Abstract
A blood clot is a meshwork of fibrin fibers built up by the systematic assembly of fibrinogen molecules proteolyzed by thrombin. Here, we describe a model of how the assembly process occurs. Five kinds of interaction are explicitly defined, including two different knob–hole interactions, an end-to-end association between γ-chains, a lateral association between γ-chains, and a hypothetical lateral interaction between β-chains. The last two of these interactions are responsible for protofibril association and are predicated on intermolecular packing arrangements observed in crystal structures of fibrin double-D fragments cocrystallized with synthetic peptides corresponding to the knobs exposed by the release of the fibrinopeptides A and B.
Keywords: blood clots, crystal packing, protofibrils, macromolecular assembly
A principal aim of structural studies on fibrinogen is to understand the details of how the units pack together in fibrin. The recent publication of several high-resolution crystal structures of major fragments from fibrinogen and fibrin represents significant progress toward meeting that goal (1–5), especially with regard to the interaction of “knobs,” exposed on the thrombin-catalyzed release of fibrinopeptides A, with “holes” located near the ends of other fibrinogen molecules.
It is generally accepted that the thrombin-catalyzed removal of the fibrinopeptides A leads to the formation of intermediate protofibrils composed of staggered, half-molecule overlaps, the end results of which are two-molecule thick, end-to-end, noncovalent oligomers (6). It is also part of current dogma that the subsequent release of fibrinopeptide B from the units in these oligomers encourages the lateral association that leads to the thick fibers that constitute natural clots (7). These conclusions were reached well before there was any structural information available about the relative locations of the sites involved. Gradually, it became clear that the release of the fibrinopeptide A leads to the exposure of centrally located Gly-Pro-Arg- “knobs” that can fit into “holes” on the distally located γ-chain carboxyl domains (γC domains). Extension of the process leads to the formation of the intermediate protofibrils. A tremendous body of biochemical evidence was accumulated to this end, including the identification of many variant human fibrinogens with amino acid substitutions in the critical areas (8), as well as ligand-binding studies with synthetic peptides (9) and the photo-affinity labeling of γ-chain residue Tyr363 by Gly-Pro-Arg derivatives (10).
Evidence for the role of the “B knob,” which is exposed by the release of fibrinopeptide B, which begins with the sequence Gly-His-Arg-, has been less direct. Indeed, the necessity of B-knob involvement has been questioned (11), and the location or even the existence of a corresponding “hole” for B knobs has never been indicated by variant fibrinogens. And, although synthetic A-knob peptides (GPR derivatives) are effective inhibitors of fibrin polymerization, synthetic B-knobs peptides (GHR derivatives) are not, even though they bind to fibrinogen and fragment D (9). Remarkably, GHR peptides actually accelerate fibrin formation (9). Moreover, fibrin can be formed by the removal of fibrinopeptide A only, as can be demonstrated with various snake venom enzymes (12). Nonetheless, such fibrin is not of the same quality as fibrin generated by the thrombin-catalyzed removal of both fibrinopeptides A and B, and the physiological role of removing the latter must be significant.
Here, we describe a detailed model for each of the steps in fibrin formation, including especially the lateral association of protofibrils, the least understood step in the process. Our proposal for how this step occurs is mainly based on packing interactions observed in crystal structures of the core fragments D (from fibrinogen) and DD (from crosslinked fibrin) complexed with the two different synthetic peptides that correspond to the natural A and B knobs (3–5). These considerations have been used in conjunction with a recently reported low-resolution structure of native chicken fibrinogen (13) as the framework model for the units being bundled together.
Methods
Extensive use was made of the o modeling program (14); superposition of double-D structures and that of native chicken fibrinogen was accomplished with the least squares function (lsq) in that package. The linear growth of model structures was managed with the pdbset routine contained in the CCP4 package (15); symmetry operations were based on the chicken fibrinogen structure (13). Estimates of the strengths of interaction at each step of the polymerization process were obtained from measures of lost solvent accessibility (16); in particular, the method of Lee and Richards (17) as provided in the CCP4 package (15) was used with a probe radius 1.4 Å.
Illustrations were prepared with molscript (18) and rendered with RASTER3D (19, 20). The program xtalview (21) was used to determine the orientation of objects in the illustrations. Some molecular surface depictions were obtained with grasp (22) and then combined with other features in RASTER3D (19, 20).
Basis for the Model.
The model is based on a consideration of packing arrangements in a series of previously reported crystal structures. In particular, a 170-kDa fragment derived by limited proteolysis of naturally crosslinked human fibrin, denoted “double-D,” was crystallized in the presence or absence of two synthetic peptides patterned on sequences that constitute the “knobs” in a series of “knob–hole” interactions (3–5). The synthetic knobs, A and B, which differ by only a single amino acid, fit into homologous but different holes on the γC and βC domains of the double-D, respectively (4). The surrogate A knob has the sequence Gly-Pro-Arg-Pro-amide (GPRPam) and the B knob, Gly-His-Arg-Pro-amide (GHRPam).
