Abstract
It is currently unknown how extensively the double-stranded RNA-binding protein Staufen (Stau)1 is utilized by mammalian cells to regulate gene expression. To date, Stau1 binding to the 3′-untranslated region (3′-UTR) of ADP ribosylation factor (ARF)1 mRNA has been shown to target ARF1 mRNA for Stau1-mediated mRNA decay (SMD). ARF1 SMD depends on translation and recruitment of the nonsense-mediated mRNA decay factor Upf1 to the ARF1 3′-UTR by Stau1. Here, we demonstrate that Stau1 binds to a complex structure within the ARF1 3′-UTR. We also use microarrays to show that 1.1 and 1.0% of the 11 569 HeLa-cell transcripts that were analyzed are upregulated and downregulated, respectively, at least two-fold upon Stau1 depletion in three independently performed experiments. We localize the Stau1 binding site to the 3′-UTR of four mRNAs that we define as natural SMD targets. Additionally, we provide evidence that the efficiency of SMD increases during the differentiation of C2C12 myoblasts to myotubes. We propose that Stau1 influences the expression of a wide variety of physiologic transcripts and metabolic pathways.
Keywords: myogenesis, Staufen1 binding sites, Staufen1-mediated mRNA decay, Upf1, 3′-UTRs
Introduction
Staufen (Stau)1-mediated mRNA decay (SMD) is a translation-dependent mechanism that occurs when Stau1, together with the nonsense-mediated mRNA decay (NMD) factor Upf1, is bound sufficiently downstream of a termination codon (Kim et al, 2005). The one proven physiologic target of SMD encodes ADP ribosylation factor (ARF)1, which is a G protein involved in membrane trafficking and organelle structure (Kim et al, 2005). Stau1 binds to the 3′-untranslated region (UTR) of ARF1 mRNA and triggers SMD through Upf1 when translation terminates at the normal termination codon (Kim et al, 2005).
The related pathway NMD also involves translation termination upstream of the site of Upf1 recruitment. The recruitment of Upf1 in NMD is normally mediated by the exon junction complex (EJC) of proteins that includes Upf2 and Upf3 (also called Upf3a) or Upf3X (also called Upf3b) (Maquat, 2004; Tange et al, 2004). In contrast, the recruitment of Upf1 in SMD is directly via Stau1 and does not require an EJC (Kim et al, 2005). It follows that mRNAs that are targeted for SMD generally will be distinct from mRNAs that are targeted for NMD. NMD downregulates transcripts that terminate translation more than ∼25 nt upstream of an EJC, that is, more than ∼50 nt upstream of a spliced exon–exon junction (Nagy and Maquat, 1998). In contrast, SMD appears to downregulate transcripts that terminate translation more than ∼25 nt upstream of a Stau1 binding site (SBS; Kim et al, 2005). As a rule, NMD targets derive from intron-containing genes and have undergone splicing, whereas SMD targets do not necessarily derive from intron-containing genes and are not required to undergo splicing (although many do).
NMD targets can harbor either a frameshift or a nonsense mutation (Maquat, 2004). They also include a variety of naturally occurring transcripts that contain a termination codon upstream of an exon–exon junction (Hillman et al, 2004; Mendell et al, 2004; Wittmann et al, 2006). In contrast, SMD targets are predicted to bind Stau1 within their 3′-UTR, as exemplified by ARF1 mRNA. In theory, they would also include other naturally occurring or abnormal transcripts that terminate translation sufficiently upstream of a SBS.
To exemplify another difference between SMD and NMD, NMD degrades newly synthesized mRNA that is bound by the cap-binding protein (CBP) heterodimer CBP80/20, which is also bound by EJCs (Ishigaki et al, 2001; Chiu et al, 2004; Lejeune et al, 2003; Hosoda et al, 2005). In contrast, SMD degrades both newly synthesized CBP80/20-bound mRNA and its remodeled product that is bound at the cap by eukaryotic translation initiation factor (eIF)4E (Hosoda et al, 2005) and lacks EJCs (Lejeune et al, 2002; Hosoda et al, 2005). Together, these findings suggest that SMD functions to conditionally regulate the expression of particular genes (Kim et al, 2005), whereas NMD provides a more broadly applied mechanism of quality control (Maquat, 2004; Weischenfeldt et al, 2005).
At the start of this work, our microarray studies had demonstrated that the bona fide SMD target ARF1 mRNA plus at least 22 other human transcripts bind Stau1 (Kim et al, 2005). In reality, there may be many more efficiently degraded SMD targets than those detectable by Stau1 binding considering that efficient degradation may preclude detectable binding. Furthermore, Stau1 binding is relevant to SMD only if binding is downstream of a termination codon. For example, Stau1 binding to the 5′ end of an mRNA harboring a translationally repressive structure enhances translation rather than triggers SMD (Dugre-Brisson et al, 2005). Therefore, instead of analyzing Stau1 binding, a more inclusive approach to identifying SMD targets would examine changes in cellular mRNA abundance after small interfering RNA (siRNA) had been used to reduce cellular Stau1 abundance. This approach would also lend important insight into mechanisms other than SMD by which Stau1 may regulate mRNA abundance.
Here, we undertake mutational and computational analyses of the SBS within the ARF1 3′-UTR and define structural features that are important for Stau1 binding. We also report the results of three independently performed microarray analyses that examined changes in the abundance of transcripts from 11 569 HeLa-cell genes upon Stau1 depletion. We find that ∼1% of the HeLa-cell transcriptome that was analyzed was upregulated at least two-fold and ∼1% was downregulated at least two-fold in all three transfections. Analyses of steady-state RNA using RT–PCR and primers that are specific for individual upregulated transcripts validated that depleting Stau1 increases mRNA abundance. As proof of principle, transcripts encoding (i) v-jun sarcoma virus 17 oncogene homolog (avian) (c-jun), (ii) serine (or cysteine) proteinase inhibitor clade E (nexin plasminogen activator inhibitor type 1) member 1 (Serpine1), (iii) interleukin-7 receptor (IL7R) and (iv) growth-associated protein (GAP)43 were examined in detail. The 3′-UTR of each transcript was found to bind Stau1. Additionally, each 3′-UTR was sufficient to direct an increase in the half-life of a heterologous mRNA upon Stau1 or Upf1 depletion.
From these and other results, we conclude that Stau1 regulates a wide range of physiologic transcripts and metabolic pathways in mammalian cells using SMD and, potentially, other mechanisms. In particular, we provide evidence that the efficiency of SMD increases during the differentiation of C2C12 myoblasts (MBs) to myotubes (MTs), suggesting that SMD is important for myogenesis.
Results
Stau1 binds a complex structure within the ARF1 3′-UTR
To date, the best characterized Staufen-binding site exists within Drosophila bicoid mRNA (Ferrandon et al, 1994). Linker scanning mutations that disrupt the interaction of Staufen with this mRNA mapped to three noncontiguous regions: 148 nt of stem III, 89 nt of the distal region of stem IV and 88 nt of the distal region of stem V (Ferrandon et al, 1994). Given the structural complexity of this Staufen-binding site(s), we anticipated that identifying functional features of the 300-nt human SBS within the 3′-UTR of ARF1 mRNA (Kim et al, 2005) by modeling its higher-order structure based on its nucleotide composition would be challenging. Additionally, sequence disparities between Drosophila Staufen and human Stau1 likely confer differences in RNA-binding specificity so that data pertaining to Drosophila Staufen may not be applicable to human Stau1. To complicate matters further, the binding specificity of human Stau1 could be influenced by other proteins. For example, association of the double-stranded RNA-binding domain 3 of Drosophila Staufen, which has been proposed to mediate direct binding of the protein to bicoid and oskar mRNAs (Micklem et al, 2000; Ramos et al, 2000), may be influenced by other proteins (Huynh et al, 2004). Therefore, we began to characterize the 300-nt ARF1 SBS by generating sets of deletions. Importantly, experiments were performed in vivo considering that other cellular proteins could influence Stau1-binding specificity.
