Abstract
With the recent increase in the available number of high-quality, full-length mitochondrial sequences, it is now possible to construct and analyze a comprehensive human mitochondrial consensus sequence. Using a data set of 827 carefully selected sequences, it is shown that modern humans contain extremely low levels of divergence from the mitochondrial consensus sequence, differing by a mere 21.6 nt sites on average. Fully 84.1% of the mitochondrial genome was found to be invariant and ‘private’ mutations accounted for 43.8% of the variable sites. Ninety eight percent of the variant sites had a primary nucleotide with an allele frequency of 0.90 or greater. Interestingly, the few truly ambiguous nucleotide sites could all be reliably assigned to either a purine or pyrimidine ancestral state. A comparison of this consensus sequence to several ancestral sequences derived from phylogenetic studies reveals a great deal of similarity, where, as expected, the most phylogenetically informative nucleotides in the ancestral studies tended to be the most variable nucleotides in the consensus. Allowing for this fact, the consensus approach provides variation data on the positions that do not contribute to phylogenetic reconstructions, and these data provide a baseline for measuring human mitochondrial variation in populations worldwide.
INTRODUCTION
Human mitochondrial sequence data has been used to make various inferences about the origins of modern humans, including a single ‘out of Africa’ dispersion event (1,2) (currently the most popular theory for modern human origins), multiple dispersions from Africa (3), and a multi-regional, parallel development model (4). Despite all this work, no attempt has been made to construct and analyze a worldwide human mitochondrial consensus sequence. This is partly due to the fact that a consensus sequence cannot be derived directly from current phylogenetic methodology. It is also due to the fact that large numbers of high-quality, full-length mitochondrial sequences have not been available until recently. However, with the publication of hundreds of sequences from human populations worldwide, an analysis of the full mitochondrial genome can be completed in detail for the first time.
The standard measure for all mitochondrial studies is the Revised Cambridge Reference Sequence (rCRS), but the rCRS is not a consensus sequence and should not be used as such. While the rCRS provides a uniform nucleotide numbering scheme, it is simply a reconstruction of a single European individual's mtDNA and contains several rare alleles (5). The rCRS also tells us nothing about the degree of variation on a nucleotide-by-nucleotide basis. Construction and analysis of a worldwide consensus sequence, however, allows for the study of variation within human populations at each nucleotide position. This is especially important in light of the fact that phylogenetic reconstructions focus only on the relatively few nucleotide positions which drive tree topology.
While there are several thousand full-length mitochondrial sequences that one can obtain from sources such as the National Institutes of Health's National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/), many of these are of questionable quality. The earliest studies on full-length sequences used early-generation DNA sequencing technology (6), and, because of this, these sequences have a higher-than-desired error rate. There are many other sequences that are either unprovenanced, unpublished (and therefore untested by peer review), from patents (and therefore often poorly described and often manipulated for patent purposes), from commercial genealogical databases (which have not been rigorously vetted by the scientific community), or from studies of mitochondrial-linked diseases (with a focus on aberrant sequences). In addition, there are at least five errors that are commonly found in mtDNA databases: base shifts, reference bias, phantom mutations, base misscoring and artificial recombination (7). Many sequence data sets have been flagged by latter studies as containing such errors, often years after the publication of warnings and methods that one can use to detect them (8). Some effort has been made by the original authors to go back and re-sequence problematic samples, occasionally with less-than-satisfactory results (9–11). Lastly, not all readers have access to unpublished information. For example, Kong et al. (12) flagged 13 sequences from Tanaka et al. (13) that were likely affected by artificial recombination. They excluded these from their analysis but did not report the sequence accession numbers.
All this forces the researcher to delve deeply into the literature if he or she wants to use publicly available sequences in further analyses. The problems with current mtDNA data sets have led to pleas for better quality control (8,14) and any database consisting of published sequences must be constructed with this issue in mind. The current set of 827 sequences (not including the rCRS) was carefully assembled from the literature and should represent the best human mitochondrial sequences currently available.
Network phylogenetic methods (using the reduced median (15) or the newer median-joining (16) algorithm) are often used in mitochondrial studies. In light of the high degree of homoplasy among diverse mitochondrial lines, a judicious selection of variable sites must be employed when using these techniques to keep the number of possible ancestral trees at a manageable level. A consensus approach, however, allows for the creation of a working ancestral mitochondrial model sequence despite the presence of homoplasies, polytomies or other confounding relationships. While a consensus cannot be considered an ancestral sequence, per se, if the underlying tree topology is balanced and the major clades are adequately sampled, the two approaches will point to the same ancestral sequence––or nearly so (17–19). Both approaches are less than perfect. The calculation of a consensus is highly dependent upon equal sampling of extant branches, yet it can reveal details of the ancestral sequence that cannot otherwise be seen, such as variation at positions that do not drive tree topology. On the other hand, phylogenetically derived ancestral sequences are estimations of likely ancestral states and are dependent on the reliability of underlying evolutionary models (19). While the latter is emphasized to a greater extent in the literature, the former is a valid approach simply because it provides different types of information.
