Abstract
The use of DNA polymerases to incorporate phosphorothioate linkages into DNA, and the use of exonuclease III to determine where those linkages have been incorporated, are re-examined in this work. The results presented here show that exonuclease III degrades single-stranded DNA as a substrate and digests through phosphorothioate linkages having one absolute stereochemistry, assigned (assuming inversion in the polymerase reaction) as S, but not the other absolute stereochemistry. This contrasts with a general view in the literature that exonuclease III favors double-stranded nucleic acid as a substrate and stops completely at phosphorothioate linkages. Furthermore, not all DNA polymerases appear to accept exclusively the (R) stereoisomer of nucleoside alpha-thiotriphosphates [and not the (S) diastereomer], a conclusion inferred two decades ago by examination of five Family-A polymerases and a reverse transcriptase. This suggests that caution is appropriate when extrapolating the detailed behavior of one polymerase from the behaviors of other polymerases. Furthermore, these results provide constraints on how exonuclease III–thiotriphosphate–polymerase combinations can be used to analyze the behavior of the components of a synthetic biology.
INTRODUCTION
The replacement of oxygen atoms in molecules by sulfur atoms has long been used to probe interactions between those molecules and components of biological systems (1,2). Work begun by Eckstein, Jaffe and their coworkers in the 1970s has been particularly important in this regard with respect to nucleoside phosphates. Their work was the first to prepare nucleoside triphosphates where a non-bridging oxygen on the alpha-phosphorus atom was replaced by a sulfur (3,4). This replacement makes the alpha-phosphorus atom stereogenic, creating the diastereomeric (alpha-S) and (alpha-R) thiotriphosphates. Two decades ago, Eckstein and coworkers assigned the absolute stereochemistries of these isomers and correlated their absolute stereochemistries with chromatographic mobility (5,6).
These diastereomeric nucleoside triphosphates were used to probe the stereochemical details of the interactions of triphosphates with enzymes that use them, and then to elucidate the stereochemical course of reactions where the sulfurated phosphorus serves as an electrophilic center. This included five DNA polymerases [the Klenow fragment of DNA polymerase I, as well as its native form, from Escherichia coli (7,8), the DNA polymerases from bacteriophage T4 (9), bacteriophage T7 (10) and M. luteus (11), Taq polymerase (12), and the reverse transcriptase from avian myeloblastosis virus (13), see also (14)]. These polymerases all preferred the alpha-S thiotriphosphate over the alpha-R triphosphate as substrate. Also in these cases, the polymerase reaction proceeded with inversion of stereochemistry at the phosphorus center (1,2).
Thiophosphates have also found application in the preparation and analysis of oligonucleotides. Several groups have used exonuclease treatment to generate truncated DNA fragments from oligonucleotides that contain phosphorothioate linkages, including libraries. For example, Lutz and Benkovic based their ITCHY (incremental truncation for the creation of hybrid enzymes) protocol for the generation of combinatorial libraries of hybrid proteins independent of DNA sequence homology using alpha–phosphorothioate dNTPs (15). In an application for sequencing DNA and RNA, oligonucleotides generated by PCR via the incorporation of phosphorothioate nucleotides can be selectively cleaved at a phosphorothioate linkage by treatment with iodine or alkylating reagent (16–18).
Enzymatic analysis of DNA containing phosphorothioate linkages has also been proposed. Putney et al. (19), for example, reported that a DNA fragment with a phosphorothioate nucleotide at the 3′-end was not degraded by brief treatment with exonuclease III. Labeit et al. (20,21) also used thiophosphates as delimiters to sequence DNA. Since the exonuclease substrate was prepared by a polymerase that accepted the (alpha-S) nucleoside thiotriphosphate with inversion, the stereochemistry of the phosphorothioate linkage where the exonuclease III digestion stopped was presumed to be ‘R’.
The combination of thiotriphosphates and exonuclease III is potentially attractive as a way to meet various challenges encountered while developing a synthetic biology based on artificially expanded genetic information systems (AEGIS) containing six (or more) nucleotide ‘letters’ instead of the four found in natural DNA (22) (Figure 1). A fully implemented synthetic biology might support replication, amplification and even evolution (23). Several examples are now available where AEGIS components have been incorporated into six-letter polymerase chain reactions (PCRs).
Figure 1.
One example of an ‘artifically expanded genetic information system’ (AEGIS). Nucleobase pairing in this system conforms to the Watson–Crick geometry, with large purines (or purine analogs, both indicated by ‘pu’) pairing with small pyrimidines (or pyrimidine analogs, both indicated by ‘py’). The hydrogen-bonding acceptor (A) and donor (D) groups are listed from the major to the minor groove as indicated. The heterocycles shown are current implementations of the indicated hydrogen-bonding patterns; others are conceivable. Unshared pairs of electrons (or ‘electron density’) presented to the minor groove are shown by the shaded lobes. The nucleotides implementing the pyDDA:puAAD hydrogen-bonding pattern, the subject of this article, is at the bottom right.
The analysis of PCRs with expanded genetic alphabets requires an analytical tool that can estimate how much non-standard nucleotide component remains after multiple PCR cycles. In some cases (for example, when the non-standard nucleotide is isocytidine), the intrinsic reactivity of the nucleotide allows for selective chemical cleavage at the site (24). In other cases, polymerases have been identified that pause or stop at the non-standard nucleotide (but not at standard nucleotides), allowing quantitation of the amount of non-standard nucleotide remaining at that site (25).