The double-D structure designated DD-GP was cocrystallized with GPRPam only (PDB code, 1FZB), DD-GH with only GHRPam (PDB code, 1FZF), DD-BOTH with both ligands (PDB code, 1FZC), and DD-NL with no ligands at all (PDB code, 1FZE). Although the crystal structures of the four preparations (DD-GP, DD-GH, DD-BOTH, and DD-NL) are very similar, their unit cells all differ significantly (Table 1). Our working hypothesis is that interactions that occur in the various crystals are good candidates for those that occur in fiber formation. We know this to be true in the case of the principal end-to-end association between fibrin units (5). Thus, a natural complementarity exists between the ends of neighboring fibrinogen molecules that involves γ-chain residues 270–310 (human numbering will be used throughout this paper). These end-to-end interactions are observed in all four double-D structures (Fig. 1)—where the ends are covalently crosslinked—as well as in the crystal packing of all known fragment D structures (3–5), a modified bovine fibrinogen (23), and native chicken fibrinogen (13). These end-to-end associations always have an offset nature; in human fibrinogen derivatives, γArg275 of one molecule is directed toward γTyr280 of the second, and γArg275 of the second is directed back toward γSer300 of the first (3).
Table 1.
Unit cells and space groups for four different structures of double-D
| Structure | PDB index | Space group | Unit cell
|
Resolution, Å | |||
|---|---|---|---|---|---|---|---|
| a, Å | b, Å | c, Å | β, deg | ||||
| DD-NL | 1FZE | P21 | 108.0 | 48.6 | 166.4 | 104.6 | 3.0 |
| DD-GP | 1FZB | P21 | 93.8 | 96.5 | 113.8 | 96.1 | 2.9 |
| DD-BOTH | 1FZC | P21 | 83.4 | 95.6 | 113.6 | 90.2 | 2.3 |
| DD-GH | 1FZF | P212121 | 54.8 | 149.4 | 234.7 | — | 2.7 |
Adapted from Everse et al. (5).
Figure 1.
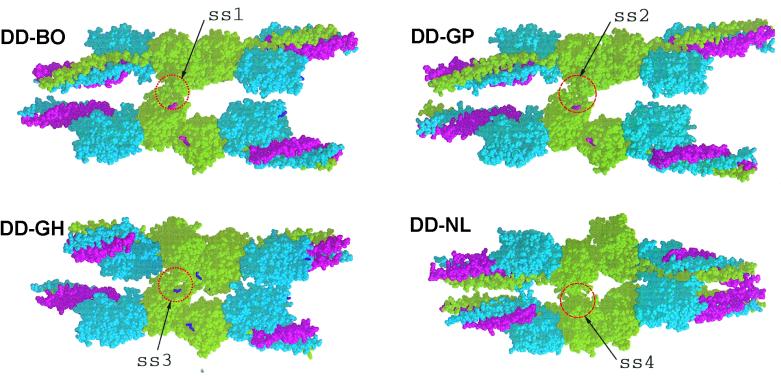
Four DD structures: DD-BO (PDB code 1FZC); DD-GP (PDB code 1FZB); DD-GH (PDB code 1FZF); and DD-NL (PDB code 1FZE) showing crystal packing with neighboring molecules. The lateral associations (ss = side-by-side) involving γC domains are circled and denoted ss1, ss2, ss3, and ss4. The two structures with GPR- knobs in the γ-chain holes (DD-BO and DD-GP) are virtually identical. When the same holes are occupied by Gly-His-Arg knobs (DD-GH), the lateral interaction is slightly shifted. When these holes are unoccupied (DD-NL), the packing is completely different (ss4), the adjacent domains being greatly rotated relative to each other compared with all of the other structures. Red, α-chains; blue, β-chains; green, γ-chains. The peptide ligand GPRPam (A knob) is shown in magenta; the B knob (GHRPam) is shown in purple.
For lateral interactions, we focused on those that accommodate the 230-Å periodicity known to exist in fibrin fibers (24), which is to say, protofibrils must be associated laterally in a fashion that keeps them in exact register. In contrast to the end-to-end situation, the nature of lateral packing arrangements in crystals depends on the presence or absence of the two synthetic knobs. Of most importance is the occupancy of the γC hole by the A knob, a situation that exists in two of the structures (DD-GP and DD-BOTH). In these cases, the crystal packing is such that a specific set of lateral interactions occurs between γC domains (Figs. 1 and 2). A similar but slightly different situation exists when only the B knob is present (DD-GH). The DD-GH is in a sense an unnatural one in that the B knob occupies the γC hole only in the absence of A knobs (5).