Deletions were generated within a derivative of pSport-ARF1 SBS (Kim et al, 2005) that lacks SBS nucleotides 250–300 but encodes mRNA that binds Stau1 (see below). For these and all subsequent experiments, nucleotide 1 is defined as the nucleotide immediately 3′ to the normal termination codon. Initially, the set consisted of progressive 50-bp deletions from the 3′ end of the SBS (Figure 1A, left). Human 293 cells were transiently co-transfected with each pSport-ARF1 SBS deletion derivative and a Stau1-HA3 expression vector. Cell extract was prepared 2 days later, and a fraction was immunopurified using anti-HA. Western blotting of immunopurified protein using anti-HA revealed uniform Stau1-HA3 immunopurification efficiencies (Figure 1A, upper right). RT–PCR of immunopurified RNA demonstrated that nucleotides 200–249 are required for Stau1 binding because anti-HA failed to immunopurify mRNA that lacks this region at a level that is above background (Figure 1A, lower right, compare lanes 1 and 2). Consistent with this finding, anti-HA failed to immunopurify all other deletion variants lacking this region (Figure 1A, lower right, compare lane 1 with lanes 3–5).
Figure 1.
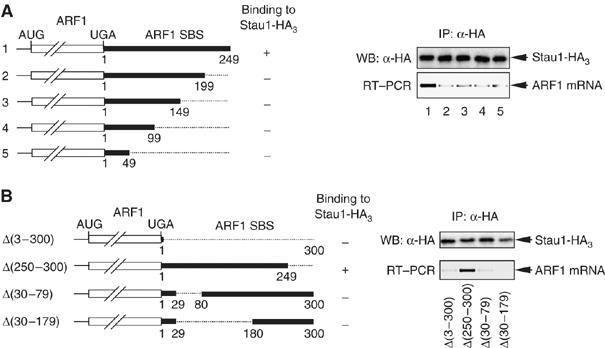
Deletions within the ARF1 SBS indicate that a central core region is required for Stau1 binding in vivo. (A) (Left) Schematic representations of the various 3′-end-deleted mRNAs that derive from pSport-ARF1 SBS derivatives. Numbering is relative to the first nucleotide following the termination codon, which is defined as 1. Plus (+) and minus (−) signs at the right of each representation indicate the ability or failure to bind Stau1, respectively. (Right) Human 293 cells were transfected with a derivative of the pSport-ARF1 SBS test plasmid and a plasmid expressing Stau1-HA3. After immunopurification (IP) using anti(α)-HA, protein was analyzed using Western blotting (WB) and anti-HA (upper), and RNA was analyzed using RT–PCR and ethidium bromide staining (lower). (B) (Left) as in (A, left) except for the pSport-ARF1 SBS derivatives that were analyzed. (Right) as in A, right. Results are representative of two independently performed experiments.
To delimit further the sequence required for Stau1 binding, additional deletions were generated within pSport-ARF1 SBS to create pSport-ARF1 SBS Δ(30–79) and pSport-ARF1 SBS Δ(30–179) (Figure 1B, left). Human 293 cells were co-transfected with each new deletion derivative or, as a positive control for Stau1 binding, pSport-ARF1 SBS Δ(250–300) and the Stau1-HA3 expression vector. RNP was immunopurified using anti-HA. Western blotting of immunopurified protein demonstrated the successful immunopurification of Stau1-HA3 (Figure 1B, upper right). RT–PCR of immunopurified RNA demonstrated that deleting nucleotides 1–300, 30–79 or 30–179 precluded Stau1 binding (Figure 1B, lower right). Therefore, sequences spanning nucleotides 30 and 79 are required for Stau1 binding (Figure 1B, lower right).
The finding that SBS nucleotides 30–79 and 200–249 are required for Stau1 binding suggests that Stau1 could associate with a folded secondary structure that involves the two nucleotide stretches instead of a contiguous sequence. To assess this possibility, the lowest energy structure of the SBS was calculated using RNAfold (Hofacker et al, 1994; Figure 2A). Strikingly, a 19-bp stem was deduced that consists of nucleotides 75–93 base-pairing to nucleotides 212–194 (Figure 2A; Supplementary Figure 1). Furthermore, this 19-bp stem is conserved among Homo sapiens, Rattus norvegicus and Mus musculus (Figure 2B). Regions of this stem overlap with the two SBS regions demonstrated by deletion mapping to be required for Stau1 binding (Figure 1).
Figure 2.
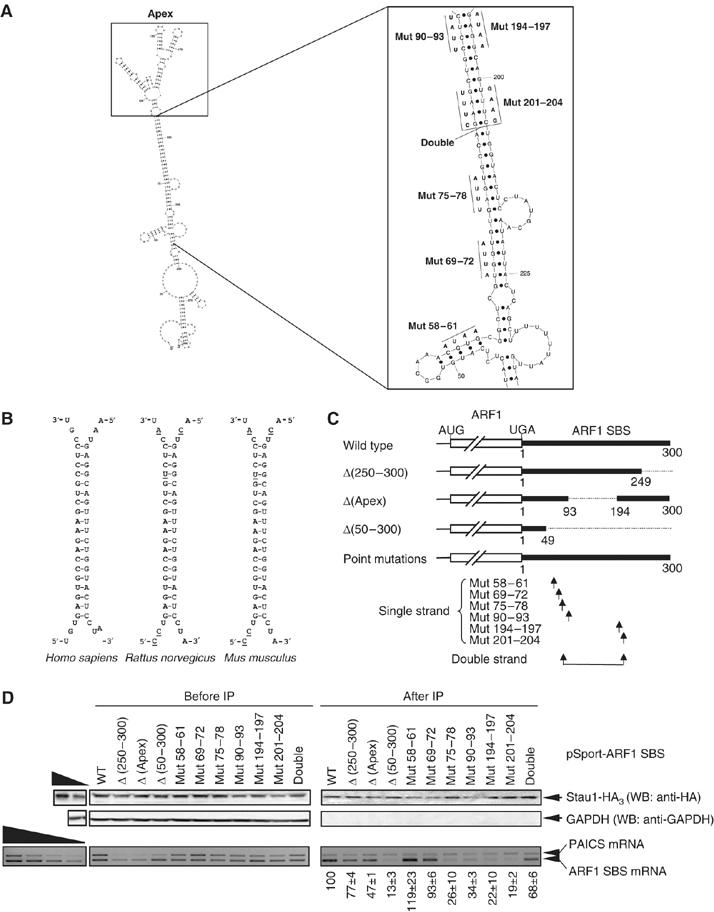
Stau1 binds in vivo to a complex structure within the ARF1 SBS. (A) Model for the secondary structure of the ARF1 SBS showing nucleotides 1 through 300. The plot was generated using Sfold v2.0 software (Wadsworth Bioinformatics Center). A larger view of the regions targeted by mutagenesis is shown in the inset to the right, along with the specific nucleotide changes that were made. See Supplementary Figure 1 for a full-page image. (B) The predicted 19-bp stem within the human ARF1 SBS is conserved in rat and mouse ARF1 mRNAs. Nucleotides that are not conserved with respect to the human sequence are underlined. (C) Schematic representation of the mutated mRNAs synthesized from pSport-ARF1 SBS derivatives. ‘Single strand' arrows indicate the relative positions of each of the 4-nt mutations, most of which individually disrupt the 19-bp stem, and ‘Double strand' arrows indicate the relative positions of the two 4-nt mutations made in cis that restore the stem. Δ(Apex) mRNA contains a replacement of nucleotides 94 through 193 that normally constitute what is predicted to be a complex structure containing a number of small loops and stems with a UGCA (see A for details). (D) As in Figure 1, except that pSport-PAICS was included in the transfections, wild-type (WT) ARF1 SBS was used as a positive control, and Δ(50–300) was used as a negative control. (Upper) Protein was analyzed before and after immunopurification (IP) using Western blotting (WB) and anti(α)-HA, or, as a negative control, anti-GAPDH. (Lower) RNA was analyzed using RT–PCR and ethidium bromide staining. The level of each ARF1-SBS-derived mRNA after IP was normalized to the level of PAICS mRNA after IP and divided by the ratio of the level of the same ARF1-SBS-derived mRNA relative to PAICS mRNA before IP. This value is defined as 100 for WT, and values for the mutated transcripts were calculated as a percentage of 100. Results are representative of three independently performed experiments that did not differ by the amount specified.