A more general approach might (in principle) exploit alpha-thionucleoside triphosphates. Once a route to the synthesis of a non-standard nucleoside triphosphate is in hand, it is a relatively simply matter to obtain the corresponding non-standard nucleoside alpha-thiotriphosphate. Schemes can be imagined that use these to determine the amount of non-standard nucleotide in an oligonucleotide. If Exo III degradation of DNA stops dead at all phosphorothioate linkages, and if standard nucleoside triphosphates did not carry a sulfur, simply by digesting the PCR oligonucleotide product with exonuclease III following the procedure of Putney et al. (19), resolving the products by electrophoresis, and quantitating the band arising from exonuclease termination, would indicate how much of the non-standard nucleotide remains after a PCR.
Other features of the exonuclease III reaction were expected to facilitate this process. For example, reports from the early 1960s from Kornberg's laboratory suggested that exonuclease III was specific for double-stranded nucleic acid (26–29). Indeed, exonuclease III was proposed some time ago to be a route for creating single-stranded DNA from duplex DNA (30), although this procedure appears to have received little use (see later).
The potential for this strategy to be used generally throughout synthetic biology caused us to re-examine the literature concerning the incorporation of phosphorothioate linkages into DNA by polymerases and the degradation of the resulting oligonucleotides by exonuclease III. Surprisingly, we found that much of the ‘common knowledge’ concerning nucleoside alpha-thiotriphosphates, polymerases and exonuclease III combinations needed updating. We report here that Exo III is not specific for double-stranded nucleic acids, exonuclease III does not stop dead at all phosphorothioate linkages, and the extent of stereospecificity of polymerases with respect to diastereomeric alpha-thiotriphosphates need not be the same for all polymerases.
MATERIALS AND METHODS
Oligonucleotides and enzymes
Oligonucleotides (Table 1), except Z-51-Temp, were synthesized by Integrated DNA Technologies (IDT) (Coralville, IA). All oligodeoxynucleotides used in this study were purified by PAGE (10–20%). The supplementary section contains files showing the purity of oligonucleotides obtained from IDT determined by capillary electrophoresis. Mass spectra showed the expected mass for the oligonucleotides containing the phosphorothioate linkages, with no detectable M-16 peak that would arise from a desulfurated degradation product. The Z-51-Temp which containing dZ was synthesized on an Expedite-8900 synthesizer employing standard β-cyanoethyl phosphoramidite chemistry. The nucleoside phosphoramidite of dZ was synthesized based on the procedure we reported recently (31) and other reagents were purchased from Glen Research (1 μmol scale, CPG 1000 column).
Table 1.
Oligonucleotides containing phosphorothioate linkages used in this study
| G*-2S-30: 5′-GCGTAATACGACTCACTAT*AGACGAG*CGTA-3′ |
|---|
| G*-2S-51: 5′-GCGTAATACGACTCACTAT*AGACGAG*CGTACTTTAGTGAGGGTTAATTCGC-3′ |
| C-51-Temp: 3′-CGCATTATGCTGAGTGATATCTGCTCGCATGAAATCACTCCCAATTAAGCG-5′ |
| C*-2S-32: 5′-GCGAATTAACCCTCACTAA*AGTACGC*TCGTCT-3′ |
| C*-2S-51: 5′-GCGAATTAACCCTCACTAA*AGTACGC*TCGTCTATAGTGAGTCGTATTACGC-3′ |
| G-51-Temp: 3′-CGCTTAATTGGGAGTGATTTCATGCGAGCAGATATCACTCAGCATAATGCG-5′ |
| Z-SS-S19: 5′-GCGTAATACGACTCACTAT*AGACGA |
| Z-51-Temp: 3′-CGCATTATGCTGAGTGATATCTGCTZGCATGAAATCACTCCCAATTAAGCG-5′ |
Note: The * indicates the position of the phosphorothioate linker.
The α-thiotriphosphate of dP was synthesized from the non-standard nucleoside by the method of Ludwig–Eckstein (32). The SP, and RP-diastereoisomers of dPTPαS were purified by DEAE-Sephadex column (HiPrep 16/10 DEAE FF) and separated by preparative rp-HPLC]Nova-Pak® HR C18 Column (7.8 × 300 mm2)] using a linear gradient of 100 mM Et3N-HOAc with an increasing content of CH3CN from 0 to 10% in 40 min (6,33). The structure of each diastereomers (S and R isomers) was proved by 31P NMR and HRMS-ESI FT-ICR, the purity of each diastereomers was checked by analytic rp-HPLC.
The Taq and 9°N (modified) DNA polymerases were purchased from New England Biolabs. Exonuclease III was purchased from Promega.
Exo III digestion of oligonucleotides with phosphorothioate linkages
5′-32P-labeled G*-2S-30, G*-2S-51, C*-2S-32 and C*-2S-51 (1 pmol, 0.025 pmol/µl) and the corresponding template, C-51-Temp or G-51-Temp (1.5 pmol) were mixed with 4 µl of 10 × exonuclease III buffer (final concentration 66 mM Tris–HCl, pH 8.0, 0.66 mM MgCl2); final volume adjusted to 40 µl with water. The solution was heated at 95°C for 5 min and allowed to cool to room temperature over 1 h. Exonuclease III (Promega, 2 units – 100 units, final concentration 0.05 U/µl – 2.5 U/µl) was added at 22°C and aliquots (5 µl) were taken from each reaction at the appropriate time (2, 5, 15 and 30 min), quenched by 2 µl of 0.5 M EDTA and then mixed with PAGE loading buffer (5 µl, formamide). Samples were resolved by electrophoresis using a 20% PAGE (7 M urea). The gel was analyzed using MolecularImager software. See Figure 2 for details.