Figure 2.
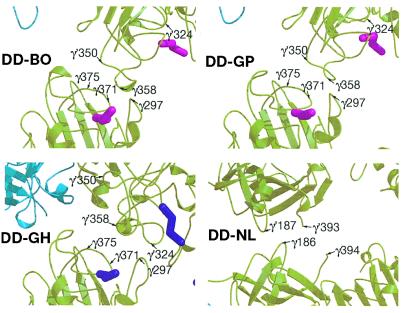
Ribbon structures of the interfacial regions involved in the side-by-side (ss) associations shown in Fig. 1. Note that the residue locations for DD-BO and DD-GP are virtually identical but are somewhat shifted in DD-GH and completely different in DD-NL. Color scheme as in Fig. 1.
The packing arrangement that occurs in the absence of ligands (DD-NL) is very different (Fig. 1). There is a lateral interaction between γC domains, but it involves a completely different face of the domain than described above (Fig. 2). We take this as evidence that occupancy of the γC hole encourages a specific lateral interaction between protofibrils.
The Model Step by Step.
Step 1: Exposure of the A knobs.
The removal of the fibrinopeptides A exposes a pair of A knobs in the central domain of fibrinogen molecules, which pins together fibrin units in an end-to-end fashion. These “A knobs” fit snugly into well-defined “holes” in the γC domains near the two ends of each molecule (Fig. 3).
Figure 3.
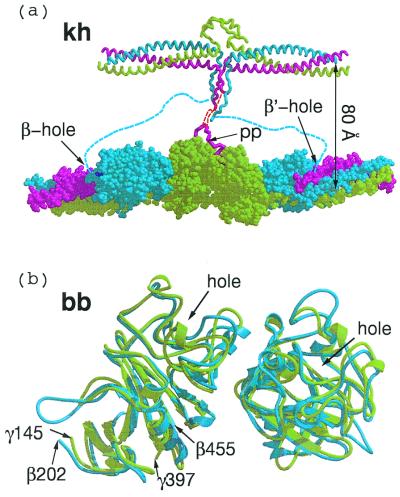
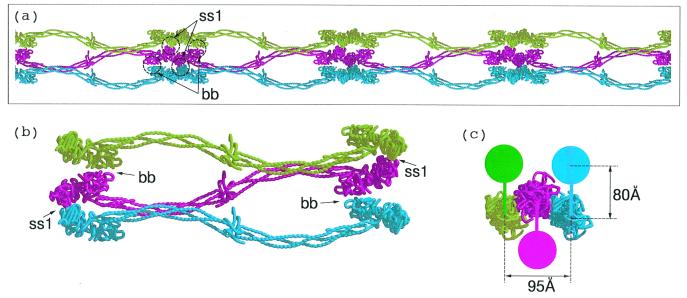
(a) Knob–hole interactions (kh) between central domain (Top) and γC and βC domains of associated molecules within a protofibril. The 80 Å-distance between the two strands of the protofibril is an approximation based on extension of α-chain residues 34 to 17, which were not clearly delineated in the chicken fibrinogen structure (red dashed line). Flexible amino-terminal regions of β chains (blue dashed line) may allow B knobs to reach β-chain holes of the same companion molecule in protofibril clasper-style. (b) Superposition of two computer-associated βC domains (blue) on homologous γC domains (light green) as they occur in crosslinked fibrin (DD-BO, PDB-code 1FZC).
The amino-terminal segments of the α-chains—including the fibrinopeptides A and the A knobs—are not visible in electron density maps of native chicken fibrinogen (PDB code 1EI3) and must be extremely mobile (13). Nonetheless, the distance from the knob to the molecular axis can be reasonably estimated; a similar estimate was apparently made in the construction of a model protofibril with modified bovine unit (23). The primary constraint on the two α-chains is a disulfide bond that joins them at residues Cys-28, 10 residues away from the Gly-Pro-Arg residues that constitute the A knob (α17–19); in the chicken structure, the disulfide is 35 Å from the molecular axis (13). If the chain is extended to include the knob, and if the knob is then superimposed on its position in a companion model in a γC hole, then the overall distance between parallel molecular axes is ≈80 Å (Fig. 3a).
The repetitive interaction of pairs of centrally located A knobs with the associated ends of companion fibrinogen molecules leads to a two-molecule thick oligomer with a half-molecule stagger. These oligomers (protofibrils) tend to average 15 units in length (25). The knob–hole interactions are largely electrostatic (positively charged knobs in negatively charged holes), although hydrogen-bonding and a water-mediated interaction are also involved (4). Surface area buried by the insertion of a single knob into one hole amounts to about 250 Å2.
Step 2: Formation of γ-γ dimers.