Current phylogenetic methods for constructing ancestral mtDNA trees depend on several key assumptions, e.g. a clock-like evolution of mtDNA, a lack of recombination and a lack of selection. Perhaps the most commonly used phylogenetic method, the median-joining algorithm (16), places the most similar sequences into nested relationships first (by taking the consensus of sequence triplets) and adds the most variable sequences last. This is a parsimonious approach, but creates an interesting situation: sequences that have changed in a non-clock-like manner will have a disproportionate influence on tree structure. Clock-like evolution has been questioned for the African L2 clades (20,21), making it difficult to accurately place L2 in the human mitochondrial phylogenetic tree—and haplogroup L2a is the most common African-specific haplogroup (22). This also raises questions about the structure of the tree in general. In their introduction, Howell et al. (21) list a series of studies that argue against the clock-like evolution of mtDNA and whether or not mtDNA fits the neutral model of evolution. These two points were raised more recently by Kivisild et al. (23).
In addition to questions about the mitochondrial clock, new data have been published that indicate that recombination may occur within mitochondrial lineages and may account for certain homoplasies in the mtDNA phylogenetic tree, although the issue has been debated for some time (24). Several studies indicate that selection may also operate on mtDNA (23,25,26). All of this indicates a need for the use of supplemental and complimentary methodologies.
The analysis reported here expands upon the results presented by several previous studies, especially that of Ingman et al. (27). That study pioneered full-length mitochondrial analysis, with an examination of 53 diverse sequences. The primary purpose of the current work was to test for the degree of conservation at each mitochondrial nucleotide position within extant human populations, using a consensus approach and employing the best sampling of worldwide mtDNA available to date. The result is a very strong consensus model that indirectly reflects upon the ancestral human mitochondrial genome and more fully informs us about the degree of human mitochondrial variation.
MATERIALS AND METHODS
Sequence selection
All full-length human mitochondrial sequences available as of November 2006 were downloaded from GenBank (http://www.ncbi.nih.gov/). For the initial analysis, poor-quality and questionable sequences were removed from the data set according to the information made available by several authors (6–7,12,28–33). Sequences excluded from the initial analysis included those from Finnila et al. (34); Ingman et al. (27); Maca-Meyer et al. (35); Tanaka et al. (13) and Rajkamur et al. (36). Also excluded were sequences that were not published in peer reviewed literature, were from patents, had a HeLa origin, or were from disease patients. As far as it is known, the resulting list of 827 sequences from 22 separate studies (6,12,21,31,32,37–52) should be free from the most common errors described above. A complete sequence list is included in Supplementary Data.
Alignment
A master sequence alignment was created in BioEdit (53) manually, with special attention being paid to problem areas noted in the literature. An alignment program (e.g. ClustalW) was not used for reasons of speed and accuracy around gaps. Throughout the alignment, a set of minimization rules was applied that produced the fewest number of changes necessary in subjective areas.
Calculations
BioPerl (54) was used for all calculations, using the rCRS as a template for nucleotide numbering. A hash table that included all variant positions was created from the master alignment with sequence name and nucleotide position as keys. All calculations were performed on this hash table and output was directed to a series of text files. These calculations included a compilation of variable positions, detected alleles, allele frequencies and transition/transversion status (Supplementary Data). The worldwide human mitochondrial consensus sequence (Supplementary Data) was reconstructed from the hash table by picking the majority allele at each variable position and adding in the invariant sites from the rCRS (there is no accession number included for the consensus sequence because GenBank does not catalog theoretical sequences or sequences that have no physical counterpart).
Rather than removing regions of recurrent length variation from the analysis, as is often the case in mitochondrial studies, eleven ‘poly-x’ sites (Table 1) were identified and dealt with separately. Each poly-x site was composed of a variable number of nucleotide repeats with at least three variants found in the data set. The starting position of each site was chosen to reflect the first variable position within the repeat (most sites had an invariant set of upstream repetitive nucleotides). The variable nucleotides from the start to the end of the poly-x site, numbered according to the rCRS, and any additional nucleotide inserts not found in the rCRS were treated as a single variant nucleotide position for all calculations. The ‘9-bp deletion’ at positions 8281-8289 is a common and well-characterized feature. It is treated as a poly-x site because individual sequences carried 1, 2 or 3 copies of this repeat (CCCCCTCTA × n; sequences with only 1 repeat carry the ‘deletion’). Pairwise sequence difference calculations included the poly-x sites.