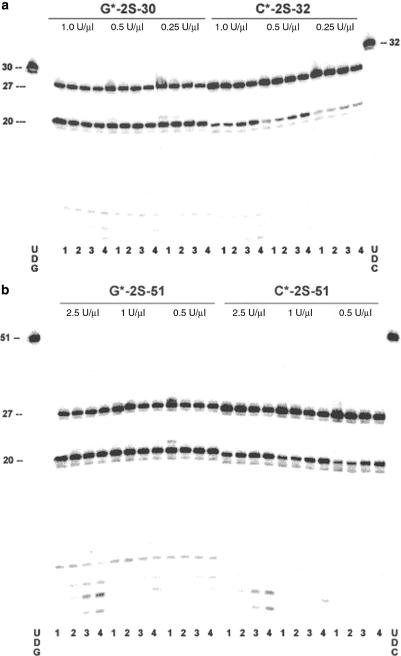
Figure 2.
Exo III digestion of oligonucleotides with phosphorothioate linkages. Denaturing (7 M urea) PAGE showing: (a) Digestion of a 5′-overhang dsDNA comprising 5′-32P-labeled G*-2S-30, C*-2S-32 and the corresponding template, C-51-Temp, G-51-Temp; negative control lanes of undigested G*-2S-30 and C*-2S-32, UDG and UDC, respectively, are indicated. The 30-mer and 32mer used as a marker is the material present at t = 0 min [see also (b)]; (b) digestion of blunt-end dsDNA comprising 5′-32P-labeled G*-2S-51, C*-2S-51 and the corresponding template, C-51-Temp, G-51-Temp; negative control lanes of undigested G*-2S-51 and C*-2S-51, UDG and UDC, respectively, are indicated. Lanes 1, 2, 3 and 4 indicate 2, 5, 15 and 30 min Exo III digestion. Units/µl (2.5, 1.0, 0.5 and 0.25) above lanes indicates the concentration of Exo III used.
Exo III digestion of single- and double-stranded oligonucleotides with phosphorothioate linkages
To prepare double-stranded substrate, 5′-32P-labeled G*-2S-51 or C*-2S-51 (2 pmol, 0.05 pmol/µl) was annealed to template, C-51-Temp or G-51-Temp (2 or 3 pmol) in 10 × Exo III buffer (final concentration, 66 mM Tris–HCl, pH 8.0, 0.66 mM MgCl2, final volume adjusted to 40 µl with water) by heating at 95°C for 5 min followed by cooling to room temperature over 1 h. A solution of single-strand substrate was prepared by following the same process, but using H2O instead of C-51-Temp or G-51-Temp.
Exonuclease III (100, 20, 5 and 1 units; final concentration: 2.5, 0.5, 0.125 and 0.025 U/µl) was then added at room temperature to the sample. Aliquots (5 µl) were taken from each reaction at time intervals (5, 10, 30 and 60 min), quenched by 2 µl of 0.5 M EDTA and mixed with PAGE loading buffer (5 µl, formamide), and resolved by electrophoresis using a 20% PAGE (7 M urea). The gel was analyzed using MolecularImager software. See Figure 3 for details.
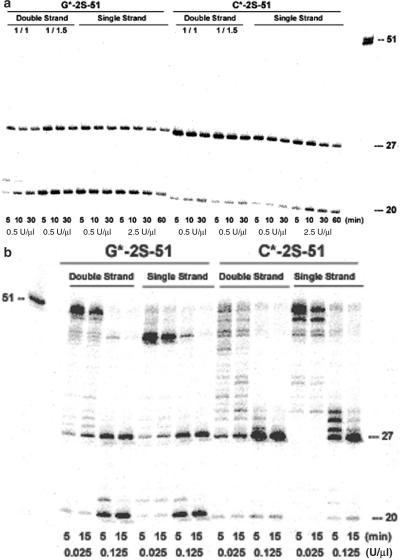
Figure 3.
Exo III digestion of single- and double-stranded DNA containing phosphorothioate linkages. Denaturing (7 M urea) PAGE showing digestion of double-stranded substrate (the duplex between 5′-32P-labeled G*-2S-51 or C*-2S-51 and the complementary C-51-Temp or G-51-Temp), and single-stranded substrate (5′-32P-labeled G*-2S-51 or C*-2S-51). (a) Digestion with high concentrations of Exo III (0.5 or 2.5 U/µl, as indicated) for the indicated times. For dsDNA substrates, the ratio of G*-2S-51/C-51-Temp or C*-2S-51/G-51-Temp was 1/1 or 1/1.5 as indicated. (b) Digestion with low concentrations of Exo III (0.025 and 0.125 U/µl, as indicated) for the indicated times. The loading of the substrate 51mer was reduced to prevent overloading of the gel; thus, the absolute intensities of these bands cannot be compared with the intensities in other lanes in the gel.
Primer extension reactions
5′-32P-labeled primer Z-SS-S19 (2 pmol, final assay concentration 100 nM) was annealed to a template sequence Z-51-Temp (3 pmol, final assay concentration 150 nM) in polymerase reaction buffer by heating (5 min at 95°C) and then slow cooling (1 h) to room temperature. The Sp or Rp diastereoisomer of dPTPαS (4 nmol, final 200 µM) were added at room temperature to give a final reaction volume of 20 µl. The reaction mixture was pre-incubated at 72°C for 30 s and followed by the addition of DNA polymerase (Taq or 9°N, 2 units). Aliquots (3 µl) were taken from each reaction at the appropriate time (30 s, 60 s, then 2 min), quenched by dilution into PAGE loading/quench buffer (7 µl, 20 mM EDTA in formamide). Samples were resolved by electrophoresis using a 20% PAGE (7 M urea). The gel was analyzed using MolecularImager software. See Figure 5a for details.