The growing protofibrils are reinforced by the factor XIII-catalyzed incorporation of ɛ-amino(γ-glutamyl)lysine crosslinks between the carboxyl-terminal segments of abutting γ-chains (26). These segments are highly exposed and flexible, and they have not been discerned in electron density maps. It is noteworthy that crosslinks are introduced before the protofibrils associate laterally and while the target sequences are still readily accessible to large macromolecules like factor XIII (27, 28).
Step 3: Initial lateral interaction.
The lateral, and necessarily staggered, association of protofibrils that gives rise to mature fibrin has not previously been defined at the level of atomic resolution, although general models have been proposed in the past (11, 29–32). There is compelling evidence that fibrinopeptide B removal, which exposes the B knob (GHRP), plays a role in the lateral association of protofibrils (33, 34), but it is also clear that lateral association can take place even when the fibrinopeptides B are not removed, as is the case when clotting is induced by certain snake venom enzymes (12). The suggestion has also been made that the flexible αC domains play a role in the association of protofibrils, shifting from an intramolecular to an intermolecular mode, perhaps as a consequence of fibrinopeptide B removal (35). Finally, the loss of electrostatic repulsion upon removal of the electronegative fibrinopeptides B may be a factor on its own, independent of any knob–hole interaction.
We now propose that the primary driving force for protofibril association is the set of lateral interactions observed between γC domains in the crystal packing of DD-GP and DD-BOTH (Fig. 1). The same packing also occurs in crystals of a modified bovine fibrinogen (23). The segments of the γ-chain involved in this lateral interaction are γ350–360 and γ370–380 (Fig. 2). In the crystal, ≈300 Å2 of previously accessible surface is buried as a result. One of the key residues is γArg375, a residue often mutated in variant human fibrinogens exhibiting defective polymerization (8). Although the amount of solvent-accessible surface area buried is relatively small on a per monomer basis, it is a significant amount when considered for the entire protofibril.
Step 4: A second lateral interaction.
A second kind of interaction may come into play upon removal of the fibrinopeptide B, the release of which would allow a pair of B knobs to fill holes in the same two companion molecules pinned together by the A knobs (Fig. 3a). The amino-terminal segments of β-chains are even more flexible and unrestrained than the corresponding regions of α-chains, the nearest constraint being imposed by a disulfide connection at residue β65 (human numbering). It is likely that the B knobs (residues β15–18) are mobile and do not become fixed until they encounter βC holes.
We propose that binding of the B knobs pulls the βC domains away from an association with the coiled coils (Fig. 4) and effects a concomitant if subsidiary lateral interaction between protofibrils that involves β-chain residues β330–375. This is the region of the β-chain that is homologous to the γ-chain region involved in end-to-end associations (Fig. 3b). Intriguingly, these regions of the βC domains are closer to each other in the crystal packing of double-D when synthetic B knobs are present than when they are not (ss3 in Fig. 3). In fact, a previous report from this laboratory noted that the coiled coils in those structures (DD-GH)—unrestrained in the fragment because they are not connected to a central domain—are pulled out of line (5). It must be emphasized that the lateral interactions involving the βC domains would be complementary to and dependent on the lateral interactions involving γC domains and, poised as they are, require only a modest twist once the proposed γ-chain associations are in place (Fig. 3).
Figure 4.
Hypothetical rearrangement of βC domains upon binding of B knobs in a way that allows the βC-βC (bb) interaction shown in Fig. 3b. As shown in the upper panel, the lateral association between γC domains (taken directly from the packing observed in DD-BO) positions the βC domains such that only a small shift is needed for a bb interaction, perhaps as a result of binding B knobs. A concomitant opening up of the region between βC domain and coiled coils occurs as a consequence.
In a given fibrin monomeric unit (which is itself a dimer), one end of the molecule will have its γC domain associated with the γC domain from another protofibril, whereas, at the other end, the other γC domain will associate with still a third protofibril (Fig. 5). The βC involvement would follow a reciprocal course involving the same three molecules.
Figure 5.
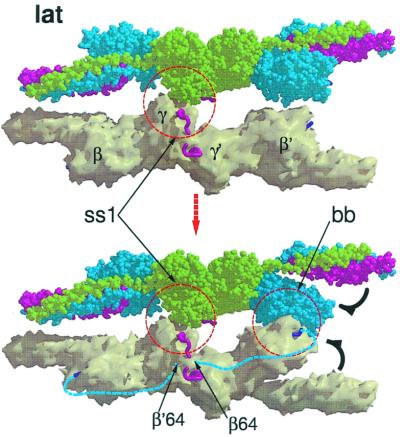
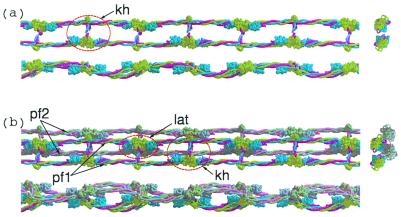
(a) Three views (top, side, and end-on) of a single protofibril formed by knob-hole (kh) interactions. Light green, γ-chains; blue, β-chains, red, α-chains. End-on view is shown at the right side. (b) Lateral association (lat) of two protofibrils (PF1 and PF2, top and side views; end-on view is shown at right).