Table 1.
‘Poly-x’ sites
| Site | Type | Number of alleles | p |
|---|---|---|---|
| 290–291 | Poly-A | 3 | 0.998 |
| 309 | Poly-C | 4 | 0.522 |
| 315 | Poly-C | 3 | 0.935 |
| 498 | Poly-C | 3 | 0.995 |
| 520–523 | CA repeat | 5 | 0.746 |
| 571–573 | Poly-C | 8 | 0.971 |
| 960 | Poly-C | 7 | 0.989 |
| 5899 | Poly-C | 4 | 0.987 |
| 8276 | Poly-C | 5 | 0.990 |
| 8281–8289 | ‘9-bp deletion’ | 6 | 0.993 |
| 16, 192–16, 193 | Poly-C | 5 | 0.945 |
RESULTS
The world wide human mtDNA consensus is composed of 16 569 nucleotides. Based on the current data set, 84.1% of the mitochondrial genome is invariant. ‘Personal mutations’ (i.e. those that occur in only one sequence within the data set) comprised 43.8% of the variable sites (7.0% of the total mitochondrial genome). Based on the current state of sequencing technology and critiques of published data sets, a small but unknown number of these are expected to be sequencing artifacts. Less than 2% of all sites were actually polymorphic (i.e. the minor allele frequency was >1%). About 0.5% of all sites had a ‘common’ minor allele (i.e. an allele found in 5% or more of the sequences).
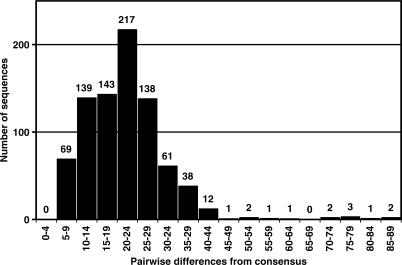
The 827 sequences within the data set contained 2631 variant positions, and on average differed from the consensus by 21.6 ± 10.0 (SD) nucleotides (Figure 1). Much of the variation observed was within specific regions of the mitochondrial genome (Table 2). This included the well-known hypervariable regions (HVS1 and HVS2) as well as the 55 non-coding nucleotides within the coding region.
Figure 1.
Pairwise nucleotide differences between all individuals within the data set and the worldwide consensus.
Table 2.
Variation within specific regions of the mitochondrial genome
| Locus | Number of variant sites | % Variant sites | |
|---|---|---|---|
| D-loop | D-loop total | 392 | 14.6 |
| HVS 1 | 181 | 6.8 | |
| HVS 2 | 114 | 4.3 | |
| 7S DNA | 238 | 8.9 | |
| Coding region | Coding region total | 2272 | 85.3 |
| Non-coding nucleotides | 23 | 0.9 | |
| tRNAs | 158 | 5.9 | |
| 12S Ribosomal RNA | 99 | 3.7 | |
| 16S Ribosomal RNA | 125 | 4.7 | |
| NADH dehydrogenase subunits | 996 | 37.4 | |
| Cytochrome c oxidase subunits | 431 | 16.2 | |
| Cytochrome b | 217 | 8.1 | |
| ATP synthase subunits | 211 | 7.9 |
Loci positions were obtained from MitoMap (www.MitoMap.org). Percent calculations included poly-x sites (Table 1) as single events in nucleotide counts. Note: The overlap between several of these categories means that the number of variant sites will be higher than reported in the text and the percentages will sum to greater than unity.
There are 12 differences between the rCRS and the human consensus (73, 263, 315 + C, 750, 1438, 2706, 4769, 7028, 8860, 11 719, 14 766, and 15 326), representing 11 transitions and one indel. This list exactly parallels the changes necessary to go from the rCRS (haplogroup H2a), through each of the intermediate haplogroups, to macrohaplogroup R according to Achilli et al. (38). The highly recurrent 315 + C indel can be inferred to reflect an historical deletion within the rCRS.