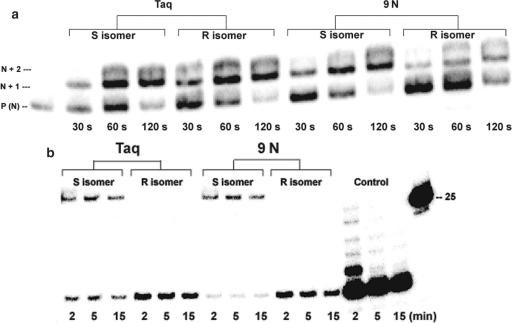
Figure 5.
(a) Extension of primers using the indicated diastereoisomers of dPTPαS with Taq and 9°N (modified) DNA polymerases for the times indicated. The positions of migration of the unextended primer, primer extended by 1 nt and the primer extended by 2 nt, are indicated by P(N), N + 1, N + 2, respectively. The amount of oligonucleotide loaded to mark the position where the primer runs (lane 1) was less than for other lanes in the gel; (b) Exo III digestion of the products of 2-min primer extension reactions using the indicated diastereomers (S or R) of alpha-thio-dPTP (from Figure 5a). Each primer extension product was purified with a QIAquick column and digested with 20 Units of Exo III for the indicated times. The control shows the degradation of 5′-32P-labeled primer Z-SS-S19 (25mer) in a duplex with unlabeled Z-51-Temp, establishing that these are degraded by Exo III.
Exo III digestion of the products of primer extension reactions
After incubation (2 min) in primer extension buffer, remaining reaction mixture (11 µl) was quenched with EDTA (3 µl, 100 mM), diluted with 150 µl of PN buffer and purified by QIAquick Nucleotide Removal Kit (Qiagen, Valencia, CA). The DNA was eluted from the spin column using 35 µl of EB buffer (10 mM Tris–HCl, pH 8.5) and mixed with 10 × Exo III buffer (4 µl, final concentration 66 mM Tris–HCl, pH 8.0, 0.66 mM MgCl2). Exonuclease III (20 U, final 0.5 U/µl) was added at room temperature to give final volume of 40 µl. Aliquots (10 µl) were taken at intervals (2, 5 and 15 min), quenched with EDTA (2 µl, 0.5 M) and mixed with PAGE loading buffer (10 µl, formamide). Samples were resolved by electrophoresis using 20% PAGE (7 M urea). The gel was analyzed using MolecularImager software. See Figure 5b for details. As a control (Figure 5b), Exo III was used to digest 5′-32P-labeled primer Z-SS-S19 (2 pmol) in a duplex with unlabeled Z-51-Temp under the same conditions.
RESULTS
Exonuclease III digests through some phosphorothioate linkages
Experiments directed towards exploiting the nuclease resistance of phosphorothioate linkages as a way of identifying sites where AEGIS nucleotides had been incorporated immediately found evidence that Exo III digests through phosphorothioate linkages. This result contrasted to classical literature reports that Exo III cannot do this. To re-examine this contrast, four sequences, G*-2S-30, G*-2S-51, C*-2S-32 and C*-2S-51 (Table 1) were prepared by IDT (Coralville, IA). These contained two phosphorothioate linkages (indicated by * in Table 1) at positions 19 and 26. The presence of phosphorothioate linkages in the synthetic oligonucleotides was confirmed by mass spectrometry (Supplementary Section). Another two sequences, C-51-Temp and G-51-Temp, were also purchased from IDT to serve as templates for the above sequences.
These sequences were then treated with Exo III and the products resolved by gel electrophoresis. The gels (Figure 2) present two prominent bands. The first migrates at a position expected for a 27mer, consistent with termination of Exo III digestion by the phosphorothioate linker joining nucleotides 26 and 27 (G*C or C*T), as expected if Exo III were to stop digestion at phosphorothioate linkers (19).
The second band, at a position expected for a 20mer, is most readily explained by assuming that Exo III digested through the 26–27 phosphorothioate linkage, continued to digest the standard internucleoside phosphate linkages, and then paused at the phosphorothioate linkage joining nucleotides 19 and 20 (T*A or A*A). Less prominent, low molecular weight bands that become stronger with longer digestion times suggest that this second phosphorothioate linkage is also susceptible to Exo III digestion, at least partially. This view is also consistent with the fact that the shorter, faster running bands, become more intense as more Exo III was used.
We considered three hypotheses to explain the apparent read-through of the 26–27 and 19–20 phosphorothioate linkages. First, we considered the possibility that the IDT oligonucleotides had suffered loss of sulfur, a process that is conceivable under oxidative conditions. The mass spectra of the oligonucleotides (Supplementary section) excluded this possibility.
Next, we hypothesized that the digestion by Exo III might slow, but not stop, at a phosphorothioate linkage. This might give some of the now-shortened duplex an opportunity to strand-separate, especially if digestion is occurring from both the 3′-end of the labeled strand and the 3′-end of the unlabeled complementary strand. Then, if literature reports were correct in suggesting that Exo III requires a double-stranded substrate (26,27), further digestion of the now single-stranded substrate would not be possible. The remaining double-stranded substrate, under this hypothesis, would continue to be digested down to the second phosphorothioate linker.
A final hypothesis was based on the expectation that the non-enzymic synthesis of phosphorothioate linkages is expected to generate both R and S diastereoisomers. If Exo III treats the diastereomers differently (not unexpected for an enzyme), degrading one but not the other, then the exonuclease would stop at only half (approximately) of the linkages. Under this hypothesis, a band containing approximately half of the total label would be present as a 27mer, while the remaining half of the labeled oligonucleotide would be digested further, with half of that stopping at the second phosphorothioate linkage at position 20.
