Abstract
The Swarm rat chondrosarcoma is a tumor tissue line derived from a tumor that arose spontaneously in a Sprague-Dawley rat. The original tissue has given rise to several tissue lines and cell lines that have been prepared in different laboratories. It has been observed that these lines differed in their growth rates and biochemical characteristics. We have characterized our Swarm rat chondrosarcoma tissue and cell lines currently in use in terms of their cytogenetic profiles and their tumorigenic properties in vivo. We found a wide variety of chromosomal abnormalities among cell lines, including translocations, deletions and polyploidy. There were also significant differences in their growth properties in vivo, giving rise to tumors of a few milligrams in the case of Ng cells, to 35 grams in the tissue line JWS. The cytogenetic complexity of the Swarm rat chondrosarcoma between and among different lines makes it very suitable to address questions about the changes that occur as a result of karyotypic abnormalities and to provide links between cytogenetic abnormalities and the dynamic oncogenic machinery.
INTRODUCTION
Chondrosarcoma represents the second most common primary malignant skeletal tumor, comprising up to 24% of all bone tumors. Despite the new advances in adjuvant therapy, surgical resection is the only effective standardized treatment. Chondrosarcomas do not respond to chemotherapy or radiation therapy, therefore, metastatic disease in these tumors is rarely amenable to curative treatment. The five-year survival for patients with grade III tumors is only 29%. To improve survival rates in patients with high-grade chondrosarcomas, a better understanding of its biology is necessary.
The Swarm rat chondrosarcoma2 is a tumor tissue line derived from a tumor that arose spontaneously in a Sprague-Dawley rat.9 It has been maintained through the years by serial subcutaneous transfer from rat to rat. Histologically, the tumor is a well-differentiated chondrosarcoma with mild-to-moderate cellular atypia. Interestingly, several tissue and cell lines derived from this primary tumor are available in a number of laboratories. These tissue and cell lines have been the subject of extensive biochemical studies on extracellular matrix molecules and chondrocyte metabolism.1–16
We have previously reported a comprehensive gene expression profiling on a tissue line (JWS) of the Swarm rat chondrosarcoma.12 This tissue line grows very aggressively when injected subcutaneously, reaching up to 35 grams about four weeks after injection. A survey of some of the groups that have been maintaining other lines of this tumor revealed that the various cell and tissue lines exhibit a wide variation in tumor growth and extracellular matrix metabolism. In this study, we evaluate the cytogenetic and in vivo tumor growth characteristics of several tissue and cell lines of Swarm rat chondrosarcoma to determine if the variation in tumor behavior can at least in part be accounted for by variations in karyotype.
MATERIAL AND METHODS
Swarm Rat Chondrosarcoma Tissue Lines and Cell Lines
Two tissue lines (TGO and JWS) and four different cell lines (LTC 86, LTC 93, Rex, and Ng)7 were used. The TGO and JWS are tissue lines derived from the Swarm rat chondrosarcoma tumor and they have been maintained by serial subcutaneous injections. The LTC line which can be propagated entirely in vitro was derived from long-term culture of rat chondrosarcoma tumor tissue with repeated selection for non-adherent or floating cells. Continued propagation of this line led to several variant cell lines as well as a few clonally derived cell lines.7 LTC 86 was from an early passage in 1986, while LTC 93 was from a passage seven years later in 1993. Rex and Ng were lines cloned from the LTC parent line.
Tumor Induction and Tumor Growth Measurement
Animal care protocol was approved by the University of Iowa. Male, four-week-old Sprague-Dawley rats were used. Tumor induction was performed by injection of the tissue line or cell line subcutaneously on both sides of the lumbar spine (2 injections per animal). For tissue lines (TGO and JWS), 40-50 ml (~5 x 105 cells) of tumor slurry6 was used. For cell lines, 1 x 107 (LTC 86, LTC 93, Rex, Ng) in 0.5 ml of Dulbecco's Modified Eagle Medium was used for each injection. The animals were euthanized 35 days post injection in the case of tumor slurries and at 70 days after injection of tissue-cultured cell lines. The tumors were removed, weighed, and processed for histology as previously described.16 A necropsy was performed to establish the presence or absence of metastases.