Summing to this point, three sets of interfacial association, including two observed in crystal packing arrangements of human double-D preparations, are proposed to play roles in the formation of fibrin fibers. These associations occur concomitantly and as a result of two different knob-hole interactions. In one, the A knobs (Gly-Pro-Arg) fit into closely situated γC holes from neighboring molecules and pin them together in an end-to-end mode. Filling these holes also induces an initial lateral interaction between γC domains of different protofibrils (Figs. 1 and 2).
The second knob-hole interaction involves the B knob (Gly-His-Arg) filling holes in widely separated βC domains in the same two molecules held together by the A-knob pair (Fig. 3a), and likely not between different protofibrils as we had previously conjectured (36). The filling of the βC holes encourages a lateral interaction between βC domains that follows a course complementary to that involved in the lateral γC interactions, but in a reciprocal manner with regard to specific units within the dimer. We hypothesize that the interaction with the B knob plays a role in the βC domains moving away from the coiled coils, exposing a region that has been implicated in the activation of tissue plasminogen activator (t-PA) by fibrin (37). The model does not preclude ancillary associations involving αC domains. Indeed, as will be shown below, the formation of factor XIII-catalyzed multimeric crosslinks between chains is readily accommodated by the model.
Symmetry Arguments.
We will now show that the interactions described above give rise to a fiber with the physical properties of natural fibrin fibers, including a solvent content of ≈80 percent, the open nature of which provides access to vital macromolecular agents, and especially t-PA, plasmin(ogen) and factor XIII. To this end, the model will be built up by the systematic addition of protofibrils to a growing bundle, care being taken that clashing between units not occur.
For simplicity, some of the depictions are diagrammatic. It must be kept in mind that the starting protofibril is a flattened ribbon (Fig. 5a), and that knob–hole interactions occur in the “flat” dimension. In some of the depictions, the two different strands of a protofibril are represented by cylinders of a given color and are numbered 1 and 2; the knob–hole connections between them have been omitted to simplify the pictures.
Two Associated Protofibrils.
The linear growth of fibrin fibers requires that protofibrils associate in a staggered overlapping fashion. Although the degree of overlap between associating protofibrils may vary, for simplicity we have assumed it to be a uniform half-stagger arrangement (this should not be confused with the half-stagger that occurs between units within the protofibril itself). In the model, the two protofibrils are associated by a uniform series of lateral interactions involving γC domains (Fig. 4). If fibrinopeptide B release occurs, the protofibrils will also have their βC holes filled with B knobs, allowing a concomitant association of βC domains to occur (Fig. 4). It is noteworthy that the lateral association of protofibrils cannot be so propagated if the γC side-by-side contacts observed in the crystal structure of DD-NL are used.
Three Associated Protofibrils.
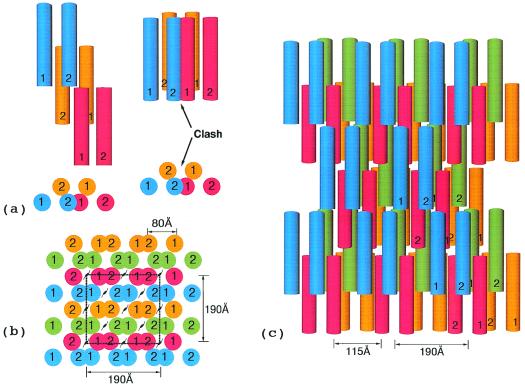
In essence, there are three different ways that a third protofibril may be added to a two-protofibril bundle, all using the same kind of interfacial associations (Fig. 6), but differing in the particular strands or regions of strands involved (Fig. 7). In one case, the new protofibril interacts with the same strand of a protofibril as is already involved in the two-protofibril bundle, but on the opposite side. Such additions are limited to growth in the x direction. In another case, the newly added protofibril interacts with the same strand and side of another protofibril, but in the unoccupied zone provided by a stagger. The resulting end-to-end association leads to growth along the z axis only.
Figure 6.
(a) Three protofibrils associated by lateral involvement of γC domains; only one of the strands from each protofibril is shown to facilitate visualization. In order for a fiber to grow the lateral interactions involving different halves of a unit must extend to different protofibrils. (b) The γC associations (denoted ss1) are depicted as blue to red and red to green. The βC associations (denoted bb) follow the same pattern except red to green and blue to red. The hypothetical interaction between βC domains uses regions that are homologous to γ-γ end-to-end faces. (c) End-on view of three associated protofibrils. The solid colored discs denote the molecular units of the protofibril not included in a and b; the colored vertical bars denote knob-hole interactions. The 80 Å-distance is taken from Fig. 3a; the 95 Å-distance between centers of associated protofibrils is taken directly from the lateral packing distances observed in DD-BO.