The remaining feature needing explanation in the gels in Figure 2 are the modest differences in the amounts of bands that arise from read-through at position 20, and the relative intensities of the 20 and 27mer degradation bands in the various substrates G*-2S-30, G*-2S-51, C*-2S-32 and C*-2S-51. The first two substrates have sequences different from the second two, and the 30mers and 51mers are likely to manage any processivity displayed by Exo III. Exo III is reported to digest at different rates depending on the sequence (C > A, T > G) (34), and this may be sufficient reason to explain the differences in the band intensities. Furthermore, Shaw and her coworkers have found similar variation in cleavage rate depending on the nucleobases flanking the modified linker with boron-modified oligonucleotides (35).
Exonuclease III does not require double-stranded DNA as substrate
Literature from three decades ago reports that exonuclease III has a double-strand specific, non-processive 3′ → 5′ exo-deoxyribonuclease activity (27). To re-examine this as the first step to test of the two hypotheses considered earlier, the following parallel experiments were performed. First, a double-stranded substrate was prepared by annealing 5′-32P-labeled G*-2S-51 or C*-2S-51 (2 pmol, 0.05 pmol/µl) to template C-51-Temp or G-51-Temp (2 or 3 pmol) in 10 × Exo III buffer. A solution of single-strand substrate was prepared by following the same process, but using water instead of C-51-Temp or G-51-Temp. Exo III was then added at room temperature.
Figure 3a shows two dominant degradation bands, corresponding to a 27mer and 20mer, for digestion of both single-stranded and double-stranded DNA. Most importantly, the absence of undigested 51mer in all samples indicates that Exo III is able to degrade single-stranded DNA.
When the concentration of Exo III was reduced greatly, intermediate degradation products were observed (Figure 3b). For example, the lowest concentration of exonuclease tested (0.025 U/µl) was not adequate to digest either the dsDNA or ssDNA significantly; increasing the enzyme concentration by only 5-fold yielded complete digestion of both samples essentially in parallel (Figure 3b). This indicates that the differential specificity of Exo III for dsDNA and ssDNA is less than a factor of five.
Curiously, a band (corresponding approximately to a 43mer) was prominent as an intermediate product at low concentrations of Exo III. We first used the self-dimer application of the OligoAnalyzer 3.0 program presented on the IDT web page to determine if the formation of an intermolecular duplex might plausibly explain this pause product at the concentrations used (25 nM). The best pair had a calculated free energy of −9.0 kcal/mol, making a weak duplex possible. However, the best explanation for the appearance of the band at position 43 that incorporates the hypothesis that Exo III prefers duplex substrates would require that the 8mer at the 3′-end forms a duplex joined by just six base pairs (three G:C and three A:T), stalls when it enters a single-strand region of just three non-matched nucleotides, and resumes in a region that has seven perfect pairs. This model would not explain why the Exo III digestion does not again stall when it encounters a single-strand region that would generate 30mer pause product, the region where the potential for duplex structure is more convincingly lost.
Secondary structure analysis did not provide a more compelling case. Analysis of the sequence using MyFolder (also from the IDT web page) suggested that the single strand might form a secondary structure having a melting temperature of 49.1°C (free energy of −7.2 kcal/mol). Here, the substrate was predicted to be single-stranded until nucleotide 40. Therefore, this hypothetical fold would not account for the 43mer pause product, under the assumption that Exo III prefers duplex DNA.
The only secondary structure that placed the 43–42 dinucleotide linker within a hypothetical duplex had a predicted melting temperature of 38.5°C. The proposed duplex from the 3′-end had only six nucleobase pairs (50% A:T) and two bulges.
This analysis suggests that while we cannot rule out an influence on duplex structure on Exo III activity, it is not dominant and is not consistent. Exo III has also been reported to degrade at differential rates depending on the nucleotide sequence (34), however. Although also ad hoc, the 43mer band might be best explained in this way.
Examination of exonuclease digestion of R and S phosphorothioate linkages
Oligonucleotides having a single diasteromeric phosphorothioate linkage are not directly obtained by standard chemical synthesis techniques. The pure diasteromer of an alpha-thiotriphosphate nucleoside is, however, obtainable via chromatographic resolution of a diastereomeric mixture. As the incorporation of an alpha-triphosphate by a DNA polymerase likely (from mechanistic principles) to always occur with inversion of stereoconfiguration, a polymerase-based synthesis of an oligonucleotide with a phosphorothioate linkage with a known stereochemistry is available. This allowed us to determine which diastereomer of the phosphorothioate linkage resists Exo III digestion, and to apply this to identify the position of incorporation.
To make a diasteromerically pure phosphorothioate linkage with a non-standard genetic information system, 2-amino-8-(1′-β-D-2′-deoxyribofuranosyl)-imidazo[1,2-a]-1,3,5-triazin-4(8H)-one and 6-amino-5-nitro-3-(1′-β-D-2′-deoxyribofuranosyl)-2(1H)-pyridinone were prepared as previously described (31). These implement two non-standard nucleotides within the artificially expanded genetic information system developed over the past decade by Benner and his coworkers. This particular purine analog (trivially called ‘P’) presents a hydrogen-bond acceptor–acceptor–donor pattern to a complementary strand; this particular pyrimidine analog (trivially called ‘Z’) presents a hydrogen-bond donor–donor–acceptor pattern to a complementary strand.