To evaluate the importance of the predicted stem to Stau1 binding, additional deletion and point-mutation variants were generated within pSport-ARF1 SBS that harbors nucleotides 1–300 (wild type, WT). Initially, 4-nt substitutions (Mut 75–78, Mut 90–93, Mut 194–197 and Mut 201–204) that disrupt the putative stem were generated (Figure 2A and C). Additionally, Mut 201–204 was placed in cis to a GAAG → CUUU mutation of nucleotides 83–86 (Double strand, also called Double; Figure 2A and C), which restores base-pairing within the stem but not the proper sequence or stacking. Finally, nucleotides 94–193 were replaced with UCGA to test whether the apical structure that is predicted to form at one end of the stem is important for Stau1 binding (Δ(Apex); Figure 2A and C).
Immunopurification of mRNA harboring Δ(50–300) served as a negative control for Stau1 binding, whereas immunopurification of WT mRNA served as a positive control for Stau1 binding. Immunopurification of mRNA that derives from pSport-PAICS (Kim et al, 2005) controlled for variations in the efficiencies of cell transfection, RNA recovery and immunopurification. Results revealed that Mut 75–78, Mut 90–93, Mut 194–197 and Mut 201–204 reduced Stau1 binding to 19–34% of WT (Figure 2D). Restoring base-pairing (Double) increased binding from 19 to 68% of WT (Figure 2D). Δ(Apex) exhibited 50% of WT binding (Figure 2D). Two other constructs, each harboring a 4-nt substitution in short predicted stems (Mut 58–61 and Mut 69–72; Figure 2A and C), had no appreciable effect on Stau1 binding (Figure 2D). These data suggest that the integrity of the 19-bp stem structure is required for Stau1 binding to the ARF1 SBS. Furthermore, sequence and/or stacking interactions within the stem contribute to binding as does the complex apical structure. Results also indicate that the levels of ARF1 SBS mRNAs that efficiently bind Stau1 are the least abundant before immunopurification, as would be expected. However, the levels of ARF1 SBS mRNAs that do not efficiently bind Stau1 are not always the most abundant before immunopurification. We conclude that some SBS mutations may variously affect Stau1 function independently of their effect on Stau1 binding by altering the binding of other trans-acting factors and/or changing RNA folding.
Identification of HeLa-cell transcripts that are regulated upon Stau1 depletion
To identify additional physiologic targets of Stau1, HeLa cells were transiently transfected with either a nonspecific control siRNA or Stau1 siRNA (Kim et al, 2005). Stau1 siRNA reduced the level of cellular Stau1 to as little as 4% of normal, where normal is defined as the level in the presence of control siRNA (data not shown). RNA from three independently performed transfections was separately hybridized to microarrays. We analyzed transcripts from 11 569 HeLa-cell genes, representing 37% of the array probe sets, in each of the three hybridization experiments. Results indicated that 124 transcripts, which correspond to 1.1% of the HeLa-cell transcriptome that was analyzed, were upregulated at least two-fold in all three transfections (Supplementary Table 1). Furthermore, 115 transcripts, which correspond to 1.0% of the HeLa-cell transcriptome that was analyzed, were downregulated at least two-fold in all three transfections (Supplementary Table 2). As expected, mRNA coding for Stau1 was among the transcripts downregulated by Stau1 siRNA.
The validity of the microarray results was tested for 12 of the upregulated transcripts and six of the downregulated transcripts using RT–PCR and a primer pair that is specific for each transcript (Supplementary Table 3). Results demonstrated that, upon Stau1 depletion, 11 of the 12 were increased in abundance by 1.5- to 8.5-fold (Supplementary Figure 2) and 6 of the 6 were decreased in abundance by 2- to 10-fold (Supplementary Figure 3). Therefore, the microarray results can be viewed as a generally reliable assessment of changes in transcript abundance upon Stau1 depletion.
We focused on transcripts that were upregulated upon Stau1 depletion. As a group, these transcripts function in a wide variety of metabolic pathways. Some produce proteins that are involved in signal transduction, cell proliferation or both (Supplementary Table 4). Others encode proteins that function in the immune response. Still others generate proteins that participate in cell adhesion, motility, the extracellular matrix or other aspects of cell structure. A number of these transcripts encode factors that regulate transcription. Others produce proteins involved in RNA metabolism, including the TIA1 cytotoxic granule-associated RNA-binding protein, which regulates the alternative splicing of pre-mRNA that encodes the human apoptotic factor Fas (Forch et al, 2002) and translationally silences mRNAs that encode inhibitors of apoptosis such as tumor necrosis factor-α (Piecyk et al, 2000; Li et al, 2004). One encodes Dcp2, which mediates global transcript decapping (Wang et al, 2002).
Stau1 or Upf1 depletion increases the abundance of c-JUN, SERPINE1 and IL7R mRNAs
Stau1 depletion could upregulate mRNA abundance directly by affecting, for example, gene transcription, pre-RNA processing, mRNA localization or mRNA half-life. Alternatively, Stau1 depletion could upregulate mRNA abundance indirectly in a mechanism that involves the product of a gene that itself is regulated transcriptionally or post-transcriptionally by Stau1. We focused on transcripts that were likely to be direct targets of SMD because ARF1 mRNA is, to date, the sole characterized SMD target. Four of the transcripts that were upregulated when Stau1 was depleted were also found in microarray analyses to be upregulated when Upf1 was depleted (Mendell et al, 2004; Supplementary Table 5). As upregulation of three of these transcripts could not be explained by the EJC-dependent rule that applies to NMD (i.e., none contain a spliced exon–exon junction situated more than ∼50 nt downstream of the termination codon and, in fact, c-JUN mRNA completely lacks exon–exon junctions), each could be an SMD target. The three transcripts encode c-jun, Serpine1 and IL7R.
To begin to determine if each transcript is an SMD target, HeLa cells were transiently transfected with one of five siRNAs (Kim et al, 2005): Stau1 or Stau1(A) siRNA, each of which targets a different Stau1 mRNA sequence; Upf1 or Upf1(A) siRNA, each of which targets a different UPF1 mRNA sequence; or a nonspecific Control siRNA. Two days later, protein and RNA were isolated and analyzed using Western blotting and RT–PCR, respectively.
Western blotting revealed that Stau1 or Stau1(A) siRNA depleted the cellular level of Stau1 to 21 or 3% of normal, respectively, and Upf1 or Upf1(A) siRNA depleted the cellular level of Upf1 to 1 or 2% of normal, respectively (Figure 3A, where normal in each case is defined as the level in the presence of Control siRNA after normalization to the level of vimentin). We found that c-JUN, SERPINE1 and IL7R transcripts were upregulated 2.1- to 9.4-fold when Stau1 or Upf1 was depleted (Figure 3B, where each transcript is normalized to the level of SMG7 mRNA). These results are consistent with the possibility that each transcript is targeted for SMD.
Figure 3.