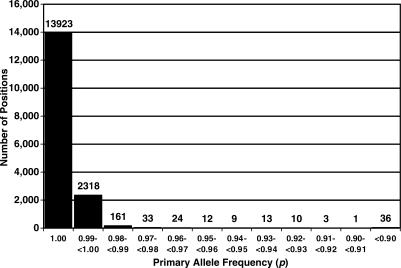
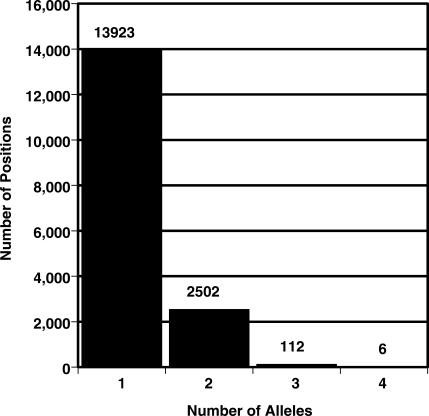
The distribution of primary allele frequencies (p) is given in Figure 2. All sites that are un-selected and freely drifting in a large population might be expected to eventually display all four nucleotides (i.e. the nucleotide could be A, T, G or C). In the entire mitochondrial genome there were only six such sites (positions 185, 13 928, 16 176, 16 265, 16 266 and 16 318), and each had a clear majority nucleotide. The average p at these 4-fold degenerate sites was 0.97 ± 0.02 (SD). Note that four of these six positions are in HVS 1 (Figure 3). There are only 36 positions where p was less than 0.90 (0.22% of the human mitochondrial genome). In every one of these cases, the ancestral nucleotide was clearly either a purine or a pyrimidine (six of these sites displayed three alleles, but in all cases the third allele had a frequency of less than 0.005). Table 3 contains a summary of the number of alleles and primary allele frequencies for the global consensus sequence.
Figure 2.
Primary allele frequency (p) distribution, where p is defined as the frequency of the most common allele at each position. Data for the 11 poly-x sites are treated separately (Table 2).
Figure 3.
Allele-count frequency histogram. The allele count is a measure of the number of alleles (i.e. A, T, G or C) found at each position. Invariant sites have an allele count of 1. Data for the 11 poly-x sites are treated separately (Table 2).
Table 3.
Summary statistics for the number of alleles and primary allele frequency (p) data on a site-by-site basis
| Number of alleles | p | |
|---|---|---|
| Average | 1.169 | 0.998 |
| SD | 0.406 | 0.012 |
| Min | 1 | 0.521 |
| Max | 8 | 1.000 |
Relative to the rCRS, there are 31 insertions within the data set that are not within the designated poly-x sites. Fourteen of these are private alleles. Reported values are the same (to three decimal places) whether or not one includes the insertions.
The most variable position (309) had a p of only 0.52. This is a poly-C region within HVS 2 that has a highly variable number of C residues. It does not represent an ambiguous nucleotide position in the normal sense. There were several similar sites (‘poly-x sites’) that were treated separately in the analysis (see Methods section). Despite the high variability in repeat length within these sites, the consensus sequence is still essentially unambiguous, with p equal to at least 0.95 for all but three of the poly-x sites (Table 1).
I tested the effects of including 491 additional full-length sequences from three older data sets (27,34,35) and one recent data set (36) plus 191 non-disease sequences from Tanaka et al. (13). Including these sequences produced no change in any consensus nucleotide and only had a trivial effect on the number of variable positions, number of alleles per site and primary allele frequencies. Due to the presence of many private and rare alleles, most changes in the dominant allele frequency stayed in the range of p = 1.00–0.99. These sequences were excluded from the primary data set only because questions have been raised concerning their accuracy (see Methods section).
I also tested the effects of including the 277 diverse coding region sequences from Kivisild et al. (23). Ignoring the lack of data for the D-loop, inclusion of these sequences added 768 new variable sites and 49 new private mutations. Despite the added diversity, the average number of alleles per site only increased slightly and no consensus nucleotide changed. The Kivisild data set is less than one-third the size of that used to generate the current consensus and their sequences do not include the D-loop. Even though this smaller data set of incomplete sequences cannot fully resolve the consensus sequence, it does include a much higher proportion of sequences from sub-Saharan Africa. Therefore, I performed a consensus analysis on just these 277 partial sequences to determine the degree to which the techniques are dependent upon the proportional representation of world regions within the included sequences. The consensus for the Kivisild data set contains five transitions (8701, 9540, 10 398, 10 873, 12 705) in addition to the ones found in the world consensus. These five transitions are putative steps either between macrohaplogroups R and N (32); or between macrohaplogroup R, macrohaplogroup N and superhaplogroup L3 (23,51). These five positions are also among the 11 positions with the lowest p in the world consensus. This indicates that final resolution of a very few sites will be subject to sample size and composition. Yet only the sites which drive the major breaks in tree topology are expected to be within this group. It is important to note that, despite the large difference in the proportions of regional representation between the world consensus and the Kivisild data sets, their consensi are 99.97% identical.