Figure 7.
(a) End view of simplest highly ordered model fibrin fiber with xy face of hypothetical unit cell outlined. The unit cell dimensions are 190 × 190 × 460 Å. The first 190 Å dimension is based on distances observed in the crystal packing in DD-BO (Table 1). The second 190-Å distance is a consequence of the two strands of any protofibril being 80 Å apart. A unit cell with the same dimensions has been reported for a neutron diffraction experiment with fibrin oriented in a magnetic field (38). (b) Top view of fiber showing 115 Å-wide cavities resulting from half-staggered overlaps; the 115-Å dimension is calculated directly from unit cell.
The most important mode has the newly added protofibril use one of its strands to interact with one protofibril and the other strand with the other protofibril. In this case, the fiber can grow both linearly and laterally (i.e., in both the y and z directions). Importantly, because of the excluded volume effect, a half-stagger is needed to avoid clashing (Fig. 7a). The result of such a stagger is a vacancy in the growing fiber; indeed, it is this forced omission that leads to the high solvent content of fibrin fibers and to access channels for enzymes like t-PA, plasmin(ogen) and factor XIII.
Whether or not mature fibers are thick or thin depends on the relative contributions of these three kinds of protofibril addition. It is well known that in vitro the lateral growth of fibrin fibers depends on environmental factors, including pH, calcium ion concentration, and ionic strength, and it is likely that the contributions of the different types of association are differently influenced by electrostatic factors. The multibundle model shown in Fig. 7 has been restricted to the last described of the interactions in the interests of simplicity; real fibrin fibers would not be expected to be so uniform.
Unit Cell Considerations.
The packing of monomers in the fibrin fiber can be considered from a pseudocrystallographic perspective. In an elegant model conceived long before detailed structural information was available, Hermans (29) invoked lateral interactions involving the fragment D portions of fibrinogen. By taking account of the known dimensions of the molecule (24), the half-staggered repeat observed in fibrin (6), and the assumption of a truly dimeric starting unit, he showed that there was a limited number of space groups that could accommodate the model fiber. Subsequently, Hudry–Clergeon and coworkers (38) conducted neutron diffraction experiments on fibrin formed in a strong magnetic field. They were able to determine a unit cell, the dimensions of which were 185 × 185 × 446 Å.
The model shown in Fig. 7 has similar dimensions (190 × 190 × 450 Å). The a = 190 Å is twice the observed packing distance in DD-GP and DD-BOTH (Table 1), whereas the b = 190 Å is a function of the 80 Å-spacing between strands in the individual protofibril. The c dimension is equal to the length of a single monomer. Because of systematic vacancies in the fiber, there are only eight molecules per unit cell. The calculated solvent content is 80 percent, in good accord with experimental measurements (39).
Discussion
Fibrin clots are well known to be permeable to a wide variety of agents. The clot itself, however, is a tangle of fibers, and the permeability of the individual fibers is more problematic. Depending on environmental conditions, fibers may range from a few to hundreds of molecules in diameter. Molecules like t-PA, plasminogen, and thrombin may be trapped within a polymerizing fiber. Whether or not these proteases have ready diffusional access from without is not so obvious. The system that presents the strongest evidence for diffusion within the fiber is the factor XIII-catalyzed formation of crosslinks between αC-domains. Thus, whereas γ-γ dimers are formed rapidly at the stage of the growing protofibril when enzyme access is not a problem, the crosslinking of α-chains is a very slow process, occurring over the course of hours or days when fiber formation occurs within seconds or minutes (40).
The question arises: can factor XIII diffuse through a model fiber that doesn't have vacancies? Factor XIII (without b subunits) has a smallest cross-sectional diameter of about 60 Å (41). If a reasonable approximation is made as to the whereabouts of the flexible αC-domains (13), then there is very little room within packed protofibrils for solvent channels unless vacancies occur during the lateral association of protofibrils. Even then, the channels are such that diffusion of the factor XIII must be greatly slowed, concomitant with the time course observed in the case of α-chain crosslinking. Given these time and distance constraints and the mobile nature of the αC domains, a wide variety of lysine and glutamine residues become involved in the crosslinking introduced by the slowly diffusing factor XIII (42, 43).