The thiotriphosphates of dP were obtained as a mixture of diastereomeric thiotriphosphates via the procedure developed by Ludwig and Eckstein. The diastereomeric mixture was first purified by DEAE-Sephadex chromatography. Then, the diastereomers of the alpha-thio-dPTP were examined by reversed phase HPLC (Figure 4). This examination showed first that they could be readily separated by rp-HPLC (Figure 4a). Furthermore, the examination showed that both diasteromers moved substantially slower than the standard dPTP (Figure 4b). The product after rp-HPLC purification, following the procedure developed by Eckstein, was >95% free of the alternative diastereomer (Figure 4c and d). The quality of each diasteromer was also confirmed by 31P NMR and mass spectrometry. The absolute stereospecificity was tentatively assigned based on the generalization that the Rp diastereomer elutes second in a rp-HPLC experiment (32).
Figure 4.
Resolution of dPTP, SP and RP-diastereoisomers of dPTPαS by rp-HPLC. Trace a shows the rp-HPLC of the dPTPαS racemate after DEAE-Sephadex chromatography. Trace b shows the purified racemate with dPTP added as a reference to indicate that any trace of dPTP can be readily separated from the diastereoisomers of dPTPαS by rp-HPLC. Traces c and d show the pure diastereomers of dPTPαS following rp-HPLC; these are the materials that were used in polymerase extension studies. Peaks: 1, S isomer; 2, R isomer; 3, dPTP.
Standing start primer extension reactions, where each polymerase was challenged to incorporate S or R isomer of dPTPαS opposite dZ in the template in 30 s, 60 s and 2 min, indicates that both S and R isomer of dPTPαS are viable substrates for Taq, a Family-A polymerase and 9°N, a Family-B polymerase (Figure 5a). The products from the 2-min extension reactions were purified and subjected to digestion with Exo III. The data, as shown in Figure 5b, indicate that the phosphorothioate linkage arising from the incorporation of the R triphosphate (S configuration in the oligonucleotide) is digested by Exo III to completion, while the linkage arising from the incorporation of S triphosphate by 9°N is not digested and the phosphorothioate linkage produced by Taq is slightly digested. In addition, the lower band in the lanes derived from the incorporation of S-thiotriphosphate by Taq may arise from partial read-through of the R-phosphorothioate linkage after long incubation times with considerable enzyme, however it is more likely to reflect incomplete primer elongations (see corresponding bands in Figure 5a).
DISCUSSION
The key results from this work, important when applying thiotriphosphate–polymerase–exonuclease III combinations to analyze oligonucleotides, are: (a) exonuclease III is not specific for double-stranded oligonucleotides, but also accepts single-stranded DNA; (b) phosphorothioate linkages having the R absolute stereochemistry are resistant to digestion by exonuclease III, but not those having the S absolute stereochemistry (if we assume that all of the polymerases examined here produce inversion, and the HPLC-based assignment of the stereochemistry of the starting thiotriphosphate is secure); (c) The 9°N polymerase, a Family-B, and Taq, a Family-A polymerase, both appear to accept both R and S alpha-thiotriphosphates.
These results update existing literatures relating to the behavior of both Exo III and DNA polymerases. For example, Richardson et al. (26) four decades ago proposed that exonuclease III was selective for double-stranded DNA based on an examination of dAT copolymers and heated bacterial DNA extracts. Given the uncertain secondary structure of such oligonucleotides, it is appropriate to update this literature using synthetic substrates.
It is similarly timely to update the literature concerning the ability of various polymerases to accept alpha-thiotriphosphates. The last time that this topic was investigated, in the early 1980s, only a few DNA polymerases and reverse transcriptases were available; the thermostable polymerases that are key tools in molecular biology today were only beginning to be described. Furthermore, when the stereospecificity of the Taq polymerase was examined in 1988, it was assumed that all polymerases would accept the Sp-diastereomer of a nucleoside triphosphate; it does not appear that the Rp-diastereomer was examined (12). Furthermore, the fact that polymerases come in distinct evolutionary families that may not share common ancestry had not yet been recognized. Given this evolutionary diversity, there is no reason that the behavior of a specific DNA polymerase could be extrapolated to the behavior of all polymerase. This is not true for many features of stereochemistry in other classes of enzymes (36).
The complications of this must be managed if we wish to apply thiotriphosphate–polymerase–exonuclease III combinations to analyze oligonucleotides containing components of a synthetic biology. Here, clearly a mixture of diastereomeric alpha triphosphates will not yield a clean Exo III degradation result. Rather, the pure S-diastereomer is recommended, and primer extension should be allowed to go to completion before beginning digestion (see left lanes of Figure 5a and b, for example) for the analysis to be reliable. Furthermore, if a new polymerase is used, it is important to determine its stereospecificity before using it as an analytical tool.