DISCUSSION
The world consensus sequence is unambiguous except for a very few nucleotide positions. In the cases where the majority nucleotide was not overwhelmingly dominant, the ancestral nucleotide could still be unambiguously classified as either a purine or a pyrimidine. Thus, the amount of mitochondrial sequence divergence within humans since the most recent common ancestor is quite low. Only a few polymorphisms separate the major lineages, and the remaining variable sites are simply ‘noise’. This noise is largely due to personal mutations. Some degree of homoplasy is also evident and recent claims that recombination can explain at least some of the homoplasy (24) within human mtDNA lineages need to be considered.
According to the newest available phylogenetic calculations (23), there are perhaps as few as 30 differences between the macrohaplogroup R consensus and the root node of all extant human mitochondrial sequences. These few positions are the most likely to change within the consensus with the addition of new data. Other potential revisions within the consensus sequence should be very rare since we have already seen that nearly all sites are either invariant or contain only very rare minor alleles. Therefore, adding new sequences, even from diverse sources such as the highly heterogeneous African-specific clades, should not significantly change the consensus. The consensus is a robust representation of the founder human mitochondrial sequence, with the exception of a few, slightly ambiguous nucleotide positions.
The phylogenetic approach and the consensus approach for discovering the ancestral sequence should produce nearly identical results if the underlying phylogeny is evenly balanced across branches (17–19). At this time, it is not possible to perfectly balance the sampling among the various people groups due to a dearth of sequences for some groups. For example, there is a preponderance of non-African mitochondrial sequences in our data set and there is a general lack of available samples from certain geographic areas (e.g. central Africa (22)). This will doubtless be corrected over time as more full-length sequences become available. Inclusion of more sequences from sub-Saharan Africa will increase the total number of variable sites. However, since the African-specific lineages are so highly heterogeneous, it is not expected that these additional sequences will have a significant affect on the consensus sequence.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This research was funded in part by FMS Foundation. I wish to thank K.R. Watson for assistance with BioPerl, T. Kivisild for providing me with a high-quality version of Figure 1 from Kivisild et al. (23), and J. Sanford for invaluable suggestions during the development of the manuscript. Funding to pay the Open Access publication charges for this article was provided by FMS Foundation.
Conflict of interest statement. None decalred
REFERENCES
- 1.Cann R.L., Stoneking M., Wilson A.C. Mitochondrial DNA and human evolution. Nature. 1987;325:31–36. doi: 10.1038/325031a0. [DOI] [PubMed] [Google Scholar]
- 2.Stoneking M., Soodyall H. Human evolution and the mitochondrial genome. Curr. Opin. Genet. Dev. 1996;6:731–736. doi: 10.1016/s0959-437x(96)80028-1. [DOI] [PubMed] [Google Scholar]
- 3.Templeton A.R. Out of Africa again and again. Nature. 2002;416:45–51. doi: 10.1038/416045a. [DOI] [PubMed] [Google Scholar]
- 4.Wolpoff M.H., Hawks J., Caspari R. Multiregional, not multiple origins. Am. J. Phys. Anthropol. 2000;112:129–136. doi: 10.1002/(SICI)1096-8644(200005)112:1<129::AID-AJPA11>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 5.Andrews R.M., Kubacka I., Chinnery P.F., Lightowlers R.N., Turnbull D.M., Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat. Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- 6.Palanichamy M.G., Chang S., Agrawal S., Bandelt H.-J., Kong Q.-P., Khan F., Wang C.-Y., Chaudhuri T.K., Palla V., et al. Phylogeny of mitochondrial DNA macrohaplogroup N in India, based on complete sequencing: implications for the peopling of South Asia. Am. J. Hum. Genet. 2004;75:966–978. doi: 10.1086/425871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bandelt H.-J., Lahermo P., Richards M., Macaulay V. Detecting errors in mtDNA data by phylogenetic analysis. Int. J. Legal. Med. 2001;115:64–69. doi: 10.1007/s004140100228. [DOI] [PubMed] [Google Scholar]