The model also explains why fibrin formed by certain snake venoms that remove only the fibrinopeptide A are not the same as fibrin formed by thrombin. The γC-γC lateral interactions allow fiber development, but the lack of ancillary βC-βC interaction limits or weakens the process. It also explains how fibrin can be formed by the exclusive or preferential release of fibrinopeptides B (“β Fibrin”), as occurs when certain heterologous thrombins are used (44, 45) or particular snake venom enzymes used (46). Although, in these situations, lateral associations may be strongly encouraged, the necessary end-to-end associations must also occur, even though the fibrinopeptides A are still present and the A-knobs remain unexposed. The same end-to-end associations must form as occur in natural fibrin, as evidenced by the factor XIII-induced formation of γ-γ crosslinks (28). It must be that the B knobs, like the A knobs, are flexibly tethered and move about until they encounter appropriate holes. There appears to be sufficient freedom and space for B knobs to stretch out clasper-like and pin together two molecules associated at the γ-γ interface, even when the fibrinopeptides A are still present.
The model also offers an interpretation of how fibrin can activate t-PA and why fibrinogen does not, the movement of the βC domain away from the coiled coil exposing a region known to be involved in the process (37). In fact, it has recently been shown that the moiety D2E can activate t-PA, whereas fragments D and double-D do not, and the authors hypothesized that the βC domains would have to move (47). On the basis of our model, we would predict that fibrin generated by a snake venom enzyme that does not release fibrinopeptide B would not be as effective in activating t-PA.
Acknowledgments
We thank Bruno Zimm and Justin Kollman for reading the manuscript and offering helpful comments. This work was supported by National Institutes of Health Grant HL26873.
Abbreviations
- GPRPam
Gly-Pro-Arg-Pro-amide
- GHRPam
Gly-His-Arg-Pro-amide
- t-PA
tissue plasminogen activator
- DD-NL
fragment double-D, no ligand
- DD-GP
fragment double-D with GPRPam
- DD-GH
fragment double-D with GHRPam
- DD-BO
fragment double-D with both GPRPam and GHRPam
References
- 1.Yee V C, Pratt K P, Cote H C, LeTrong I, Chung D W, Davie E W, Stenkamp R E, Teller D C. Structure (London) 1997;5:125–138. doi: 10.1016/s0969-2126(97)00171-8. [DOI] [PubMed] [Google Scholar]
- 2.Pratt K P, Cote H C F, Chung D W, Stenkamp R E, Davie E W. Proc Natl Acad Sci USA. 1997;94:7176–7181. doi: 10.1073/pnas.94.14.7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spraggon G, Everse S, Doolittle R F. Nature (London) 1997;389:455–462. doi: 10.1038/38947. [DOI] [PubMed] [Google Scholar]
- 4.Everse S J, Spraggon G, Veerapandian L, Riley M, Doolittle R F. Biochemistry. 1998;37:8637–8642. doi: 10.1021/bi9804129. [DOI] [PubMed] [Google Scholar]
- 5.Everse S J, Spraggon G, Veerapandian L, Doolittle R F. Biochemistry. 1999;38:2941–2946. doi: 10.1021/bi982626w. [DOI] [PubMed] [Google Scholar]
- 6.Ferry J D. Proc Natl Acad Sci USA. 1952;38:566–569. doi: 10.1073/pnas.38.7.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laurent T C, Blomback B. Acta Chem Scand. 1958;12:1875–1977. [Google Scholar]
- 8.Matsuda M. Intern J Hematol. 1996;64:167–179. doi: 10.1016/0925-5710(96)00482-3. [DOI] [PubMed] [Google Scholar]
- 9.Laudano A P, Doolittle R F. Proc Natl Acad Sci USA. 1978;75:3085–3089. doi: 10.1073/pnas.75.7.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamazumi K, Doolittle R F. Proc Natl Acad Sci USA. 1992;89:2893–2896. doi: 10.1073/pnas.89.7.2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fowler W E, Hantgan R R, Hermans J, Erickson H. Proc Natl Acad Sci USA. 1981;78:4872–4876. doi: 10.1073/pnas.78.8.4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blomback B, Blomback M, Nilsson I M. Thromb Diath Haemostasis. 1957;1:1–13. [Google Scholar]
- 13.Yang Z, Mochalkin I, Veerapandian L, Riley M, Doolittle R F. Proc Natl Acad Sci USA. 2000;97:3907–3912. doi: 10.1073/pnas.080065697. . (First Published March 28, 2000; 10.1073/pnas.080065697) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones T A, Zou J-Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 15.Collaborative Computing Project Number 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 16.Janin J, Miller S, Chothia C. J Mol Biol. 1987;204:155–164. doi: 10.1016/0022-2836(88)90606-7. [DOI] [PubMed] [Google Scholar]