Cytogenetic Analysis
The cytogenetic techniques used for chromosome analysis have been described previously.15 Aseptically collected 1- to 2-cm3 samples were mechanically and enzymatically disaggregated6 and cultured in RPMI 1640 medium supplemented with 20% fetal bovine serum, 1% penicillin/streptomycin, and 1% L-glutamine for one to ten days. Three to eight hours before harvest, cells were exposed to colcemid (0.02 mg/ml) to achieve metaphase arrest. After incubation in hypotonic solution, the preparations were fixed three times with 3:1 methanol:glacial acetic acid, and dropped onto water-rinsed slides. Chromosomes were stained by the GTW (G-bands by trypsin using Wright's stain) banding method. The chromosome number was determined by microscopic analysis and cells were examined for the presence or absence of detectable structural rearrangements. Karyotypes were prepared from digitized images of these metaphases. When possible, about 15-20 metaphases were scored (counted and checked for markers) for each specimen. However, a detailed analysis and complete karyotype were only performed on a small number of the cells (minimum of two) from each sample. In some cases, there was only a limited characterization of structural abnormalities, due to the complexity of the karyotypes. Normal male rat cells were used as controls. Karyotypes were based on the nomenclature rules for rat chromosome G-bands.8 The system for rat chromosome nomenclature closely follows the international system for human cytogenetic nomenclature.10
RESULTS
Karyotype Analysis
The results of the cytogenetic analysis from the different Swarm rat chondrosarcoma tissue and cell lines are presented in Tables 1 and 2. Abnormal karyotypes were observed in all cases. No cultures contained any de-differentiated cells.
TABLE 1. Chromosome numbers in Swarm rat chondrosarcoma.
| chromosome count |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| source | <42 | modal# | diploid | 43 | 70 | <75 | 84 | 78-89 | <82 | 74-82 | 82-84 | >84 | 86-90 | 90-100 | ~160 | total |
| TGO | 1 | 4 | 1 | 1 | 2 | 9 | ||||||||||
| JWS | 5 | 26 | 4 | 6 | 12 | 53 | ||||||||||
| Rex | 15 | 3 | 2 | 20 | ||||||||||||
| Ng | 6 | 9 | 5 | 20 | ||||||||||||
| LTC86 | 96 | 3 | 3 | 19 | 25 | |||||||||||
| LTC93 | 77 | 4 | 11 | 5 | 20 | |||||||||||
TABLE 2. Chromosomal rearrangements in Swarm rat chondrosarcoma.
| Chromosome | TGO | JWS | Ng | Rex | LTC86 | LTC93 |
|---|---|---|---|---|---|---|
| 1 | -1, -1 | der(1) | der(1) | -1, t(1q;11) | ||
| 2 | +2 | -2, add(2p) | -2, t(1q;2) | |||
| 3 | -3, i(3q), t(3q;11)x2 |
-3, -3, -3 | -3, -3, i(3q) | add(3p), (3q;11), +t(3q;11)x2 |
-3,-3, ?add(3q) |
|
| 4 | +i(4q), | +4 | +add(4q), add(4q)x2 |
+4, +add(4q)x2 |
+4, +add(4q), +t(4q;11) |
-4, ?der(4), i(4q) i(4q) |
| 5 | -5, -5 | -5, add(5p)x2 |
-5, -5, -5, t(5q;8) |
add(5p), t(5q;8) |
-5, -5, -5, t(5q;8) |
|
| 6 | -6, i(6q) | add(6q) | -6, i(6q) | -6 | -6 | |
| 7 | add(7p)x2 | -7 | -7, t(7q;9) | +7, +add(7p) |
+t(7q;8), add(7p), t(7q;?9) |
|
| 8 | add(8p), t(8q;10) |
-8 | -8, t(8q;X) | -8, -8 | +8 | -8, -8, -8, 8? |
| 9 | -9 | -9 | -9 | t(9q;11) | -9, -9 | -9,-9, 9? |
| 10 | -10, -10, -10 | -10 | +10 | -10 | t(10q;11) | |
| 11 | -11 | +11, +add(11q) |
-11, -11, add(11p) |
add(11p) | +add(11q) x2 |
|
| 12 | -12 | -12, -12 | -12, -12 | add(12q) | -12, | |
| 13 | i(13q) | -13 | -13, add (13p) |