- 8.Yao Y.-G., Bravi C.M., Bandelt H.-J. A call for mtDNA data quality control in forensic science. Forensic Sci. Int. 2004;141:1–6. doi: 10.1016/j.forsciint.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 9.Yao Y.-G., Macaulay V., Kivisild T., Zhang Y.-P., Bandelt H.-J. To trust or not to trust an idiosyncratic mitochondrial data set. Am. J. Hum. Genet. 2003a;72:1341–1346. doi: 10.1086/375119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Silva W.A., Bonatto S.L., Holanda A.J., Ribeiro-dos-Santos A.K., Paixão B.M., Goldman G.H., Abe-Sandes K., Rodriguez-Delfin L., Barbosa M., et al. Correction: mitochondrial variation in Amerindians. Am. J. Hum. Genet. 2003;72:1346–1348. [Google Scholar]
- 11.Yao Y.-G., Macaulay V., Kivisild T., Zhang Y.-P., Bandelt H.-J. Reply to Silva et al. Am. J. Hum. Genet. 2003b;72:1348–1349. doi: 10.1086/375119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kong Q.-P., Bandelt H.-J., Sun C., Yao Y.-G., Salas A., Achilli A., Wang C.-Y., Zhong L., Zhu C.-L., et al. Updating the East Asian mtDNA phylogeny: a prerequisite for the identification of pathogenic mutations. Hum. Mol. Genet. 2006;15:2076–2086. doi: 10.1093/hmg/ddl130. [DOI] [PubMed] [Google Scholar]
- 13.Tanaka M., Cabrera V.M., González A.M., Larruga J.M., Takeyasu T., Fuku N., Guo L.-J, Hirose R., Fujita Y., et al. Mitochondrial genome variation in eastern Asia and the peopling of Japan. Genome Res. 2004;14:1832–1850. doi: 10.1101/gr.2286304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yao Y.-G., Salas A., Bravi C.M., Bandelt H.-J. A reappraisal of complete mtDNA variation in East Asian families with hearing impairment. Hum. Genet. 2006;119:505–515. doi: 10.1007/s00439-006-0154-9. [DOI] [PubMed] [Google Scholar]
- 15.Bandelt H.-J., Forster P., Sykes B.C., Richards M.B. Mitochondrial portraits of human populations using median networks. Genetics. 1995;141:743–753. doi: 10.1093/genetics/141.2.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bandelt H.-J., Forster P., Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- 17.Gaschen B., Taylor J., Yusim K., Foley B., Gao F., Lang D., Novitsky V., Haynes B., Hahn B.H., et al. Diversity considerations in HIV-1 vaccine selection. Science. 2002;296:2354–2360. doi: 10.1126/science.1070441. [DOI] [PubMed] [Google Scholar]
- 18.Nickle D.C., Jensen M.A., Gottlieb G.S., Shriner D., Learn G.H., Rodrigo A.G., Mullins J.I. Consensus and ancestral state HIV vaccines. Science. 2003;299:1515–1517. doi: 10.1126/science.299.5612.1515c. [DOI] [PubMed] [Google Scholar]
- 19.Gao F., Bhattacharya T., Gaschen B., Taylor J., Moore J.P., Novitsky V., Yusim K., Lang D., Foley B., et al. Response to Nickle et al. 2003. Science. 2003;299:1517–1518. [Google Scholar]
- 20.Torroni T., Rengo C., Guida V., Cruciani F., Sellitto D., Coppa A., Luna Calderon F., Simionati B., Valle G., et al. Do the four clades of the mtDNA haplogroup L2 evolve at different rates? Am. J. Hum. Genet. 2001;69:1348–1356. doi: 10.1086/324511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Howell N., Elson J.L., Turnbull D.M., Herrnstadt C. African haplogroup L mtDNA sequences show violations of clock-like evolution. Mol. Biol. Evol. 2004;21:1843–1854. doi: 10.1093/molbev/msh184. [DOI] [PubMed] [Google Scholar]