If these precautions are taken, then thiotriphosphate–polymerase–exonuclease III combinations can be used to estimate the amounts and placement of other non-standard nucleotides, including those that are being developed by Minakawa et al. (37), Kool (38), Romesberg (39), Schultz and others (40). Results from measuring the fidelity of six-letter PCR (four dNTPs, dZTP and dPTP) by the above approach will be reported shortly.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This work was supported by the John Templeton Foundation, the National Institutes of Health (HG3581), the National Aeronautics and Space Administration under its Exobiology program and the National Science Foundation under its Chemical Bonding Program, under a collaboration between the Foundation for Applied Molecular Evolution, the Harvard Medical School, the Howard Hughes Medical Institute, and the Scripps Research Institute (La Jolla). Funding to pay the Open Access publication charges for this article was provided by the National Science Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Eckstein F. Phosphorothioate analogues of nucleotides. Tools for the investigation of biochemical processes. Angew. Chem. Int. Ed. Engl. 1983;22:423–439. [Google Scholar]
- 2.Eckstein F. Nucleoside phosphorothioates. Ann. Rev. Biochem. 1985;54:367–402. doi: 10.1146/annurev.bi.54.070185.002055. [DOI] [PubMed] [Google Scholar]
- 3.Eckstein F., Gindl H. Polyribonucleotides containing a phosphorothioate backbone. Eur. J. Biochem. 1970;13:558–564. doi: 10.1111/j.1432-1033.1970.tb00961.x. [DOI] [PubMed] [Google Scholar]
- 4.Jaffe E.K., Cohn M. Divalent cation-dependent stereospecificity of adenosine 5′-o-(2-thiotriphosphate) in hexokinase and pyruvate-kinase reactions. Absolute stereochemistry of diastereoisomers of adenosine 5′-O-(2-thiotriphosphate) J. Biol. Chem. 1978;253:4823–4825. [PubMed] [Google Scholar]
- 5.Burgers P.M., Eckstein F. Absolute configuration of the diastereomers of adenosine 5′-O-(1-thiotriphosphate): consequences for the stereochemistry of polymerization by DNA-dependent RNA polymerase from Escherichia coli. Proc. Natl. Acad. Sci. USA. 1978;88:4798–4800. doi: 10.1073/pnas.75.10.4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Connolly B.A., Romaniuk P.J., Eckstein F. Synthesis and characterization of diastereomers of guanosine 5′-O-(1-thiotriphosphate) and guanosine 5′-O-(2-thiotriphosphate) Biochemistry. 1982;21:1983–1989. doi: 10.1021/bi00538a002. [DOI] [PubMed] [Google Scholar]
- 7.Burgers P.M.J., Eckstein F. A study of the mechanism of DNA polymerase I from Escherichia coli with diastereomeric phosphorothioate analogs of deoxyadenosine triphosphate. J. Biol. Chem. 1979;254:6889–6893. [PubMed] [Google Scholar]
- 8.Brody R.S., Frey P.A. Unambiguous determination of the stereochemistry of nucleotidyl transfer catalyzed by DNA polymerase I from Escherichia coli. Biochemistry. 1981;20:1245–1252. doi: 10.1021/bi00508a030. [DOI] [PubMed] [Google Scholar]
- 9.Romaniuk P.J., Eckstein F. A study of the mechanism of T4 DNA polymerase with diastereomeric phosphorothioate analogues of deoxyadenosine triphosphate. J. Biol. Chem. 1982;257:7684–7688. [PubMed] [Google Scholar]
- 10.Brody R.S., Adler S., Modrich P., Stec W.J., Leznikowski Z.J., Frey P.A. Stereochemical course of nucleotidyl transfer catalyzed by bacteriophage T7 induced DNA polymerase. Biochemistry. 1982;21:2570–2572. doi: 10.1021/bi00539a042. [DOI] [PubMed] [Google Scholar]
- 11.Eckstein F., Jovin T.M. Assignment of resonances in the phosphorus-31 nuclear magnetic resonance spectrum of poly[d(A-T)] from phosphorothioate substitution. Biochemistry. 1983;22:4546–4550. doi: 10.1021/bi00288a030. [DOI] [PubMed] [Google Scholar]
- 12.Nakamaye K., Gish G., Eckstein F., Vosberg H.P. Direct sequencing of polymerase chain reaction amplified DNA fragments through the incorporation of deoxynucleoside alpha-thiotriphosphates. Nucleic Acids Res. 1988;21:9947–9959. doi: 10.1093/nar/16.21.9947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartlett P.A., Eckstein F. Stereochemical course of polymerization catalyzed by avian myeloblastosis virus reverse transcriptase. J. Biol. Chem. 1982;257:8879–8884. [PubMed] [Google Scholar]
- 14.Kunkel T.A., Eckstein F., Mildvan A.S., Koplitz R.M., Loeb L.A. Deoxynucleoside [1-thio]triphosphates prevent proofreading during in vitro DNA synthesis. Proc. Natl Acad. Sci. USA. 1981;78:6734–6738. doi: 10.1073/pnas.78.11.6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lutz S., Ostermeier M., Benkovic S.J. Rapid generation of incremental truncation libraries for protein engineering using α–phosphothioate nucleotides. Nucleic Acids Res. 2001;29(4):e16. doi: 10.1093/nar/29.4.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schatz D., Leberman R., Eckstein F. Interaction of Escherichia coli tRNAser with its cognate aminoacyl-tRNA synthetase as determined by footprinting with phosphorothioate-containing tRNA transcripts. Proc. Natl Acad. Sci. USA. 1991;88:6132–6136. doi: 10.1073/pnas.88.14.6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alexeeva E.V., Shpanchenko O.V., Dontsova O.A., Bogdanov A.A., Nierhaus K.H. Interaction of mRNA with the Escherichia coli ribosome. Accessibility of phosphorothioate containing mRNA bound to ribosomes for iodine cleavage. Nucleic Acids Res. 1996;24:2228–2235. doi: 10.1093/nar/24.12.2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gish G., Eckstein F. DNA and RNA sequence determination based on phosphorothioate chemistry. Science. 1988;240:1520–1522. doi: 10.1126/science.2453926. [DOI] [PubMed] [Google Scholar]