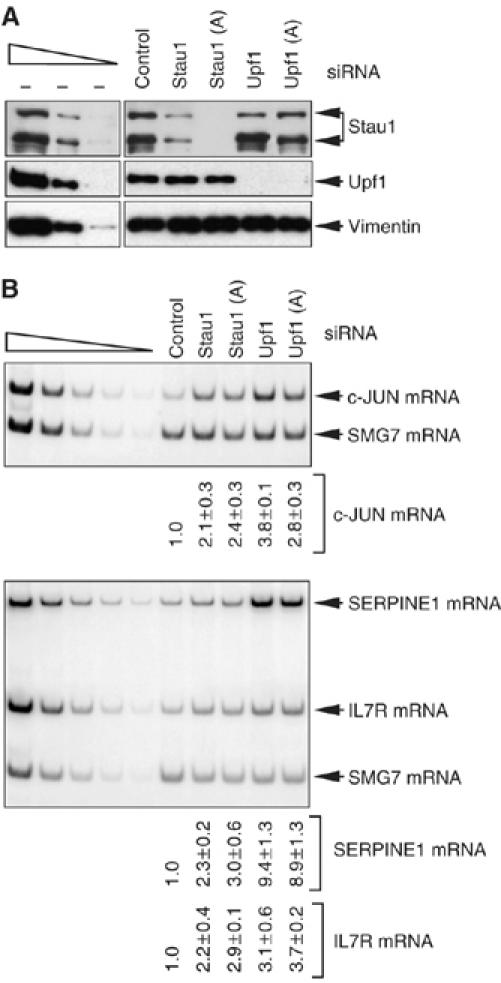
c-JUN, SERPINE1 and IL7R mRNAs are increased in abundance in human cells depleted of either Stau1 or Upf1. HeLa cells were transiently transfected with Stau1, Stau1(A), Upf1, Upf1(A) or, to Control for nonspecific depletion, Control siRNA. Three days later, protein and RNA were purified. (A) Western blot analysis using anti-Stau1, anti-Upf1 or, to control for variations in protein loading, anti-vimentin. The normal level of Stau1 or Upf1 is defined as the level in the presence of Control siRNA after normalization to the level of vimentin. (B) RT–PCR analysis of the level of endogenous c-JUN mRNA (upper), SERPINE1 mRNA or IL7R mRNA (lower). The level of each mRNA is normalized to the level of endogenous SMG7 mRNA, which is insensitive to Stau1 and Upf1 siRNAs (data not shown). The normalized level of each mRNA in the presence of Control siRNA is defined as 1. RT–PCR results are representative of three independently performed experiments that did not differ by the amount specified.
Stau1 binds the 3′-UTR of c-JUN, SERPINE1 and IL7R mRNAs
To investigate further whether the three transcripts are SMD targets, 3′-UTR sequences from each were inserted immediately downstream of the Firefly (F) luciferase (Luc) translation termination codon within pcFLuc (Kim et al, 2005; see Materials and methods). These sequences consist of (i) nucleotides 482–693 of the c-JUN 3′-UTR, which contains the 151-nt class III (i.e., non-AUUUA-containing) AU-rich element (ARE; Peng et al, 1996) plus 41 flanking nucleotides, (ii) nucleotides 1–1592 of the SERPINE1 3′-UTR or (iii) nucleotides 1–340 of the IL7R 3′-UTR. The encoded hybrid transcripts were tested for Stau1-HA3 binding.
Cos cells were transfected with the four test plasmids: pcFLuc-c-JUN 3′-UTR, pcFLuc-SERPINE1 3′-UTR, pcFLuc-IL7R 3′-UTR and pcFLuc-ARF1 SBS (Figure 4A), the latter of which serves as a positive control for Stau1-HA3 binding (Kim et al, 2005). Cells were simultaneously transfected with the Stau1-HA3 expression vector and phCMV-MUP, the latter of which serves as a negative control for Stau1-HA3 binding (Kim et al, 2005). In cells producing Stau1-HA3 (Figure 4B), anti-HA immunopurified FLuc-ARF1 SBS mRNA as well as FLuc-c-JUN 3′-UTR, FLuc-SERPINE1 3′-UTR and FLuc-IL7R 3′-UTR mRNAs but not MUP mRNA (Figure 4C). In contrast, rat (r) IgG, which controls for nonspecific immunopurification, failed to immunopurify any of the transcripts (Figure 4C). Furthermore, anti-HA failed to immunopurify FLuc mRNA that harbors the FLuc 3′-UTR (data not shown). We conclude that the 3′-UTRs of c-JUN, SERPINE1 and IL7R mRNAs bind Stau1-HA3. Notably, a larger fraction of FLuc-ARF1 SBS mRNA was bound by Stau1-HA3 relative to FLuc-c-JUN 3′-UTR, FLuc-SERPINE1 3′-UTR or FLuc-IL7R 3′-UTR mRNA (Figure 4C). This finding is consistent with detection of ARF1 mRNA but not c-JUN, SERPINE1 or IL7R mRNA in our earlier microarray analyses of transcripts that bind Stau1-HA3 (Kim et al, 2005).
Figure 4.
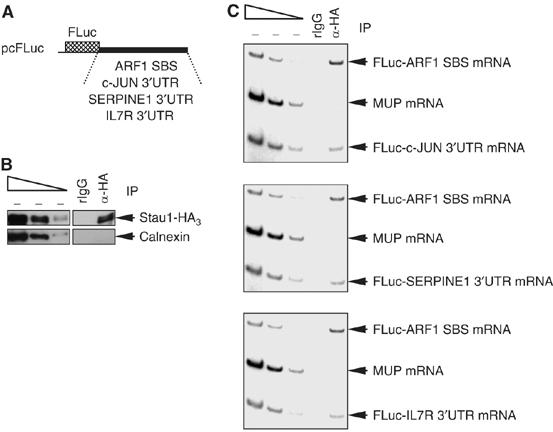
Stau1 binds within the 3′-UTR of c-JUN, SERPINE1 and IL7R mRNAs. Cos cells were transiently transfected with six plasmids: (i) pcFLuc-c-JUN 3′-UTR, pcFLuc-SERPINE1 3′-UTR and pcFLuc-IL7R 3′-UTR test plasmids; (ii) the Stau1-HA3 expression vector; (iii) the pcFLuc-ARF1 SBS test plasmid that serves as a positive control for Stau1-HA3 binding; and (iv) phCMV-MUP, which encodes MUP mRNA that serves as a negative control for Stau1-HA3 binding. Two days later, cells were lysed, and a fraction of each lysate was immunopurified using anti-HA or, as a control for nonspecific immunopurification (IP), rat (r)IgG. (A) Schematic representations of the pcFLuc-ARF1 SBS, pcFLuc-c-JUN 3′-UTR, pcFLuc-SERPINE1 3′-UTR and pcFLuc-IL7R 3′-UTR test plasmids. (B) Western blot analysis using anti(α)-HA or anti-Calnexin demonstrates that Stau1-HA3 but, as expected, not Calnexin was immunopurified. (C) RT–PCR analysis demonstrates that c-JUN, SERPINE1 and IL7R 3′-UTRs, like ARF1 SBS, bind Stau1-HA3, whereas MUP mRNA does not. Results are representative of three independently performed experiments.
c-JUN, SERPINE1 and IL7R 3′-UTRs trigger SMD
To determine if each 3′-UTR sequence is sufficient to elicit SMD, the effect of depleting Stau1 or Upf1 on the half-life of fos-FLuc-c-JUN 3′-UTR, fos-FLuc-SERPINE1 3′-UTR or fos-FLuc-IL7R 3′-UTR mRNA or, as a positive control, fos-FLuc-ARF1 SBS mRNA was tested. Production of each mRNA was driven by the fos promoter (Figure 5A). This promoter is transiently inducible upon the addition of serum to serum-deprived mouse L cells and therefore provides a way to analyze mRNA half-life (Lejeune et al, 2003; Kim et al, 2005). L cells were transfected with a mixture of two mouse (m)Stau1 siRNAs, mUpf1 siRNA or a nonspecific Control siRNA (Kim et al, 2005) and, 2 days later, with one of the four pfos-FLuc test plasmids and the phCMV-MUP reference plasmid. One day later, serum was added. Protein was purified at 0 min, and RNA was purified after 30, 45 and 60 min. Under conditions where either mStau1 or mUpf1 was depleted (Figure 5B), the half-life of each fos-FLuc mRNA was increased in each of two or three independently performed experiments (Figure 5C). Therefore, Stau1 and Upf1 together with the 3′-UTR of c-JUN, SERPINE1 or IL7R mRNA mediate mRNA decay, indicating that c-JUN, SERPINE1 and IL7R mRNAs are bona fide SMD targets. Notably, c-JUN mRNA derives from an intronless gene, consistent with SMD occurring independently of splicing (Kim et al, 2005).
Figure 5.
