sala C, Niethammer M, Sheng M, Zhang M (2003) Supramodular structure and synergistic target binding of the N-terminal tandem PDZ domains of PSD-95. J Mol Biol 327: 203–214 [DOI] [PubMed] [Google Scholar]
- Macara IG (2004) Par proteins: partners in polarization. Curr Biol 14: R160–R162 [PubMed] [Google Scholar]
- Margolis B, Borg JP (2005) Apicobasal polarity complexes. J Cell Sci 118: 5157–5159 [DOI] [PubMed] [Google Scholar]
- Mizuno K, Suzuki A, Hirose T, Kitamura K, Kutsuzawa K, Futaki M, Amano Y, Ohno S (2003) Self-association of PAR-3-mediated by the conserved N-terminal domain contributes to the development of epithelial tight junctions. J Biol Chem 278: 31240–31250 [DOI] [PubMed] [Google Scholar]
- Nicholls A (1992) GRASP: Graphical Representation and Analysis of Surface Properties. Columbia University: New York [Google Scholar]
- Nishimura T, Yamaguchi T, Kato K, Yoshizawa M, Nabeshima Y, Ohno S, Hoshino M, Kaibuchi K (2005) PAR-6–PAR-3 mediates Cdc42-induced Rac activation through the Rac GEFs STEF/Tiam1. Nat Cell Biol 7: 270–277 [DOI] [PubMed] [Google Scholar]
- Pintacuda G, Moshref A, Leonchiks A, Sharipo A, Otting G (2004) Site-specific labelling with a metal chelator for protein-structure refinement. J Biomol NMR 29: 351–361 [DOI] [PubMed] [Google Scholar]
- Plant PJ, Fawcett JP, Lin DC, Holdorf AD, Binns K, Kulkarni S, Pawson T (2003) A polarity complex of mPar-6 and atypical PKC binds, phosphorylates and regulates mammalian Lgl. Nat Cell Biol 5: 301–308 [DOI] [PubMed] [Google Scholar]
- Shi SH, Jan LY, Jan YN (2003) Hippocampal neuronal polarity specified by spatially localized mPar3/mPar6 and PI 3-kinase activity. Cell 112: 63–75 [DOI] [PubMed] [Google Scholar]
- Su XC, Huber T, Dixon NE, Otting G (2006) Site-specific labelling of proteins with a rigid lanthanide-binding tag. Chembiochem 7: 1599–1604 [DOI] [PubMed] [Google Scholar]
- Suzuki A, Ohno S (2006) The PAR–aPKC system: lessons in polarity. J Cell Sci 119: 979–987 [DOI] [PubMed] [Google Scholar]
- Tabuse Y, Izumi Y, Piano F, Kemphues KJ, Miwa J, Ohno S (1998) Atypical protein kinase C cooperates with PAR-3 to establish embryonic polarity in Caenorhabditis elegans. Development 125: 3607–3614 [DOI] [PubMed] [Google Scholar]
- Terasawa H, Noda Y, Ito T, Hatanaka H, Ichikawa S, Ogura K, Sumimoto H, Inagaki F (2001) Structure and ligand recognition of the PB1 domain: a novel protein module binding to the PC motif. EMBO J 20: 3947–3956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Hurd TW, Margolis B (2004) Tight junction protein Par6 interacts with an evolutionarily conserved region in the amino terminus of PALS1/stardust. J Biol Chem 279: 30715–30721 [DOI] [PubMed] [Google Scholar]
- Watts JL, Etemad-Moghadam B, Guo S, Boyd L, Draper BW, Mello CC, Priess JR, Kemphues KJ (1996) par-6, a gene involved in the establishment of asymmetry in early C. elegans embryos, mediates the asymmetric localization of PAR-3. Development 122: 3133–3140 [DOI] [PubMed] [Google Scholar]
- Wilson MI, Gill DJ, Perisic O, Quinn MT, Williams RL (2003) PB1 domain-mediated heterodimerization in NADPH oxidase and signaling complexes of atypical protein kinase C with Par6 and p62. Mol Cell 12: 39–50 [DOI] [PubMed] [Google Scholar]
- Wodarz A, Ramrath A, Kuchinke U, Knust E (1999) Bazooka provides an apical cue for Inscuteable localization in Drosophila neuroblasts. Nature 402: 544–547 [DOI] [PubMed] [Google Scholar]
- Wüthrich K (1986) NMR of Proteins and Nucleic Acids. John Wiley: New York [Google Scholar]
- Zhang H, Macara IG (2006) The polarity protein PAR-3 and TIAM1 cooperate in dendritic spine morphogenesis. Nat Cell Biol 8: 227–237 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures and Legends