- 19.Putney S.D., Benkovic S.J., Schimmel P.R. A DNA fragment with an α-phosphorothioate nucleotide at one end is asymmetrically blocked from digestion by exonuclease III and can be replicated in vivo. Proc. Natl Acad. Sci. USA. 1981;78:7350–7354. doi: 10.1073/pnas.78.12.7350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Labeit S., Lehrach H., Goody R.S. A new method of DNA sequencing using deoxynucleoside alpha-thiotriphosphates. DNA-A J. Mol. Cell. Biol. 1986;5:173–177. doi: 10.1089/dna.1986.5.173. [DOI] [PubMed] [Google Scholar]
- 21.Labeit S., Lehrach H., Goody R.S. DNA sequencing using alpha-thiodeoxynucleotides. Methods Enzymol. 1987;155:166–177. doi: 10.1016/0076-6879(87)55015-7. [DOI] [PubMed] [Google Scholar]
- 22.Benner S.A. Understanding nucleic acids using synthetic chemistry. Acc. Chem. Res. 2004;37:784–797. doi: 10.1021/ar040004z. [DOI] [PubMed] [Google Scholar]
- 23.Sismour A.M., Benner S.A. The use of thymidine analogs to improve the replication of an extra DNA base pair. A synthetic biological system. Nucleic Acids Res. 2005;33:5640–5646. doi: 10.1093/nar/gki873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson S.C., Sherrill C.B., Marshall D.J., Moser M.J., Prudent J.R. A third base pair for the polymerase chain reaction. Inserting isoC and isoG. Nucleic Acids Res. 2004;32:1937–1941. doi: 10.1093/nar/gkh522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sismour A.M., Lutz S., Park J.-H., Lutz M.J., Boyer P.L., Hughes S.H., Benner S.A. PCR amplification of DNA containing non-standard base pairs by variants of reverse transcriptase from human immunodeficiency virus-1. Nucleic Acids Res. 2004;32:728–735. doi: 10.1093/nar/gkh241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richardson C.C., Lehman I.R., Kornberg A. A deoxyribonucleic acid phosphatase-exonuclease from Escherichia coli II. Characterization of the exonuclease activity. J. Biol. Chem. 1964;239:251–258. [PubMed] [Google Scholar]
- 27.Weiss B. Endonuclease II of Escherichia coli is Exonuclease III. J. Biol. Chem. 1976;251:1896–1901. [PubMed] [Google Scholar]
- 28.Rogers S.G., Weiss B. Exonuclease III of Escherichia coli K-12, an AP endonuclease. Methods Enzymol. 1980;65:201–211. doi: 10.1016/s0076-6879(80)65028-9. [DOI] [PubMed] [Google Scholar]
- 29.Hoheisel J.D. On the activities of Escherichia coli Exonuclease III. Anal. Biochem. 1993;209:238–246. doi: 10.1006/abio.1993.1114. [DOI] [PubMed] [Google Scholar]
- 30.Smith A.J.H. The use of exonuclease Ill for preparing single stranded DNA for use as a template in the chain terminator sequencing method. Nucleic Acids Res. 1979;6:831–848. doi: 10.1093/nar/6.3.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Z., Hutter D., Sheng P., Sismour A.M., Benner S.A. Artificially expanded genetic information system. A new base pair with an alternative hydrogen bonding pattern. Nucleic Acids Res. 2006;34:6095–6101. doi: 10.1093/nar/gkl633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ludwig J., Eckstein F. Rapid and efficient synthesis of nucleoside 5′-O (1-thiotriphosphates), 5′-triphosphates and 2′,3′-cyclophosphorothioates using 2-chloro-4H- 1,3,2-benzodioxaphosphorin-4-one. J. Org. Chem. 1989;54:631–635. [Google Scholar]
- 33.Taylor J.W., Schmidt W., Cosstick R., Okruszek A., Eckstein F. The use of phosphorothioate-modified DNA in restriction enzyme reactions to prepare nicked DNA. Nucleic Acids Res. 1985;13:8749–8764. doi: 10.1093/nar/13.24.8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Linxweiler W., Horz W. Sequence specificity of exonuclease III from E. coli. Nucleic Acids Res. 1982;10:4845–4859. doi: 10.1093/nar/10.16.4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He K.Z., Porter K.W., Hasan A., Briley D.J., Shaw B.R. Synthesis of 5-substituted 2′-deoxycytidine 5′-(alpha-P-borano)triphosphates, their incorporationinto DNA and effects on exonuclease. Nucleic Acids Res. 1999;27:1788–1794. doi: 10.1093/nar/27.8.1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benner S.A., Glasfeld A., Piccirilli J.A. Stereospecificity in enzymology. Its place in evolution. Topics Stereochem. 1989;19:127–207. [Google Scholar]
- 37.Minakawa N., Kojima N., Hikishima S., Sasaki T., Kiyosue A., Atsumi N., Ueno Y., Matsuda A. New base pairing motifs. The synthesis and thermal stability of oligodeoxynucleotides containing imidazopyridopyrimidine nucleosides with the ability to form four hydrogen bonds. J. Am. Chem. Soc. 2003;125:9970–9982. doi: 10.1021/ja0347686. [DOI] [PubMed] [Google Scholar]
- 38.Liu H.B., Gao J.M., Maynard L., Saito Y.D., Kool E.T. Toward a new genetic system with expanded dimensions: Size-expanded analogues of deoxyadenosine and thymidine. J. Am. Chem. Soc. 2004;126:1102–1109. doi: 10.1021/ja038384r. [DOI] [PubMed] [Google Scholar]
- 39.Tae E.L., Wu Y.Q., Xia G., Schultz P.G., Romesberg F.E. Efforts toward expansion of the genetic alphabet: replication of DNA with three base pairs. J. Am. Chem. Soc. 2001;123:7439–7440. doi: 10.1021/ja010731e. [DOI] [PubMed] [Google Scholar]
- 40.Henry A.A., Romesberg F. Beyond A, C, G and T: Augmenting Nature's alphabet. Curr. Opin. Chem. Biol. 2003;7:727–733. doi: 10.1016/j.cbpa.2003.10.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.