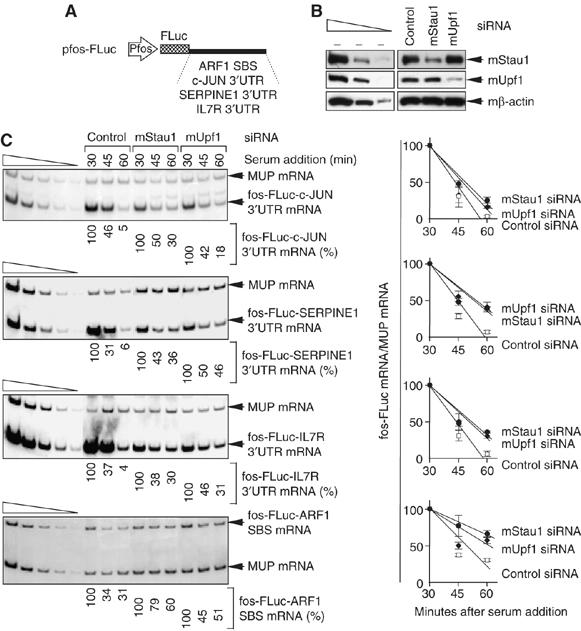
Depleting cells of Stau1 or Upf1 increases the half-life of FLuc mRNA when the 3′-UTR consists of c-JUN, SERPINE1 or IL7R 3′-UTR sequences that bind Stau1. L cells were transfected with mouse (m)Stau1, mUpf1 or Control siRNA. Two days later, cells were re-transfected with the pfos-FLuc-ARF1 SBS, pfos-FLuc-c-JUN 3′-UTR, pfos-FLuc-SERPINE1 3′-UTR and pfos-FLuc-IL7R 3′-UTR test plasmids and the phCMV-MUP reference plasmid in the absence of serum. Serum was added to 15% after an additional 24 h. Protein was immediately purified from the cytoplasmic fraction (at 0 min) for Western blot analysis. RNA was purified from the nuclear fraction for RT–PCR analysis at the specified times. (A) Schematic representations of each test plasmid. (B) Western blotting of mStau1 or mUpf1, where the level of mβ-actin serves to control for variations in protein loading. mStau1 was depleted to at least 35% of its normal level, and mUpf1 was depleted to at least 30% of its normal level. (C) (Left) RT–PCR analysis of the SMD candidate mRNAs. For each time point, the level of each mRNA transcribed from the fos promoter is normalized to the level of MUP mRNA. Normalized levels are calculated as a percentage of the normalized level of each mRNA transcribed at 30 min in the presence of each siRNA, which is defined as 100%. (Right) These levels are plotted as a function of time after serum addition. Error bars specify the extent of variation among two or three independently performed experiments.
GAP43 mRNA is an SMD target
Our data indicate that SMD is conferred by 192 nt of the c-JUN 3′-UTR, 151 nt of which constitute the c-JUN class III ARE. This finding, together with microarray data indicating the class III ARE-containing mRNA for GAP43 is also upregulated when Stau1 is depleted (Supplementary Table 4; Supplementary Figure 2), directed us to test if GAP43 mRNA is another SMD target.
Using RNA from samples analyzed in Figure 3, depleting Stau1 or Upf1 was found to increase the cellular abundance of GAP43 mRNA 7.4- or 4.1-fold, respectively (Figure 6A).
Figure 6.
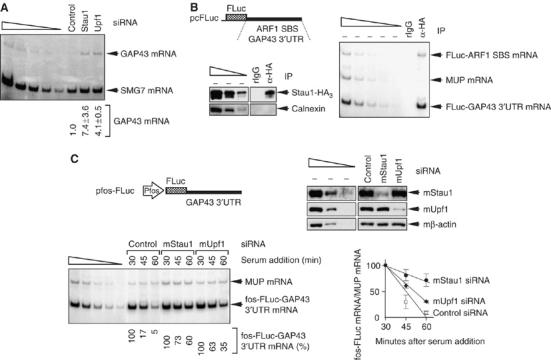
GAP43 mRNA is an SMD target. (A) RT–PCR demonstrating that depleting cells of Stau1 or Upf1 increases the cellular abundance of GAP43 mRNA. RNA derived from samples used in Figure 3. (B) As in Figure 4, except that FLuc-GAP43 3′-UTR and FLuc-ARF1 SBS mRNAs were analyzed. (C) As in Figure 5, except that fos-FLuc-GAP43 3′-UTR mRNA was analyzed. Results are representative of two independently performed experiments.
In cells producing Stau1-HA3 (Figure 6B, left), anti-HA immunopurified FLuc-GAP43 3′-UTR mRNA (Figure 6B, right), in which nucleotides 1–423 of the 3′-UTR of GAP43 mRNA were inserted immediately downstream of the FLuc translation termination codon within pcFLuc (Figure 6B, left). As expected, anti-HA also immunopurified FLuc-ARF1 SBS mRNA (Figure 6B, right). In contrast, rIgG immunopurified neither mRNA. As a control for nonspecific immunopurification, anti-HA did not immunopurify MUP mRNA (Figure 6B, right).
Finally, in experiments that utilized pfos-FLuc-GAP43 3′-UTR (Figure 6C, upper left), depleting L cells of mStau1 or mUpf1 (Figure 6C, upper right) increased the half-life of fos-FLuc-GAP43 3′-UTR mRNA (Figure 6C, lower left and right). We conclude that GAP43 mRNA is another SMD target.
Class III AREs have been defined simply as elements that signal mRNA decay via U-rich sequences rather than the AUUUA pentamer typical of class I and class II AREs (Brennan and Steitz, 2001). Other human mRNAs that contain a class III ARE within their 3′-UTRs encode β-adrenergic receptor, N-Myc and neurofibromin (Brennan and Steitz, 2001). Using mRNA-specific primers and RT–PCR, it was not possible to detect HeLa-cell mRNA for N-Myc, and neither β-adrenergic receptor nor neurofibromin mRNA appeared to be upregulated upon Stau1 depletion (Kim YK and Maquat LE, unpublished data). Thus, there is no reason to believe that class III AREs generally direct SMD.
Evidence for increased efficiency of SMD during differentiation of C2C12 MBs to MTs
In view of data indicating that SMD may affect a wide range of cellular targets, we aimed to define a physiologic circumstance under which SMD is differentially regulated. Considering that the cellular abundance of Stau1 increases during differentiation of the mouse skeletal C2C12 cell line from MBs to multinucleated MTs (Bélanger et al, 2003), we tested if the efficiency of SMD concomitantly increases. C2C12 MBs were propagated in medium containing 15% fetal bovine serum, which maintains MB morphology, or in medium containing 5% horse serum or 5% horse serum plus 200 μg/ml of heregulin, which results in differentiation to MTs (Figure 7A). Western blotting revealed that differentiation was accompanied by an increase in the level of the two mouse (m)Stau1 isoforms (Figure 7B, where the level mβ-actin controlled for variations in protein loading), as expected (Bélanger et al, 2003). Also as expected (Blais et al, 2005 and references therein), differentiation was accompanied by an increase in the levels of mMyogenin and mMyoglobin (Figure 7B). Remarkably, differentiation was further characterized by an increase in the level of Upf1 (Figure 7B).
Figure 7.