- 22.Salas A., Richards M., Lareu M.-V., Scozzari R., Coppa A., Torroni A., Macaulay V., Carracedo A. The African diaspora: mitochondrial DNA and the Atlantic slave trade. Am. J. Hum. Genet. 2004;74:454–465. doi: 10.1086/382194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kivisild T., Shen P., Wall D.P., Do B., Sung R., Davis K., Passarino G., Underhill P.A., Scharfe C., et al. The Role of Selection in the Evolution of Human Mitochondrial Genomes. Genetics. 2006;172:373–387. doi: 10.1534/genetics.105.043901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zsurka G., Hampel K.G., Kudina T., Kornblum C., Kraytsberg Y., Elger C.E., Khrapko K., Kunz W.S. Inheritance of mitochondrial DNA recombinants in double-heteroplasmic families: potential implications for phylogenetic analysis. Am. J. Hum Genet. 2007;80:298–305. doi: 10.1086/511282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mishmar D., Ruiz-Pesini E., Golik P., Macaulay V., Clark A.G., Hosseini S., Brandon M., Easley K., Chen E., et al. Natural selection shaped regional mtDNA variation in humans. Proc. Natl. Acad Sci. USA. 2003;100:171–176. doi: 10.1073/pnas.0136972100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruiz-Pesini E., Mishmar D., Brandon M., Procaccio V., Wallace D.C. Effects of purifying and adaptive selection on regional variation in human mtDNA. Science. 2004;303:223–226. doi: 10.1126/science.1088434. [DOI] [PubMed] [Google Scholar]
- 27.Ingman M., Kaessmann H., Pääbo S., Gyllensten U. Mitochondrial genome variation and the origin of modern humans. Nature. 2000;408:708–713. doi: 10.1038/35047064. [DOI] [PubMed] [Google Scholar]
- 28.Bandelt H.-J., Quintana-Murci L., Salas A., Macaulay V. The fingerprint of phantom mutations in mitochondrial DNA data. Am. J. Hum. Genet. 2002;71:1150–1160. doi: 10.1086/344397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bandelt H.-J., Salas A., Lutz-Bonengel S. Artificial recombination in forensic mtDNA population databases. Int. J. Legal. Med. 2004;118:267–273. doi: 10.1007/s00414-004-0455-2. [DOI] [PubMed] [Google Scholar]
- 30.Bandelt H.-J., Achilli A., Kong Q.-P., Salas A., Lutz-Bonengel S., Sun C., Zhang Y.-P., Torroni A., Yao Y.-G. Low “penetrance” of phylogenetic knowledge in mitochondrial disease studies. Biochem. Biophys. Res. Commun. 2005;333:122–130. doi: 10.1016/j.bbrc.2005.04.055. [DOI] [PubMed] [Google Scholar]
- 31.Kong Q.-P., Yao Y.-G., Sun C., Bandelt H.-J., Zhu C.L., Zhang Y.-P. Phylogeny of East Asian mitochondrial DNA lineages inferred from complete sequences. Am. J. Hum. Genet. 2003;73:671–676. doi: 10.1086/377718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun C., Kong Q.-P., Palanichamy M.G., Agrawal S., Bandelt H.-J., Yao Y.-G., Khan F., Zhu C.-L., Chaudhuri T.K., et al. The dazzling array of basal branches in the mtDNA macrohaplogroup m from India as inferred from complete genomes. Mol. Biol. Evol. 2006;23:683–690. doi: 10.1093/molbev/msj078. [DOI] [PubMed] [Google Scholar]
- 33.Yao Y.-G., Macaulay V., Kivisild T., Zhang Y.-P., Bandelt H.-J. To Trust or not to trust an idiosyncratic mitochondrial data set. Am. J. Hum. Genet. 2003;72:1341–1346. doi: 10.1086/375119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Finnila S., Lehtonen M.S., Majamaa K. Phylogenetic network for European mtDNA. Am. J. Hum. Genet. 2001;68:1475–1484. doi: 10.1086/320591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maca-Meyer N., González A.M., Larrugo J.M., Flores C., Cabrera V.M. Major genomic mitochondrial lineages delineate early human expansions. BMC Genet. 2001;2:13. doi: 10.1186/1471-2156-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rajkumar R., Banerjee J., Gunturi H.B., Trivedi R., Kashyap V.K. Phylogeny and antiquity of M macrohaplogroup inferred from complete mtDNA sequence specific lineages. BMC Evol. Biol. 2005;5:26. doi: 10.1186/1471-2148-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Achilli A., Rengo C., Magri C., Battaglia V., Olivieri A., Scozzari R., Cruciani F., Zeviani M., Briem E., et al. The molecular dissection of mtDNA haplogroup H confirms that the Franco-Cantabrian glacial refuge was a major source for the European gene pool. Am. J. Hum. Genet. 2004;75:910–918. doi: 10.1086/425590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Achilli A., Rengo C., Battaglia V., Pala M., Olivieri A., Fornarino S., Magri C., Scozzari R., Babudri N., et al. Saami and Berbers-an unexpected mitochondrial DNA link. Am. J. Hum. Genet. 2005;76:883–886. doi: 10.1086/430073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Behar D.M., Metspalu E., Kivisild T., Achilli A., Hadid Y., Tzur S., Pereira L., Amorim A., Quintana-Murci L., et al. The matrilineal ancestry of Ashkenazi Jews: portrait of a recent founder event. Am. J. Hum. Genet. 2006;78:487–497. doi: 10.1086/500307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coble M.D., Just R.S., O’Callaghan J.E., Letmanyi I.H., Peterson C.T., Irwin J.A., Parsons T.J. Single nucleotide polymorphisms over the entire mtDNA genome that increase the power of forensic testing in Caucasians. Int. J. Legal Med. 2004;118:137–146. doi: 10.1007/s00414-004-0427-6. [DOI] [PubMed] [Google Scholar]