del(13q) | -13, -13, ?add (13p), add (13q) |
|
| 14 | -14,-14,-14 | add(14q) | -14, -14 | i(14p)x3 | add(14q) | |
| 15 | -15,-15 | -15, -15, add(15p) | -15, -15 | -15, i(15q)x2 | -15, add(15q), i(15q) |
|
| 16 | -16,-16 | -16 | -16, -16 | |||
| 17 | -17,-17 | -17, -17 | -17, -17 | +17 | -17, -17, -17 | |
| 18 | -18,-18 | -18 | -18, -18 | -18, -18 | ||
| 19 | +19,+19 | -19 | +19, +19 | -19, -19, -19 | ||
| 20 | -20, -20 | add(20q) | -20 | |||
| markers | 5 | 0 | 16 | 16 | 13 | 15 |
| X | XXXX, t(X;8) | XX | XX | XX | XX | X, t(Xq;4) |
| Y | Y | YY | YY | YY | YY | Y?Y? |
TGO cells were the only cells demonstrating a very slow growth when cultured for cytogenetic studies. Five cultured flasks were used for analysis. Only a limited number of cells were available for analysis after multiple attempts to obtain dividing cells. Nine cells were counted; five cells were in the diploid range (37-42 chromosomes); and four cells were in the hypertriploid to tetraploid range (70-84 chromosomes). Several chromosomal abnormalities were observed in this tissue line, including deletion of chromosomes 1, 5, 9, 10, 11, 12, 14-18; i(3q); t(3q;11)x2; +i (4q); -6, i(6q); add (7p)x2; t(8q;10); i(13q); +19; and XXXX , t(X;8).
JWS cells (53 examined) demonstrated that the majority of cells (26) were diploid, with 42 chromosomes, or near-diploid (nine cells), with 40-43 chromosomes. Six cells were in the tetraploid range, with 78-89 chromosomes. The remainder of the cells were in the hypertetraploid to hypopentaploid (90-100 chromosomes) range. There was no evidence of structural abnormalities.
LTC 86 cells (25 examined) demonstrated 12 cells in the near-tetraploid range, with 74 to 94 chromosomes. The remaining 13 cells were in the hypopentaploid range, with 95 to 99 chromosomes. The modal chromosomal number was 96. Numerous structural rearrangements and marker chromosomes were present (Table 2).
LTC 93 cells (20 examined) demonstrated four cells in hypertriploid range, with 70 to 73 chromosomes. The remaining 16 cells were in the near-tetraploid range, with 75 to 84 chromosomes. The modal chromosomal number was 77. Numerous structural rearrangements and marker chromosomes were present (Table 2).
Rex cells (20 examined) demonstrated 18 cells in the near-tetraploid range, with 76-82 chromosomes. Two cells were octaploid, with approximately 160 chromosomes. Numerous structural rearrangements and marker chromosomes were present (Table 2).
Ng cells (20 examined) had chromosome counts in the hypertriploid to hypertetraploid range, with 69-86 chromosomes. Numerous structural rearrangements and marker chromosomes were present (Table 2).
In vivo Tumor Growth
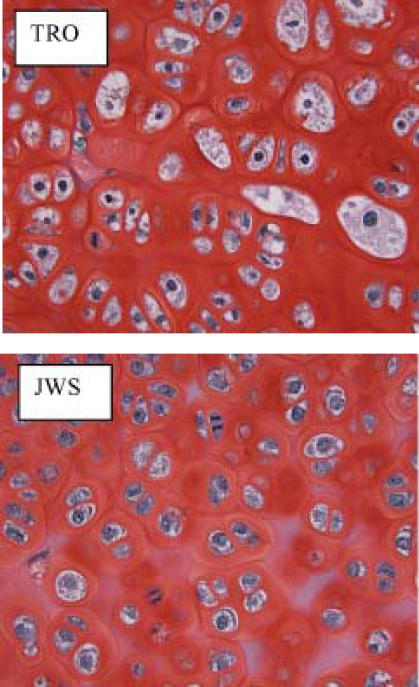
Tissue lines and cell lines were injected subcutaneously into both sides of the lumbar spine. The animals were sacrificed 35 or 70 days post injection. The tumor lines were of a well-differentiated chondrosarcoma with mild cellular atypia (Figure 1). The tumors were weighed. It was found that there was a great variability in tumor growth among cell lines (Figure 2). JWS grew up to 35 grams of tumor in most cases, more than doubling the size of the TGO-induced tumors. Both LTC and Rex grew moderately-sized tumors (3 grams). However, Ng cells did not result in much tumor growth (about 60 mg). There was no evidence of metastases in any of the tissue or cell lines.