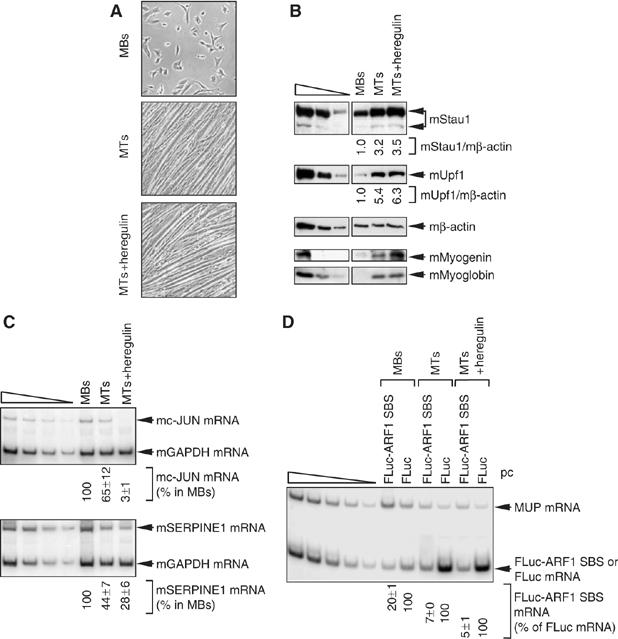
Evidence that the efficiency of SMD is upregulated during the differentiation of mouse C2C12 MBs to MTs. C2C12 cells at 20–30% confluency were maintained as MBs or induced to differentiate to MTs by exposure to 5% horse serum with or without 200 ng/ml of heregulin, a type of growth factor important for muscle development and function (Gramolini et al, 1999). (A) Light microscopy of C2C12 cells as MBs, MTs or MTs+heregulin. (B) Western blotting of mouse (m)Stau1, mUpf1, mMyogenin, mMyoglobin or, as a control for protein loading, mβ-actin. (C) RT–PCR analysis of the level of endogenous c-JUN mRNA (upper) or mSERPINE1 mRNA (lower), each of which is normalized to the level of endogenous mGAPDH mRNA. The normalized level of mc-JUN mRNA or mSERPINE1 mRNA in MBs is defined as 100%. (D) Using RNA from cells that had been transiently transfected with pcFLuc-ARF1 SBS or pcFLuc and phCMV-MUP, RT–PCR analysis of FLuc-ARF1 SBS or FLuc mRNA, each of which is normalized to the level of MUP mRNA. The normalized level of FLuc mRNA under each of the three conditions is defined as 100%. All RT–PCR results are representative of two independently performed experiments that did not differ by the amount specified.
The increased abundance of mStau1 and mUpf1 could indicate an increase in the efficiency of SMD. RT–PCR revealed that differentiation was indeed accompanied by a 33- and 4-fold decrease, respectively, in the levels of cellular mc-JUN and mSERPINE1 mRNAs (Figure 7C, where the level of cellular mGAPDH mRNA controlled for variations in RNA loading), which are proven SMD targets in HeLa cells (Figures 3, 4, 5 and 6). To more specifically measure SMD, cells were transiently transfected with two plasmids: (i) the pcLuc-ARF1 SBS test plasmid or, as a negative control for Stau1 function, pcFLuc and (ii) the phCMV-MUP reference plasmid. Upon differentiation, the level of FLuc-ARF1 SBS mRNA decreased 3- to 4-fold relative to the level of SMD-insensitive FLuc mRNA (Figure 7D, where the level of MUP mRNA controlled for variations in transfection efficiencies and RNA loading). Taken together, these data suggest that the efficiency of SMD increases during myogenesis.
Discussion
In this work, we present the analysis of transcripts from 11 569 HeLa-cell genes and report that 1.1% were upregulated and 1.0% were downregulated in three independently performed experiments when the cellular abundance of Stau1 was depleted (Supplementary Tables 1 and 2). These results suggest that Stau1 regulates a wide variety of transcripts by affecting their synthesis, processing, transport, localization or decay. Of those transcripts that are upregulated upon Stau1 depletion, we demonstrate that human c-JUN, SERPINE1, IL7R and GAP43 mRNAs, in addition to ARF1 mRNA (Kim et al, 2005), are targeted for SMD by a mechanism that depends on Stau1 binding to 3′-UTR sequences (Figures 3, 4, 5 and 6). At least in theory, some of the other transcripts that are upregulated upon Stau1 depletion could be regulated independently of Upf1. Transcripts that are downregulated upon Stau1 depletion could be controlled by transcriptional or post-transcriptional factors that are encoded by an SMD target. As another possibility, these transcripts could be protected from degradation through Stau1-mediated enhanced translation (Dugre-Brisson et al, 2005).
It is very likely that ARF1, c-JUN, SERPINE1, IL7R and GAP43 mRNAs are not the only transcripts upregulated upon Stau1 depletion that are SMD targets. For example, CYR61 mRNA, which encodes the cysteine-rich angiogenic inducer 61, was upregulated upon Stau1 depletion in only two of our three microarray analyses and, thus, fell below the stringent criteria we used to define a candidate SMD target. However, this mRNA may very well be targeted for SMD as it was also among the transcripts upregulated upon Upf1 depletion (Mendell et al, 2004). Furthermore, of 21 transcripts that were upregulated upon Stau1 depletion in all three of our microarray analyses but not studied further, 10 were present in two and 11 were present in all three new microarray analyses that assayed for Stau1-HA3 binding (Supplementary Table 6). Nevertheless, there are five features that we advise should be observed, as we have done here, before considering an mRNA to be a bona fide SMD target. First, the mRNA must be upregulated when the cellular abundance of Stau1 is reduced. Second, the mRNA must be upregulated when the cellular abundance of Upf1 is reduced. Third, the mRNA must bind Stau1 downstream of the termination codon. Fourth, sequences downstream of the termination codon must confer a lengthened mRNA half-life upon the depletion of Stau1. Fifth, sequences downstream of the termination codon must confer a lengthened mRNA half-life upon the depletion of Upf1.
We also provide evidence that the efficiency of SMD increases during the differentiation of mouse C2C12 MBs to MTs (Figure 7). First, the cellular abundance of Stau1 and Upf1 increases during C2C12 myogenesis. Second, the cellular abundance of endogenous c-JUN and SERPINE1 mRNAs decreases during C2C12 myogenesis. Third, providing a more specific assay for SMD, the cellular abundance of transiently expressed FLuc-ARF1 SBS mRNA, relative to the level of SMD-insensitive FLuc mRNA, decreases during C2C12 myogenesis. Despite progress toward understanding myogenesis as a transcriptionally regulated process (see, e.g., Blais et al, 2005 and references therein), a more complete understanding of myogenesis will surely derive from future studies of post-transcriptional control in general and SMD in particular.
The best characterized binding site for Drosophila Staufen resides within the 3′-UTR of bicoid mRNA. Deletion and linker scanning analyses suggest that binding requires three noncontiguous regions of bicoid mRNA that correspond to stem III and distal portions of stems IV and V (Ferrandon et al, 1994). Each bicoid mRNA region forms stems that are interrupted by bulges and interior loops of different sizes (Ferrandon et al, 1994). The finding that an RNA stem alone is insufficient for Drosophila Staufen binding suggested that the same may also be true for human Stau1 binding.
We show here that a 19-bp stem within the human ARF1 SBS is required for human Stau1 binding in vivo in a way that depends on nucleotide composition (Figures 1 and 2). In support of its importance to Stau1 binding, this stem is conserved within the 3′-UTRs of rat and mouse ARF1 mRNAs: the one nucleotide difference that typifies rat and mouse sequences as compared to the human sequence converts a C-G base-pair to a U-G base-pair (Figure 2B). Our in vivo binding data indicate that disrupting the 19-bp stem within the human ARF1 SBS can reduce Stau1 binding to as low as 19% of the WT level (Figure 2). Furthermore, restoring the stem but not its WT sequence rescues Stau1 binding, although to only 68% of the WT level (Figure 2). This suggests that nucleotide sequence and/or stacking interactions influence Stau1 binding. Additionally, the 14-bp stem that remains in ARF1 SBS Δ(30–79) mRNA does not support Stau1 binding, indicating that other features in addition to the stem may be required for binding efficacy (Figure 1B). One of these features appears to reside within the 100-nt apex of the stem, which when deleted results in only 50% of WT binding (Figure 2). Computational modeling of the SBSs within c-JUN, SERPINE1, IL7R and GAP43 mRNAs is consistent with the view that fully functional SBSs are generally much more complex than a single long stem. None of the four SBSs is predicted to contain an uninterrupted stem that is more than 12 bp (data not shown).