- 41.Friedlaender J., Schurr T., Gentz F., Koki G., Friedlaender F., Horvat G., Babb P., Cerchio S., Kaestle F., et al. Expanding Southwest Pacific mitochondrial haplogroups P and Q. Mol. Biol. Evol. 2005;22:1506–1517. doi: 10.1093/molbev/msi142. [DOI] [PubMed] [Google Scholar]
- 42.Gonzalez A.M., Garcia O., Larruga J.M., Cabrera V.M. The mitochondrial lineage U8a reveals a Paleolithic settlement in the Basque country. BMC Genomics. 2006;7:124–130. doi: 10.1186/1471-2164-7-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ingman M., Gyllensten U. Mitochondrial genome variation and evolutionary history of Australian and New Guinean aborigines. Genome Res. 2003;13:1600–1606. doi: 10.1101/gr.686603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Macaulay V., Hill C., Achilli A., Rengo C., Clarke D., Meehan W., Blackburn J., Semino O., Scozzari R., et al. Single, rapid coastal settlement of Asia revealed by analysis of complete mitochondrial genomes. Science. 2005;308:1034–1036. doi: 10.1126/science.1109792. [DOI] [PubMed] [Google Scholar]
- 45.Maca-Meyer N., González A.M., Larruga J.M., Flores C., Cabrera V.M. Mitochondrial DNA transit between West Asia and North Africa inferred from U6 phylogeography. BMC Genet. 2003;4:15. doi: 10.1186/1471-2156-4-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Merriwether D.A., Hodgson J.A., Friedlaender F.R., Allaby R., Cerchio S., Koki G., Friedlaender J.S. Ancient mitochondrial M haplogroups identified in the Southwest Pacific. Proc. Natl. Acad. Sci. USA. 2005;102:13034–13039. doi: 10.1073/pnas.0506195102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pierson M.J., Martinez-Arias R., Holland B.R., Gemmell N.J., Hurles M.E., Penny D. Deciphering Past Human Population Movements in Oceania: Provably Optimal Trees of 127 mtDNA Genomes. Mol. Biol. Evol. 2006;23:1966–1975. doi: 10.1093/molbev/msl063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Starikovskaya E.B., Sukernik R.I., Derbeneva O.A., Volodko N.V., Ruiz-Pesini E., Torroni A., Brown M.D., Lott M.T., Hosseini S.H., et al. Mitochondrial diversity in indigenous populations of the southern extent of Siberia, and the origins of Native American haplogroups. Ann. Hum. Genet. 2005;69:67–89. doi: 10.1046/j.1529-8817.2003.00127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thangaraj K., Chaubey G., Kivisild T., Reddy A.G., Singh V.K., Rasalkar A.A., Singh L. Reconstructing the origin of Andaman Islanders. Science. 2005;308:996. doi: 10.1126/science.1109987. [DOI] [PubMed] [Google Scholar]
- 50.Thangaraj K., Chaubey G., Singh V.K., Vanniarajan A., Thanseem I., Reddy A.G., Singh L. In situ origin of deep rooting lineages of mitochondrial Macrohaplogroup M in India. BMC Genomics. 2006;7:151. doi: 10.1186/1471-2164-7-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Torroni A., Achilli A., Macaulay V., Richards M., Bandelt H.-J. Harvesting the fruit of the human mtDNA tree. Trends Genet. 2006;22:339–345. doi: 10.1016/j.tig.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 52.van Holst Pellekaan S.M., Ingman M., Roberts-Thomson J., Harding R.M. Mitochondrial genomics identifies major haplogroups in Aboriginal Australians. Am. J. Phys. Anthropol. 2006;131:282–294. doi: 10.1002/ajpa.20426. [DOI] [PubMed] [Google Scholar]
- 53.Hall T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- 54.Stajich J.E., Block D., Boulez K., Brenner S.E., Chervitz S.A., Dagdigian C., Fuellen G., Gilbert J.G.R., Korf I., et al. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 2002;12:1611–8. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.