- 17.Lee B K, Richards F M. J Mol Biol. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- 18.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 19.Bacon D J, Anderson W F. J Mol Graphics. 1988;6:219–220. [Google Scholar]
- 20.Merritt E A, Murphy M E P. Acta Crystallogr D. 1994;50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- 21.McRee D E. J Mol Graphics. 1992;10:44–46. [Google Scholar]
- 22.Nicholls A, Sharp K, Honig B. Proteins Struct Funct Genet. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 23.Brown J H, Volkmann N, Jun G, Henschen A H, Cohen C. Proc Natl Acad Sci USA. 2000;97:85–90. doi: 10.1073/pnas.97.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hall C E, Slayter H S. J Biophys Biochem Cytol. 1959;5:11–16. doi: 10.1083/jcb.5.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bale M D, Jamney P A, Ferry J D. Biopolymers. 1982;21:2265–2277. doi: 10.1002/bip.360211113. [DOI] [PubMed] [Google Scholar]
- 26.Chen R, Doolittle R F. Biochemistry. 1971;10:4486–4491. doi: 10.1021/bi00800a021. [DOI] [PubMed] [Google Scholar]
- 27.Doolittle R F. Adv Protein Chem. 1973;27:1–109. doi: 10.1016/s0065-3233(08)60446-5. [DOI] [PubMed] [Google Scholar]
- 28.Dyr J E, Blomback B, Hessel B, Kornalik F. Biochim Biophys Acta. 1989;990:18–24. doi: 10.1016/s0304-4165(89)80006-6. [DOI] [PubMed] [Google Scholar]
- 29.Hermans J. Proc Natl Acad Sci USA. 1979;76:1189–1193. doi: 10.1073/pnas.76.3.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weisel J W, Phillips G N, Jr, Cohen C. Nature (London) 1981;289:263–267. doi: 10.1038/289263a0. [DOI] [PubMed] [Google Scholar]
- 31.Rao S P S, Poojary D, Elliot B W, Melanson L A, Oriel B, Cohen C. J Mol Biol. 1991;222:89–98. doi: 10.1016/0022-2836(91)90739-s. [DOI] [PubMed] [Google Scholar]
- 32.Weisel J W, Nagaswami C. Biophys J. 1992;63:111–128. doi: 10.1016/S0006-3495(92)81594-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hurlet-Jensen A, Cummins H Z, Nossel H L, Liu C Y. Thromb Res. 1982;27:419–427. doi: 10.1016/0049-3848(82)90059-7. [DOI] [PubMed] [Google Scholar]
- 34.Hantgan R R, Hermans J. J Biol Chem. 1979;254:11272–11282. [PubMed] [Google Scholar]
- 35.Gorkun O V, Veklich Y I, Medved L V, Henschen A H, Weisel J W. Biochemistry. 1994;33:6986–6997. doi: 10.1021/bi00188a031. [DOI] [PubMed] [Google Scholar]
- 36.Doolittle R F. Jpn J Thromb Hemostasis. 2000;11:123–130. [Google Scholar]
- 37.Schielen W J G, Adams H P H M, Voskuilen M, Tesser G J, Nieuwenhuizen W. Biochem J. 1991;276:655–659. doi: 10.1042/bj2760655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hudry-Clergeon G, Freyssinet J-M, Torbet J, Marx J. Ann NY Acad Sci. 1983;408:380–387. doi: 10.1111/j.1749-6632.1983.tb23258.x. [DOI] [PubMed] [Google Scholar]
- 39.Carr M E, Jr, Hermans J. Macromolecules. 1978;11:46–50. doi: 10.1021/ma60061a009. [DOI] [PubMed] [Google Scholar]
- 40.McKee P A, Mattock P, Hill R L. Proc Natl Acad Sci USA. 1970;66:738–744. doi: 10.1073/pnas.66.3.738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yee V C, Pedersen L C, Truong I L, Bishop P D, Stenkamp R E, Teller D C. Biochemistry. 1994;91:7296–7300. doi: 10.1073/pnas.91.15.7296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sobel J H, Gawinowicz M A. J Biol Chem. 1996;271:19288–19297. doi: 10.1074/jbc.271.32.19288. [DOI] [PubMed] [Google Scholar]
- 43.Matsuka Y V, Medved L V, Migliorini M M, Ingham K C. Biochemistry. 1996;35:5810–5816. doi: 10.1021/bi952294k. [DOI] [PubMed] [Google Scholar]
- 44.Doolittle R F. Biochem J. 1965;94:735–741. doi: 10.1042/bj0940735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cottrell B A, Doolittle R F. Biochim Biophys Acta. 1976;453:426–438. doi: 10.1016/0005-2795(76)90138-0. [DOI] [PubMed] [Google Scholar]
- 46.Shainoff J R, Dardik B N. Science. 1979;204:200–204. doi: 10.1126/science.155308. [DOI] [PubMed] [Google Scholar]
- 47.Yakovlev, S., Makogonenko, E., Kurochkina, N., Nievwenhuizen, W., Ingham & Medved, L. Biochemistry, in press. [DOI] [PubMed]