Figure 1.
Histopathological features of the Swarm rat chondrosarcoma. Note a well-differentiated chondrosarcoma with mild to moderate cellular atypia. Safranin O staining, 400x
Figure 2.
In vivo tumor growth of the different tumors and cell lines.
DISCUSSION
Swarm rat chondrosarcoma is one of the most useful cell culture systems for the study of cartilage metabolism. The cells grow very reliably under many culture conditions and they produce great amounts of extracellular matrix. In addition, when injected into the subcutaneous tissue or into the bone, they behave in a manner closely resembling the behavior of human chondrosarcoma.
Since the isolation of the tumor in the 1960's, many laboratories have maintained tissue lines and cell lines for experimental work. Due to investigator preferences and to differences in experimental designs, a selection in cell populations has occurred over the years. Since many of the biochemical extracellular matrix characteristics have been very consistent among laboratories, and because the cells came from the same original tumor, it has been assumed that these tissue and cell lines were similar.
However, in this study, we observed that the karyotypes and in vivo tumor growth of several tissue lines and cell lines were very different. The cytogenetic analysis also demonstrated a wide range of numerical and structural abnormalities among the lines. While caution must be used in comparing the significance of chromosomal differences among the cell lines due to the limited number of cells that were completely karyotyped because of the complex chromosome constitution, some differences were found. For instance, the two tissue lines (TGO and JWS) have complex cell populations with cells in hypodiploidy and other cells in pentaploidy (cells with >100 chromosomes). However, TGO cells demonstrated several chromosome deletions and numerous structural abnormalities that were not present in JWS. Interestingly, the karyotypes of the LTC cell subclones (Ng, Rex) indicate that these cells are related, with the later LTC (1993) clone having fewer chromosomes but more stable cell counts than the earlier clone (1987).
The in vivo tumor growth varied from just a few milligrams in the case of the Ng cells up to 20 grams of tumor in JWS. Interestingly, the in vitrogrowth of the different tissue and cell lines demonstrate the same growth curves (data not shown). There was no evidence of metastases in any of the cell lines when injected subcutaneously.
The present view of malignant transformation and tumor progression requires the accumulation of multiple genetic alterations such as chromosomal abnormalities, oncogene activation, loss of tumor suppressor genes, or abnormalities in genes that control DNA repair and genetic stability. This view correlates with the increasing complexity of karyotypes seen during tumor progression. Importantly, in malignant human cartilage tumors, a strong cytogenetic-pathologic correlation between complex karyotypes and high-grade chondrogenic tumors has been reported. The cytogenetic complexity of the Swarm rat chondrosarcoma model, therefore, makes it very suitable for experimental work designed to address questions on the changes that occur as a result of the DNA abnormalities and to provide links between genetic abnormalities and the dynamic oncogenic machinery. Further studies will take advantage of micro-array hybridization technology and SAGE technology to increase our understanding of the biology of chondrosarcoma, and that might allow the development of more specific and targeted therapies.
ACKNOWLEDGMENTS
We acknowledge Gail Kurriger for the technical support in this study, Dr. Jerry A. Maynard for his assistance with microscopy, and Dr. Theodore R. Oegema, Jr. for the Swarm chondrosarcoma samples.