If parallels can be drawn using another member of the family of dsRNA-binding proteins, ADAR1, Stau1-specific binding is likely to depend on the positions and lengths of bulges and interior loops relative to a stem structure (Lehmann and Bass, 1999). Irregularities in RNA helices due to bulged nucleotides or distortions of grooves are generally required to provide specific determinants for protein binding (Draper, 1999; Hallegger et al, 2006). Thus, there are undoubtedly other features of RNA beside a stem that are recognized by Stau1 and, possibly, one or more other proteins that comprise the Stau1-binding complex (Saunders and Barber, 2003). Furthermore, the possibility of protein-induced stem formation cannot be excluded considering that RNA binding proteins are known to induce higher-order RNA structures (Wozniak et al, 2005). Given the complexity of characterized RNA structures that bind proteins, including the ARF1 SBS and the Staufen-binding site within bicoid mRNA, future work that aims to define the exact SBS nucleotides required for Stau1 binding will be quite challenging.
In summary, our results suggest that Stau1 influences the expression of a wide range of physiologic transcripts. While an understanding of the extent of its influence will require further studies, SMD can be added to the growing list of homeostatic gene control mechanisms.
Materials and methods
Plasmid constructions
Details are available in Supplementary data.
Cell culture and transfections, and protein and RNA purification
Human HeLa or 293 cells, monkey Cos cells or mouse L cells were propagated in Dulbecco's modified Eagle's medium (DMEM; Gibco-BRL) containing 10% fetal bovine serum (Gibco-BRL). Where specified, 2 × 106 HeLa or 293 cells were transiently transfected with plasmid DNA, in vitro-synthesized siRNA, or both as described (Kim et al, 2005). Monkey Cos cells (4 × 107), which were used in experiments involving the immunopurification of c-JUN, SERPINE1, IL7R and GAP43 transcripts, were transfected using calcium phosphate and 6 μg of pcFLuc-ARF1 SBS, 6 μg of a pcFLuc derivative and 10 μg of pCDNA3-hStau1-HA3. Mouse L cells were transfected and the fos-promoter was induced as described (Kim et al, 2005). Mouse C2C12 cells (ATCC) were propagated as MBs in DMEM containing 15% fetal bovine serum and induced to differentiate to MTs (Gramolini et al, 1998, 1999; Bélanger et al, 2003) using DMEM containing 5% horse serum (Gibco-BRL) with or without 200 ng/ml of heregulin (Sigma). When indicated, cells (4 × 106) were transiently transfected with 0.6 μg of pcFLuc-ARF1 SBS or pcFLuc and 0.1 μg of phCMV-MUP using Lipofectamine Plus (Invitrogen) 2 days before cell lysis.
Total cell protein or RNA or nuclear RNA were prepared as reported (Kim et al, 2005).
Confocal microscopy
C2C12 cells were visualized using an MRC-1024 laser scanning microscope (Bio-Rad Laboratories) equipped with an Axiovert 100 microscope (Zeiss) at an excitation wavelength of 488 nm.
Western blotting and RNA analysis
Western blotting was as described previously (Kim et al, 2005).
In the immunopurification of ARF1 transcripts shown in Figures 1 and 2, RNA was recovered and digested with Turbo DNAse (Ambion), which was subsequently removed with DNAse Removal Reagent (Ambion). To amplify PAICS or ARF1 transcripts simultaneously, multiplex RT–PCR was performed using the One-Step RT–PCR kit (Qiagen), the common sense primer 5′-AGGCTGGTACCGGTCCGGAATTC-3′ and 5′-GCAGCAATCAGCTGTTCCTCAGACC-3′ (antisense) or 5′-CTCTGTCATTGCTGTCCACCACG-3′ (antisense), respectively.
The RT–PCR of SMG7 and MUP mRNAs was as noted previously (Lejeune et al, 2003; Chiu et al, 2004). FLuc-c-JUN 3′-UTR, FLuc-SERPINE1 3′-UTR, FLuc-IL7R 3′-UTR, FLuc-GAP43 3′-UTR or FLuc-ARF1 SBS mRNA was amplified using 5′-AATACGACTCACTATAGGGA-3′ (sense, which annealed to the T7 promoter that resides downstream of the CMV promoter) and, 5′-AGGCAGGCCAGAAAGAGTTC-3′ (antisense), 5′-TGAAGGCGTCTTTCCCCAGG-3′ (antisense), 5′-TCAGTCTGGGTTTCTTACAC-3′ (antisense), 5′-TGGAAAGCCATTTCTTAGAG-3′or 5′-GCCTGGCCGCAGGCTGCGTC-3′ (antisense), respectively.
fos-FLuc-c-JUN 3′-UTR, fos-FLuc-SERPINE1 3′-UTR, fos-FLuc-IL7R 3′-UTR, fos-FLuc-GAP43 3′-UTR or fos-FLuc-ARF1 SBS mRNA that derived from pfos-FLuc constructs was amplified using the common primer 5′-AGACTGAGCCGATCCCGCGC-3′ (sense) and the corresponding antisense primer described above.
Primer pairs for human c-JUN, SERPINE1, IL7R and GAP43 mRNAs and the 21 additional transcripts that were amplified to test the validity of microarray results are provided (Supplementary Table 3).
Primer pairs for mouse c-JUN, SERPINE1 and GAPDH mRNAs were: 5′-CTGCAAAGATGGAAACGACC-3′ (sense) and 5′-CGGAGGCTCACTGTGCAGGC-3′ (antisense), 5′-TTGCTTGCCTCATCCTGGGC-3′ (sense) and 5′-GTCATTGATCATACCTTTGG-3′ (antisense), and 5′-GGTGTGAACGGATTTGGCCG-3′ (sense) and 5′-CCCCGGCCTTCTCCATGGTG-3′ (antisense), respectively.
Microarray analyses
HeLa-cell RNA was purified using TriZol reagent (Invitrogen) and deemed to be intact using an RNA 6000 Nano LabChip (Agilent) together with a Bioanalyser 2100 and Biosizing software (Agilent). Biotin-labeled cRNAs were generated and hybridized to U133 Plus 2.0 Array human gene chips (comprising 52 245 probe sets that correspond to 29 555 unique genes). Hybridized chips were scanned using an Affymetrix GeneChip 3000 Scanner. Results were recorded using the GeneChip Operating Software platform, which includes the GeneChip Scanner 3000 high-resolution scanning patch that enables feature extraction (Affymetrix). Notably, the Affymetrix Gene Expression Assay identifies changes that are greater than two-fold with 98% accuracy (Wodicka et al, 1997). Arrays were undertaken using three independently generated RNA samples. Transcripts that showed at least a two-fold increase in abundance with a P-value of less than 0.05 in each of the three analyses were scored as potential SMD targets. Transcripts that showed at least a two-fold decrease in abundance with a P-value of less than 0.05 in each of the three analyses were scored as downregulated upon Stau1 depletion. Microarray data have been deposited in the GEO database and are available through the accession number GSE6679.
Bioinformatics prediction of RNA secondary structures
Optimal secondary structure predictions were obtained by using the RNAfold computer program (Hofacker et al, 1994) of the Vienna RNA package (Hofacker et al, 1994). Statistical samples of optimal and suboptimal secondary structures were evaluated using the Sfold algorithm (Ding et al, 2004).
Supplementary Material
Supplementary Figures
Supplementary Table 1
Supplementary Table 2
Supplementary Table 3
Supplementary Table 4
Supplementary Table 5
Supplementary Table 6
Supplementary Material
Acknowledgments
We thank the Genome Quebec Innovation, in particular André Ponton, for microarray screening and analysis, Juan Ortín for anti-human Stau1, Tom Gelehrter and Ann-Bin Shyu for helpful conversations and Chris Proschel for assistance with light microscopy. We are also grateful to Maquat laboratory members Liz Wolcott and Alma Muharemagic for technical assistance and Daiki Matsuda and Holly Kuzmiak for help in preparing the manuscript. This work was supported by National Institutes of Health grant GM074593 to LEM and the Natural Sciences and Engineering Research Council of Canada (NSERC) to LDG. LF was supported by scholarships from Fonds de la recherche en santé du Québec (FRSQ) and NSERC. FM is a Canadian Institutes of Health Research Investigator.
References
- Bélanger G, Stocksley MA, Vandromme M, Schaeffer L, Furic L, DesGroseillers L, Jasmin BJ (2003) Localization of the RNA-binding proteins Staufen1 and Staufen2 at the mammalian neuromuscular junction. J Neurochem 86: 669–677 [DOI] [PubMed] [Google Scholar]
- Blais A, Tsikitis M, Acosta-Alvear D, Sharan R, Kluger Y, Dynlacht BD (2005) An initial blueprint for myogenic differentiation. Genes Dev 19: 553–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan CM, Steitz JA (2001) HuR and mRNA stability. Cell Mol Life Sci 58: 266–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu SY, Lejeune F, Ranganathan AC, Maquat LE (2004) The pioneer translation initiation complex is functionally distinct from but structurally overlaps with the steady-state translation initiation complex. Genes Dev 18: 745–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Chan CY, Lawrence CE (2004) Sfold web server for statistical folding and rational design of nucleic acids. Nucleic Acids Res 32: W135–W141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper DE (1999) Themes in RNA-protein recognition. J Mol Biol 293: 255–270 [DOI] [PubMed] [Google Scholar]
- Dugre-Brisson S, Elvira G, Boulay K, Chatel-Chaix L, Mouland AJ, DesGroseillers L (2005) Interaction of Staufen1 with the 5′ end of mRNA facilitates translation of these RNAs. Nucleic Acids Res 33: 4797–4812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrandon D, Elphick L, Nusslein-Volhard C, St Johnston D (1994) Staufen protein associates with the 3′UTR of bicoid mRNA to form particles that move in a microtubule-dependent manner. Cell 79: 1221–1232 [DOI] [PubMed] [Google Scholar]
- Forch P, Puig O, Martinez C, Séraphin B, Valcárcel J (2002) The splicing regulator TIA-1 interacts with U1-C to promote U1 snRNP recruitment to 5′ splice sites. EMBO J 21: 6882–6892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramolini AO, Angus LM, Schaeffer L, Burton EA, Tinsley JM, Davies KE, Changeux JP, Jasmin BJ (1999) Induction of utrophin gene expression by heregulin in skeletal muscle cells: role of the N-box motif and GA binding protein. Proc Natl Acad Sci USA 96: 3223–3227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramolini AO, Burton EA, Tinsley JM, Ferns MJ, Cartaud A, Cartaud J, Davies KE, Lunde JA, Jasmin BJ (1998) Muscle and neural isoforms of agrin increase utrophin expression in cultured myotubes via a transcriptional regulatory mechanism. J Biol Chem 273: 736–743 [DOI] [PubMed] [Google Scholar]
- Hallegger M, Taschner A, Jantsch MF (2006) RNA aptamers binding the double-stranded RNA-binding domain. RNA 12: 1993–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillman RT, Green RE, Brenner SE (2004) An unappreciated role for RNA surveillance. Genome Biol 5: R8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker IL, Fontana W, Stadler PF, Bonhoeffer S, Tacker M, Schuster P (1994) Fast folding and comparison of RNA secondary structures. Monatshefte Chemie 125: 167–188 [Google Scholar]
- Hosoda N, Kim YK, Lejeune F, Maquat LE (2005) CBP80 promotes interaction of Upf1 with Upf2 during nonsense-mediated mRNA decay in mammalian cells. Nat Struct Mol Biol 12: 893–901 [DOI] [PubMed] [Google Scholar]
- Huynh JR, Munro TP, Smith-Litiere K, Lepesant JA, St Johnston D (2004) The Drosophila hnRNPA/B homolog, Hrp48, is specifically required for a distinct step in osk mRNA localization. Dev Cell 6: 625–635 [DOI] [PubMed] [Google Scholar]
- Ishigaki Y, Li X, Serin G, Maquat LE (2001) Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell 106: 607–617 [DOI] [PubMed] [Google Scholar]
- Kim YK, Furic L, Desgroseillers L, Maquat LE (2005) Mammalian Staufen1 recruits Upf1 to specific mRNA 3′UTRs so as to elicit mRNA decay. Cell 120: 195–208 [DOI] [PubMed] [Google Scholar]
- Lehmann KA, Bass BL (1999) The importance of internal loops within RNA substrates of ADAR1. J Mol Biol 291: 1–13 [DOI] [PubMed] [Google Scholar]
- Lejeune F, Ishigaki Y, Li X, Maquat LE (2002) The exon junction complex is detected on CBP80-bound but not eIF4E-bound mRNA in mammalian cells: dynamics of mRNP remodeling. EMBO J 21: 3536–3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune F, Li X, Maquat LE (2003) Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol Cell 12: 675–687 [DOI] [PubMed] [Google Scholar]
- Lejeune F, Maquat LE (2004) Immunopurification and analysis of protein and RNA components of mRNP in mammalian cells. Methods Mol Biol 257: 115–124 [DOI] [PubMed] [Google Scholar]
- Li W, Simarro M, Kedersha N, Anderson P (2004) FAST is a survival protein that senses mitochondrial stress and modulates TIA-1-regulated changes in protein expression. Mol Cell Biol 24: 10718–10732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat LE (2004) Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol 5: 89–99 [DOI] [PubMed] [Google Scholar]
- Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC (2004) Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet 36: 1073–1078 [DOI] [PubMed] [Google Scholar]
- Micklem DR, Adams J, Grunert S, St Johnston D (2000) Distinct roles of two conserved Staufen domains in oskar mRNA localization and translation. EMBO J 19: 1366–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy E, Maquat LE (1998) A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem Sci 23: 198–199 [DOI] [PubMed] [Google Scholar]
- Peng SS, Chen CY, Shyu AB (1996) Functional characterization of a non-AUUUA AU-rich element from the c-jun proto-oncogene mRNA: evidence for a novel class of AU-rich elements. Mol Cell Biol 16: 1490–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piecyk M, Wax S, Beck AR, Kedersha N, Gupta M, Maritim B, Chen S, Gueydan C, Kruys V, Streuli M, Anderson P (2000) TIA-1 is a translational silencer that selectively regulates the expression of TNF-alpha. EMBO J 19: 4154–4163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos A, Grunert S, Adams J, Micklem DR, Proctor MR, Freund S, Bycroft M, St Johnston D, Varani G (2000) RNA recognition by a Staufen double-stranded RNA-binding domain. EMBO J 19: 997–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders LR, Barber GN (2003) The dsRNA binding protein family: critical roles, diverse cellular functions. FASEB J 17: 961–983 [DOI] [PubMed] [Google Scholar]
- Tange TO, Nott A, Moore MJ (2004) The ever-increasing complexities of the exon junction complex. Curr Opin Cell Biol 16: 279–284 [DOI] [PubMed] [Google Scholar]
- Wang Z, Jiao X, Carr-Schmid A, Kiledjian M (2002) The hDcp2 protein is a mammalian mRNA decapping enzyme. Proc Natl Acad Sci USA 99: 12663–12668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weischenfeldt J, Lykke-Andersen J, Porse B (2005) Messenger RNA surveillance: neutralizing natural nonsense. Curr Biol 15: R559–R562 [DOI] [PubMed] [Google Scholar]
- Wittmann J, Hol EM, Jack HM (2006) hUPF2 silencing identifies physiologic substrates of mammalian nonsense-mediated mRNA decay. Mol Cell Biol 26: 1272–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodicka L, Dong H, Mittmann M, Ho MH, Lockhart DJ (1997) Genome-wide expression monitoring in Saccharomyces cerevisiae. Nat Biotechnol 15: 1359–1367 [DOI] [PubMed] [Google Scholar]
- Wozniak AK, Nottrott S, Kuhn-Holsken E, Schroder GF, Grubmuller H, Luhrmann R, Seidel CA, Oesterhelt F (2005) Detecting protein-induced folding of the U4 snRNA kink-turn by single-molecule multiparameter FRET measurements. RNA 11: 1545–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures
Supplementary Table 1
Supplementary Table 2
Supplementary Table 3
Supplementary Table 4
Supplementary Table 5
Supplementary Table 6
Supplementary Material