References
- 1.Caterson B, Baker JR. The link proteins as specific components of cartilage proteoglycan aggregates in vivo. Associative extraction of proteoglycan aggregate from Swarm rat chondrosarcoma. J Biol Chem. 1979;254:2394–2399. [PubMed] [Google Scholar]
- 2.Choi HU, Meyer K, Swarm R. Mucopolysaccharide and protein-polysaccharide of a transplantable rat chondrosarcoma. Proc Natl Acad Sci U S A. 1971;68:877–879. doi: 10.1073/pnas.68.5.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Faltz LL, Caputo CB, Kimura JH, Schrode J, Hascall VC. Structure of the complex between hyaluronic acid, the hyaluronic acid-binding region, and the link protein of proteoglycan aggregates from the Swarm rat chondrosarcoma. J Biol Chem. 1979;254:1381–1387. [PubMed] [Google Scholar]
- 4.Fernandes RJ, Schmid TM, Harkey MA, Eyre DR. Incomplete processing of type II procollagen by a rat chondrosarcoma cell line. Eur J Biochem. 1997;247:620–624. doi: 10.1111/j.1432-1033.1997.00620.x. [DOI] [PubMed] [Google Scholar]
- 5.Kimata K, Hascall VC, Kimura JH. Mechanisms for dissociating proteoglycan aggregates. J Biol Chem. 1982;257:3827–3832. [PubMed] [Google Scholar]
- 6.Kimura JH, Hardingham TE, Hascall VC, Solursh M. Biosynthesis of proteoglycans and their assembly into aggregates in cultures of chondrocytes from the Swarm rat chondrosarcoma. J Biol Chem. 1979;254:2600–2609. [PubMed] [Google Scholar]
- 7.King KB, Kimura JH. The establishment and characterization of an immortal cell line with a stable chondrocytic phenotype. J Cell Biochem. 2003;89:992–1004. doi: 10.1002/jcb.10571. [DOI] [PubMed] [Google Scholar]
- 8.Levan G. Nomenclature for G-bands in rat chromosomes. Hereditas. 1974;77:37–52. doi: 10.1111/j.1601-5223.1974.tb01352.x. [DOI] [PubMed] [Google Scholar]
- 9.Maibenco HC, Krehbiel RH, Nelson D. Transplantable osteogenic tumor in the rat. Cancer Res. 1967;27:362–366. [PubMed] [Google Scholar]
- 10.Mitelman F, editor. An international system for human cytogenetic nomenclature. Basel: S Karger; 1995. [Google Scholar]
- 11.Mason RM, Bansal MK. Different growth rates of Swarm chondrosarcoma in Lewis and Wistar rats correlate with different thyroid hormone levels. Connect Tissue Res. 1987;16:177–185. doi: 10.3109/03008208709002005. [DOI] [PubMed] [Google Scholar]
- 12.Morcuende JA, Huang XD, Stevens J, Kucaba TA, Brown B, Abdulkawy H, Sheetz TE, Malchenko S, Bonalda F, Casavant TL, Soares B. Identification and initial characterization of 6,000 expressed sequenced tags (ESTs) from rat normal-growing cartilage and swarm rat chondrosarcoma cDNA libraries. Iowa Orthop J. 2002;22:28–34. [PMC free article] [PubMed] [Google Scholar]
- 13.Oegema TR, Jr, Hascall VC, Dzwiewiatkowski DD. Isolation and characterization of proteoglycans from the Swarm rat chondrosarcoma. J Biol Chem. 1975;250:6151–6159. [PubMed] [Google Scholar]
- 14.Oegema TR, Jr, Parzych SM. Effect of the retinoic acid analog Ro 11-1430 on proteoglycans of Swarm rat chondrosarcoma. J Natl Cancer Inst. 1981;67:99–106. [PubMed] [Google Scholar]
- 15.Priest JH. General cell culture principles and fibroblast culture. In: Barch MJ, Knusten T, Spurbeck JL, editors. The AGT cytogenetics laboratory manual. Third edition. Philadelphia: Lippincott-Raven Publishers; 1997. pp. 173–197. [Google Scholar]
- 16.Stevens JW, Kurriger GL, Carter AS, Maynard JA. CD44s expression in the developing and growing rat intervertebral disc. Dev Dynamics. 2000;219:381–390. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1060>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]